Abstract
Introduction
Glioblastoma is the most malignant brain tumor in adults and is associated with poor survival despite multimodal treatments. Glioma stem-like cells (GSCs) are cells functionally defined by their self-renewal potential and the ability to reconstitute the original tumor upon orthotopic implantation. They have been postulated to be the culprit of glioma chemo- and radio-resistance ultimately leading to relapse. Understanding the molecular circuits governing the GSC compartment is essential. SOX2, a critical transcription regulator of embryonic and neural stem cell function, is deregulated in GSCs however; the precise molecular pathways regulated by this gene in GSCs remain poorly understood.
Results
We performed a genome-wide analysis of SOX2-regulated transcripts in GSCs, using a microarray. We identified a total of 2048 differentially expressed coding transcripts and 261 non-coding transcripts. Cell adhesion and cell-cell signaling are among the most enriched terms using Gene Ontology (GO) classification. The pathways altered after SOX2 down-modulation includes multiple cellular processes such as amino-acid metabolism and intercellular signaling cascades. We also defined and classified the set of non-coding transcripts differentially expressed regulated by SOX2 in GSCs, and validated two of them.
Conclusions
We present a comprehensive analysis of the transcriptome controlled by SOX2 in GSCs, gaining insights in the understanding of the potential roles of SOX2 in glioblastoma.
Introduction
Glioblastoma is the most common and deadly primary brain tumors in adults and despite multiple treatments, the survival of glioma patients remains poor, with a median time between 12–15 months [1–3]
Glioblastoma contains a subpopulation of tumor propagating stem-like cells, known as glioma stem-like cells (GSCs) [4], which display the ability to self-renew, to differentiate into distinct lineages and to efficiently initiate and propagate tumors in xenografts models that recapitulate the phenotypic characteristics of the initial tumor from which they were derived [5,6]. Moreover, GSCs have been shown to increase resistance to radio-and chemotherapy [7,8], explaining in part the poor overall survival despite multiple treatments.
SOX2, a member of the SRY gene family, is a key transcription factor in the regulation of stemness properties, and it is essential in early embryonic development [9]. SOX2 has been reported to be deregulated in several human cancers [10–12] including glioblastoma where is over expressed due to several mechanisms such as amplification and promoter hypomethylation [13]. SOX2 is enriched in human-derived GSCs where it sustains stemness, migration, invasion and maintenance of tumorigenicity [13,14]. Although SOX2 response program in a glioblastoma cell line has been analyzed [15], to the best of our knowledge, an exhaustive analysis of SOX2-regulated molecular circuitries in GSCs has not been performed. Deciphering the molecular circuitries controlled by SOX2 in GSCs will provide insights about glioma development, biology and possible novel molecular therapies.
Besides coding genes, long non-coding RNAs (lncRNAs) are an emerging class of RNAs with no functional protein-coding ability that consists of more than 200 nucleotides [16]. Recent discoveries have proven that they play important roles regulating gene expression and function. These non-coding RNAs actively participate in many pathological processes in human malignancies [17–19] including cancer where a number of lncRNAs have been shown to act as oncogenes or tumor suppressors [20]. Recently, different groups published a signature of lncRNAs with aberrant expression in glioblastoma [21] and a set of prognostic lncRNAs that could have clinical implications in the sub-classification of this disease [22]; though the functional effect of lncRNAs in glioblastoma is not well understood.
Given that SOX2 is predominantly expressed in the GSCs compartment, which plays prominent roles in driving the growth, treatment resistance and recurrence of glioblastoma, the elucidation of the transcriptome and the molecular pathways involved in the generation and maintenance of this recalcitrant population is critical to understand the molecular underpinnings of glioblastoma malignancy. The aim of this work was to characterize the transcriptome regulated by SOX2 in GSCs. We set out to describe not only the coding genes but also the lncRNAs, which have been shown to play predominant roles in cancer. In this study we present a comprehensive analysis of the transcriptome controlled by SOX2 in GSCs, gaining insights in the understanding of the potential roles of SOX2 in glioblastoma.
Materials and Methods
Cell Lines, culture and transfection
Neurosphere cultures (GSC11 and GSC23), a kind gift of Dr. Lang at UT MD Anderson Cancer Center, were established from acute cell dissociation of human glioblastoma surgical specimens and maintained in Dulbecco's modified Eagle's medium/nutrient mixture F12 supplemented with B27 (Invitrogen, Carlsbad, CA), epidermal growth factor, and basic fibroblast growth factor (20 ng/mL each; Sigma-Aldrich, St Louis, MO).
To inhibit SOX2 expression, transient transfection assays were performed using two commercially available, specific siRNA against human SOX2 (si-SOX2, s13295 and s13294, Ambion) and a non-targeting control siRNA (si-scramble) (Ambion) in four independent experiments. The siRNA transfections were performed according to the manufacturer's instructions using Lipofectamine 2000 (Invitrogen). The cells were then cultured for 72 h after transfections and subjected to different analysis.
RNA extraction and Real Time PCR analysis
Total cellular RNA was isolated from the cultured cells using a Trizol reagent (Ambion) according to the manufacturers' protocols. For lncRNAs analysis, total RNA was subjected to DNase I treatment to digest the DNA. RNA quantity and quality were measured by NanoDrop ND-1000. The RNA samples were then reversely transcribed into cDNA using the Taqman miRNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's instructions. Real Time PCR (RT-PCR) was performed using the Sybr Green Fast Master Mix (Applied Biosystems) in the ABI 7700 sequence detection system (Applied Biosystems, Foster City, CA). The quality of the products was controlled by the melting curve. Transcript levels were normalized against human GAPDH. Transcripts expression levels relative to GAPDH were calculated using the ddCt method. Primers for lncRNA detection and quantification were designed at Universal ProbeLibrary Assay Design Center (http://www.roche-applied-science.com/). All primer sequences are listed below (Tables 1 and 2):
Table 1. Primers of lncRNAs for qRT-PCR.
| lncRNA position | TCONs | Forward Primer | Reverse Primer |
|---|---|---|---|
| chr19:28281401–28284848 | TCONS_00027256 | GCCCAAAGTTTGATTTCTCG | CGAGGTCTAACCCAGGTGTG |
| chr11:121899032–121899389 | TCONS_00020142 | GCTGAGCCTTCCATGAAAAT | GTGCAAATCACTCCAGTCACA |
Table 2. Primers of genes for qRT-PCR.
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| GAPDH | AGCCACATCGCTCAGACAC | GCCCAATACGACCAAATCC |
| SOX2 | AGCTCGCAGACCTACATGAA | CCGGGGAGATACATGCTGAT |
| PLP1 | ACCTATGCCCTGACCGTTG | TGCTGGGGAAGGCAATAGACT |
| COL2A1 | TGGACGCCATGAAGGTTTTCT | TGGGAGCCAGATTGTCATCTC |
| ATP8B1 | ACGACATTTGACGAGGATTCTC | GGTTTTGTTCTGGTTCAACAGC |
| PPP1R1B | CAAGTCGAAGAGACCCAACCC | GCCTGGTTCTCATTCAAATTGCT |
| CMTM5 | GGAGGACCACATCCGCTAGAT | CCAGGGAGTGGAAGCAGAT |
| GALNT14 | CACTGCTGGTGTATTGCACG | CGGATCAGATGCGTAGGGG |
| F11R | GTGCCTACTCGGGCTTTTCTT | GTCACCCGGTCCTCATAGGAA |
| SYT4 | ATGGGATACCCTACACCCAAAT | TCCCGAGAGAGGAATTAGAACTT |
| SLC18A1 | GTGGTGGTATTCGTCGCTTTG | CCGAGGTGCAGAGAAGAGT |
| ITLN2 | GCAGGGCAACAAAGCAGACTA | CAGGGCGCTGTTTCTCCAA |
Immunoblotting Assay
For the western blot assay, cells were lysed in RIPA buffer (Triton and PBS) for 30 min on ice. Samples containing identical amounts of protein (30 μg) were resolved in a 12% polyacrylamide gel, transferred to polyvinylidene membranes, and blocked in 5% nonfat milk in phosphate-buffered saline/Tween-20. Membranes were incubated with the following antibodies: SOX2 (Cell Signaling, Danvers, MA) and α-Tubulin (Sigma-Aldrich) using 1:1000 dilution. The membranes were developed according to the protocol for enhanced chemiluminiscence from Perkin Elmer.
Microarray expression analysis
Total RNA was isolated from scrambled and SOX2-siRNA GSC11 cells using Trizol extraction. RNA was purified by the QIAGEN RNAeasy mini kit (QIAGEN) according to the manufacturer´s protocol. One-color Cy3 RNA labeling, array hybridization to Agilent SurePrint G3 8 × 60 K Human Gene Expression Arrays (Agilent Technologies), data collection, and analysis were performed at the Bioinformatics Unit (Fundación para la Investigación Médica Aplicada, CIMA, Pamplona, Spain). Normalization of microarray data was performed using quantile algorithm. After quality assessment a filtering process was carried out to eliminate low expression probe sets. Applying the criterion of an expression value greater than 64 in 2 samples of at least one of the experimental conditions, 40986 probe sets were selected for statistical analysis. LIMMA (Linear Models for Microarray Data) [23] was used to find out the probe sets that showed significant differential expression between experimental conditions. Genes were selected as significant using a B statistic cut off B>0. Data processing and statistical analyses were performed with R and Bioconductor [24]. The microarray data from this study are publicly available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE79302.
Functional group analysis
In this study we applied Gene Ontology (GO) analysis to find the primary function of the differential expression of mRNAs regulated by SOX2, using online software DAVID (Database for Annotation, Visualization and Integrated Discovery, http://david.abcc.ncifcrf.gov/). GO analysis can organized genes into hierarchical categories (Gene Ontology Consortium). To find out the significant pathway of the differential genes participating we performed gene regulatory network analysis using Ingenuity Pathway Analysis (IPA) software (http://www.ingenuity.com), which can integrate gene-expression data with other molecular databases to facilitate the development of new and more complete pathway maps. Fisher's exact test was used to select the significant GO categories. The threshold of significance was defined by P value with a cut-off set in 0.05.
Statistical analysis
Experimental data are represented as the mean ± SD of three biologic replicates and were compared using Student's t-test. Significant P-values are indicated with asterisks as follows: *P < 0.05, **P < 0.01.
Results
Transcripts regulated by SOX2
Since SOX2 is a key driver in the maintenance of the GSCs phenotype and therefore in the perpetuation of this devastating tumor we down-regulated the expression of this gene in the GSC11 cell line in four independent experiments, using a SOX2 specific siRNA (Fig 1A). The efficiency of SOX2 knockdown was assessed by real-time PCR and western blot (Fig 1B) and was also confirmed in our array results. Microarray data identified a total of 2048 differentially expressed coding transcripts and 261 non-coding transcripts (B value >0) (Fig 1A and S1 Table).
Fig 1. Transcripts regulated by SOX2.
(A) Schematic representation of the research design employed to uncover the SOX2 transcriptome in GSC11 cells. (B) qRT-PCR and western blot confirmation of SOX2 inhibition in GSC11 cells after 72h of si-SOX2 or si-Scramble (si-Scrbl) transfection. SOX2 relative mRNA levels are presented as 2-ΔΔCt standardized with their constitutive gene GAPDH. Each bar represents the mean ± SD. For western blot tubulin was used as housekeeping control and shown as a representative blot of four independent experiments.
SOX2 controls a wide spectrum of protein-coding genes and pathways in GSCs
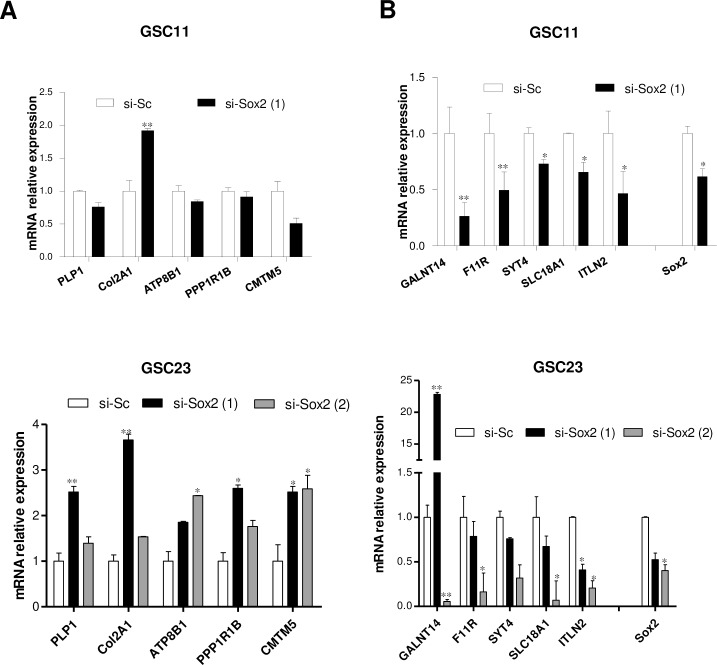
To further narrow the coding transcripts data a cut-off 1 logarithmic fold difference between SOX2 knockdown and scrambled GSC11 cells was set, identifying 35 up-regulated and 100 down-regulated genes, which suggest that SOX2 act primarily as a transcriptional activator. In Table 3 we showed the top-10 up or down-regulated protein coding-genes, and select the top 5 candidates of each group for further validation by qRT-PCR using the GSC11 and GSC23 cell lines. We confirmed the observed microarray expression changes in 5 out of 5 down-regulated coding-genes (Fig 2B) and in one out of 5 up-regulated coding genes in GSC11 cells (Fig 2A). Regarding GSC-23 cells, we down-modulate SOX2 expression using two different siRNAs against human SOX2, and we validate the expression of 5 out of 5 up-regulated coding-genes (Fig 2A) and of 4 out of 5 down-regulated coding genes (Fig 2B), partially validating the microarray results.
Table 3. The top 10 up- and down-regulated genes in Sox2-downmodulated GSC-11 cells, organized by logFC.
| GeneName | logFC | P.Value | B |
|---|---|---|---|
| PLP1 | 2,443 | 4,73E-05 | 2,488 |
| COL2A1 | 2,122 | 2,18E-05 | 3,281 |
| ATP8B1 | 1,954 | 1,50E-06 | 5,943 |
| PPP1R1B | 1,925 | 7,27E-06 | 4,393 |
| CMTM5 | 1,702 | 1,12E-04 | 1,586 |
| ELMO1 | 1,66 | 5,97E-05 | 2,246 |
| ITIH5L | 1,555 | 6,67E-05 | 2,133 |
| IGFBP5 | 1,522 | 1,82E-06 | 5,759 |
| SCARNA9 | 1,498 | 7,10E-05 | 2,067 |
| SCARNA17 | 1,435 | 4,46E-06 | 4,879 |
| GALNT14 | -3,006 | 0,000299 | 0,569 |
| F11R | -2,824 | 1,13E-07 | 8,308 |
| SYT4 | -2,664 | 2,61E-06 | 5,405 |
| SLC18A1 | -2,335 | 4,01E-06 | 4,984 |
| ITLN2 | -2,329 | 3,57E-07 | 7,288 |
| RASEF | -2,294 | 9,38E-07 | 6,39 |
| GADD45G | -2,231 | 3,55E-08 | 9,277 |
| CYP26A1 | -2,056 | 7,66E-10 | 11,938 |
| KRTAP21-1 | -2,028 | 0,00021 | 0,94 |
| PNLIPRP2 | -1,992 | 0,000495 | 0,042 |
Fig 2.
Analysis by qRT-PCR of the top 5 (A) up- and (B) down-regulated coding transcripts in SOX2 downmodulated GSC11 and GSC23 cells. Total RNA was extracted after 72h of si-Sc or si-SOX2 transfection in GSC11 and GSC23 cells. In GSC23 cells we used two different siRNAs against human SOX2, si-SOX2 (1) is referred to s13295 and si-SOX2 (2) is referred to s13294 from Ambion. Values are normalized to GAPDH and each bar represents the mean ± SD.
To understand the significance of differential gene expression, bioinformatics analysis related to Gene Ontology (GO) Classification and pathway analysis were performed. GO classifications using the DAVID web tool and pathway analysis using Ingenuity Pathway Analysis (IPA) was performed. For these analysis, gene lists were classified based upon decreased (logFC < -1) or increased (logFC > 1) expression relative to control and analyzed altogether as a single list.
Enrichment analysis of GO categories including biological process (BP), molecular function (MF), and cellular component (CC) were obtained using DAVID web tool (Fig 3). We observed the highest enrichment in the categories related to “cell adhesion”, “biological adhesion”, “cell-cell signaling”, “extracellular region” and “calcium ion binding”.
Fig 3. Top-10 GO Biological Processes analysis of protein-coding genes regulated by SOX2 in GSC11 cells.
Bar chart represents classification of GO Biological Processes, Cellular Component or Molecular Function as determined by DAVID web tool. Bars represent the number of genes in the specified category, organized by p-value.
We used IPA analysis to undercover the canonical pathways regulated by SOX2 in GSCs. Our results showed 13 pathways significantly altered (Table 4). Most of them related with amino-acid metabolism pathways, such as “histamine biosynthesis”, where histamine is an important regulator of numerous physiological processes including neurotransmission in the central nervous system (CNS) [25]; “L-cysteine degradation process” where cystathionine γ-lyase (CTH) activity has been related with glioblastoma treatment [26] and “serotonin receptor signaling pathway” being serotonin an important neurotransmitter in the CNS during neuronal development [27]. Other enriched pathways were “hematopoiesis from multipotent stem cells”, where KITLG has been reported to regulate neoplastic processes such as growth and invasion [28]; apoptosis [29] and cell adhesion [30]. Role of JAK2 in Hormone-like Cytokine Signaling stood out because GHR (growth hormone receptor) and IRS1 (insulin receptor substrate 1) has been linked with glioma progression [31,32]. A well-characterized pathway frequently altered in tumors is the NOTCH signaling cascade, which was also enriched in our analysis. The NOTCH pathway is a conserved intercellular signaling route that has been implicated in different developmental processes. Interestingly, NOTCH pathway is deregulated in human glioblastoma and plays a key role in maintaining the growth, the undifferentiated state of glioma cells and tumorigenesis [33–35]. The integrated analysis of SOX2 enriched canonical pathways revealed the link between this transcription factor and multiple cellular processes such as amino-acid metabolism and intercellular signaling cascades, like NOTCH pathway.
Table 4. List of top-13 canonical pathways identified by IPA software.
| Pathway | -log(p-value) | Ratio | Molecules |
|---|---|---|---|
| Glycine Betaine Degradation | 2,63E+00 | 2,50E-01 | DMGDH,PIPOX |
| Hepatic Stellate Cell Activation | 2,38E+00 | 4,42E-02 | LY96,COL2A1,COL22A1,IGFBP5,COL28A1 |
| Histamine Biosynthesis | 2,03E+00 | 1,00E+00 | HDC |
| L-cysteine Degradation II | 2,03E+00 | 1,00E+00 | CTH |
| Triacylglycerol Degradation | 2,02E+00 | 1,25E-01 | PNLIPRP2,CES1 |
| Retinol Biosynthesis | 2,02E+00 | 1,25E-01 | PNLIPRP2,CES1 |
| Hematopoiesis from Multipotent Stem Cells | 1,73E+00 | 5,00E-01 | KITLG |
| Cysteine Biosynthesis/Homocysteine Degradation | 1,73E+00 | 5,00E-01 | CTH |
| Role of JAK2 in Hormone-like Cytokine Signaling | 1,61E+00 | 7,69E-02 | GHR,IRS1 |
| Serotonin Receptor Signaling | 1,58E+00 | 7,41E-02 | SLC18A1,HTR1D |
| Phenylethylamine Degradation I | 1,56E+00 | 3,33E-01 | AOC3 |
| Notch Signaling | 1,35E+00 | 5,56E-02 | HES5,HEY1 |
| Lysine Degradation V | 1,34E+00 | 2,00E-01 | PIPOX |
This selection is organized by the negative logarithm of p-values (Fisher Test), calculated by IPA ([-Log (0.05) = 1.3]).
The IPA analysis also showed the most relevant biological functions and diseases in our data set. The most significant bio-functions altered following SOX2 down-modulation are showed in Table 5. The set of SOX2-associated genes were assigned mainly to the following networks: “cancer”, “organismal injury and abnormalities”, “cellular movement”, “tissue morphology”, “cellular development” and “hematopoiesis”. Interestingly, most of these networks involved very well-known functions of SOX2 such as morphology determination [36], development [37] and cellular proliferation and migration in glioma [13,13]. Fig 4 shows the most relevant selection of bio-function categories: disease and disorders, molecular and cellular functions and physiological system development and function, obtained by using IPA software organized by p-value.
Table 5. The top ten significant Bio-Functions altered following Sox2 down-modulation in the GSC11 cell line.
| Category | p-value | Number of Targets |
|---|---|---|
| Dermatological Diseases and Conditions | 1,63 x 10−08–9,35 x 10−03 | 74 |
| Cancer | 2,93 x 10−08–9,35 x 10−03 | 93 |
| Organismal Injury and Abnormalities | 2,93 x 10−08–9,35 x 10−03 | 100 |
| Cellular Movement | 3,98 x 10−06–9,35 x 10−03 | 41 |
| Connective Tissue Development and Function | 1,71 x 10−05–9,35 x 10−03 | 23 |
| Tissue Morphology | 1,71 x 10−05–9,35 x 10−03 | 39 |
| Reproductive System Disease | 2,52 x 10−05–9,35 x 10−03 | 16 |
| Cellular Development | 2,72 x 10−05–9,35 x 10−03 | 51 |
| Hematological System Development and Function | 2,72 x 10−05–9,35 x 10−03 | 31 |
| Hematopoiesis | 2,72 x 10−05–9,35 x 10−03 | 9 |
The p-value range indicates the p-values of the various pathways and processes belonging to that category. The number of targets indicates the total number of genes associated with the functional category.
Fig 4. 10-Top Bio Functions categories altered following SOX2 inhibition.
The categories listed are Physiological System Development and Function, Molecular and cellular Functions and Disease and Disorders, identified using IPA software. Bars represent the number of genes in the specified category, organized by p-value.
These results established a signature of protein coding-genes regulated by SOX2 in GSCs with biological functions relevant to glioblastoma growth and maintenance of its malignant phenotype. The tight overlap between the existing literature and our enrichment analysis highlights the robustness of our results and predicts that this approach will be an excellent discovery platform to identify novel SOX2 targets.
SOX2–regulated non-coding RNAs in GSC
Reprogramming transcription factors, including SOX2, have been shown to regulate both coding and non-coding RNAs [38]. LncRNAs are emerging as key regulators of biological processes and disease [39] therefore, seems reasonable to hypothesize that SOX2 will regulate this class of genes as well. The strength of our data-sets allowed us to identify potential non-coding transcripts differentially expressed (B value > 0) regulated by SOX2 in GSCs. After biotype distribution analysis we identify protein coding RNAs (44% for up-regulated and 41% for down-regulated), while the rest were classified as different types of non-coding transcripts. Out of the total number of transcripts differentially expressed we identify 80 upregulated and 181 down-regulated and we classify them as intergenic RNAs, antisense, processed transcripts, transcripts derived from pseudogenes and unassigned transcripts (Fig 5). The transcripts classified as “others” correspond to transcripts derived from miRNAs, rRNAs, sense-overlaping and sense intronic transcripts. The lncRNA annotation was performed with the Bioconductor package ChIPpeakAnno [40] and using Gencode v19 as reference [41]. The gene type corresponding to the gene that overlaps with the lincRNA locus was assigned to each lincRNA. Table 6 shows the top 25 non-coding transcripts regulated by SOX2 in GSCs.
Fig 5. SOX2 regulated non-coding transcripts.
(A) A total of 261 transcripts were found differentially expressed (B > 0), which were distributed in 80 upregulated and 181 downregulated transcripts. (B) Biotype distribution of the differentially expressed transcripts following SOX2 down-modulation in GSC11 cells.
Table 6. List of the top 25 non-coding transcripts regulated by Sox2, organized by B value.
| Probe | GeneName | Classification | logFC | B |
|---|---|---|---|---|
| A_19_P00320471 | lincRNA:chr9:2535671–2536375_R | antisense | -1,28 | 11,75 |
| A_19_P00315804 | lincRNA:chr9:2530903–2539456_R | antisense | -1,15 | 10,77 |
| A_19_P00320469 | lincRNA:chr9:2535671–2536375_R | antisense | -1,27 | 9,70 |
| A_19_P00811613 | lincRNA:chr9:2452800–2552025_R | antisense | -1,17 | 7,94 |
| A_33_P3397743 | LOC100128088 | pseudogene | -1,83 | 7,42 |
| A_19_P00321203 | lincRNA:chr6:72126155–72129954_R | lincRNA | -0,92 | 7,33 |
| A_19_P00322118 | lincRNA:chr2:39745746–39826668_F | antisense | -0,86 | 6,90 |
| A_23_P3552 | LOC730092 | pseudogene | -0,65 | 6,50 |
| A_19_P00322220 | lincRNA:chr20:37055062–37063887_R | processed_transcript | -0,69 | 6,28 |
| A_33_P3392460 | LOC100128077 | processed_transcript | -1,47 | 6,18 |
| A_19_P00322149 | lincRNA:chr6:72126142–72129923_R | lincRNA | -0,91 | 6,16 |
| A_19_P00317793 | lincRNA:chr20:37055062–37063916_R | processed_transcript | -0,68 | 6,10 |
| A_19_P00808846 | lincRNA:chr21:17992729–18010729_F | lincRNA | -0,64 | 6,09 |
| A_19_P00318304 | lincRNA:chr20:37050986–37063998_R | processed_transcript | -0,67 | 5,94 |
| A_19_P00316341 | lincRNA:chr7:130600800–130606702_F | lincRNA | -0,86 | 5,93 |
| A_19_P00316985 | lincRNA:chr6:72126162–72129969_R | lincRNA | -0,91 | 5,93 |
| A_19_P00322967 | lincRNA:chr20:37050934–37057222_R | processed_transcript | -0,69 | 5,54 |
| A_19_P00802098 | lincRNA:chr2:3579550–3585150_R | lincRNA | -0,58 | 5,15 |
| A_24_P756289 | SOX2OT | other | -0,86 | 5,08 |
| A_33_P3613516 | LOC254057 | antisense | -1,10 | 4,94 |
| A_19_P00318174 | lincRNA:chr2:3579840–3584422_R | lincRNA | -0,73 | 4,52 |
| A_33_P3287710 | chr10:79,686,570–79,689,583 | unassigned | -0,67 | 4,46 |
| A_33_P3405043 | LOC100133264 | unassigned | -0,72 | 4,43 |
| A_32_P88349 | LOC730256 | pseudogene | -0,48 | 4,36 |
| A_33_P3705884 | chr19:28,281,401–28,284,848 | lincRNA | -0,89 | 4,27 |
| A_19_P00321044 | lincRNA:chr16:50682543–50683160_F | lincRNA | 1,04 | 6,09 |
| A_19_P00315647 | lincRNA:chr11:121899032–121899389_R | other | 0,65 | 5,63 |
| A_32_P63013 | LOC283174 | unassigned | 1,32 | 5,08 |
| A_32_P47157 | LOC92973 | unassigned | 0,74 | 4,95 |
| A_19_P00317484 | lincRNA:chr3:112315643–112316945_R | lincRNA | 0,55 | 4,11 |
| A_19_P00809440 | lincRNA:chr11:133765815–133774297_R | other | 1,23 | 4,06 |
| A_33_P3789382 | chr10:65,224,989–65,226,322 | antisense | 0,57 | 3,98 |
| A_19_P00321420 | lincRNA:chr11:133766329–133767054_R | unassigned | 1,34 | 3,74 |
| A_19_P00332120 | lincRNA:chr3:156455706–156471081_R | lincRNA | 0,59 | 3,57 |
| A_19_P00320101 | lincRNA:chr11:133767609–133771496_R | other | 1,06 | 3,51 |
| A_19_P00812924 | lincRNA:chr11:121895965–121904065_R | other | 0,53 | 3,46 |
| A_19_P00326763 | lincRNA:chr3:112308735–112318605_R | lincRNA | 0,48 | 2,73 |
| A_33_P3753757 | LOC158402 | other | 0,53 | 2,46 |
| A_33_P3393679 | LOC645323 | lincRNA | 0,47 | 2,24 |
| A_19_P00315649 | lincRNA:chr11:121899032–121899389_R | other | 0,55 | 2,23 |
| A_19_P00809838 | lincRNA:chrX:100247844–100257469_R | unassigned | 0,79 | 2,20 |
| A_19_P00331576 | lincRNA:chr3:114043485–114052926_F | unassigned | 0,36 | 2,00 |
| A_33_P3259557 | LOC440104 | pseudogene | 0,45 | 1,67 |
| A_19_P00319347 | lincRNA:chr2:168149680–168414843_F | lincRNA | 0,77 | 1,67 |
| A_19_P00320212 | lincRNA:chr9:114795825–114797203_R | other | 0,41 | 1,63 |
| A_24_P93703 | LOC440104 | pseudogene | 0,40 | 1,50 |
| A_19_P00318878 | lincRNA:chr1:247350513–247352101_R | lincRNA | 0,38 | 1,44 |
| A_19_P00316010 | lincRNA:chr17:67547498–67549996_F | lincRNA | 0,57 | 1,43 |
| A_24_P349207 | ENST00000380727 | pseudogene | 0,29 | 1,31 |
| A_19_P00802064 | lincRNA:chr8:2522118–2527693_R | lincRNA | 0,38 | 1,04 |
The top four differentially expressed lncRNAs (Table 7) that presented chromatin marks and high abundance in brain were selected using GRCh37/hg19 assembly in UCSC Genome Browser, and their expression was validated using qRT-PCR in GSC11 cells, comparing SOX2-siRNA versus scrambled control. Our data indicated that the expression of chr19:28,281,401–28,284,848 (TCONS_00027256) was significantly down-regulated (p value = 0,018), while chr11:121899032–121899389 (TCONS_00020142) was significantly up-regulated (p value = 0.042) after SOX2 inhibition (Fig 6A). The results were consistent with the microarray data (Fig 6B).
Table 7. List of the top-four lncRNAs regulated by Sox2, organized by p-value.
| lncRNA | logFC | P.Value | B |
|---|---|---|---|
| lincRNA:chr6:72126155–72129954 | -0,9188744 | 3,40E-07 | 7,3327381 |
| lincRNA:chr6:29701971–29740296 | -0,78724372 | 3,93E-07 | 7,2007586 |
| lincRNA: chr19:28,281,401–28,284,848 | -0,89247792 | 8,26E-06 | 4,265306 |
| lincRNA:chr11:121899032–121899389 | 0,64633267 | 2,07E-06 | 5,6334606 |
This selection was evaluated by the presence of histone modifications and high abundance in brain according to UCSF genome browser tool.
Fig 6. Validation of two lncRNAs regulated by SOX2 in GSC11 cells.
(A) The expression of the transcripts located in chr11:121899032–121899389 (TCONS_00020142) and chr19:28,281,401–28,284,848 (TCONS_00027256) were assessed. In both cases GSC11 cells were transfected with siRNA control or siRNA against SOX2 and three days later RNA was extracted and subject to RT-PCR. Values are normalized to GAPDH and are the mean ± SD of three replicates. (B) Comparison between microarray and qRT-PCR results. The height of each column in this graph represents the log-transformed mean fold changes in the expression of lncRNA between Scramble and siSOX2 transfected cell line.
Altogether, these results identified and confirmed the non-coding transcript profile controlled by SOX2 in GSCs. Characterizing the functional relevance of these lncRNAs will undoubtedly impact our understanding of glioblastoma biology.
Discussion
Our work provides a comprehensive view of the genome wide SOX2 regulated transcripts in GSCs, illustrating a complex scenario where SOX2 is the central player regulating different molecules and pathways in glioblastoma.
In this study, we used state-of-the art microarray technology to query the SOX2 coding and non-coding RNA transcriptome in GSCs. It is interesting to note that among the down-regulated genes following SOX2 knockdown, F11R has been shown to be overexpressed in glioblastoma cells [42,43]. F11R is necessary and sufficient for GSC maintenance and self-renewal and of clinical significance is associated with increased malignancy and poor patient prognosis [42,43]. On the other hand, we found several interesting over-expressed candidates controlled by SOX2; for example, PPP1R1B is a well-known striatal projection neuron signature marker [44]. The fact that its expression increases following SOX2 inhibition is in line with its role in neuronal differentiation.
In a previous work where the SOX2 response program in a glioblastoma cell line was analyzed [15], authors identified 489 genes whose expression were altered in response to SOX2 knockdown, using several genomic technologies. Interestingly several of these genes are also differentially expressed in our array data, such as NGFR, CEBPA, BCL2, BNIP3, EBF4, ALCAM, protocadherins and solute carrier family members. Overall these results highlight the strength of our array data and the link between SOX2 and GSCs biology. The work of Fang and colleagues exhibits some similarities with our study, such as the analysis of SOX2 regulated-coding genes. However, we focused in the molecular circuitries controlled by SOX2 in GSCs, meanwhile they performed their study in an established glioma cell line. Additionally, our study addressed the SOX2-regulated lncRNAs in GSCs. Altogether both works provide clues regarding SOX2 functions in glioblastoma.
Gene-set enrichment analysis shows SOX2 is involved in regulating “cell adhesion”, “biological adhesion”, “cell-cell signaling”, and “calcium ion binding” pathways, undercovering its key function as a driver of the glioma stem-like phenotype [45–48].
We also analyzed the canonical pathways regulated by SOX2 in GSCs. Pathways related with amino-acid metabolism were among the most deregulated, illustrating that SOX2 expression is critical for maintaining metabolic homeostasis in the GSC population, and plays important role in different tumor microenvironment conditions, such as hypoxic stress conditions [49]. Other enriched pathway altered in our analysis was the NOTCH pathway, where Hes5 and Hey1 had the most significantly down-modulated expression. Hes5 is a marker of neural multipotent progenitors with stem cell properties [50] where it sustains progenitors proliferative state inhibiting their differentiation into neurons [51]. On the other hand Hey1 has been related to a subset of molecules directly associated with hypoxia in glioblastoma tumors [52]; and might be used as a marker to distinguish glioblastoma patients with a relative good prognosis (negative Hey1 expression) [53]. Furthermore, Hey 1 is up-regulated in glioma samples correlating with tumor grade, and functionally its down-regulation results in a proliferation reduction [54], suggesting a role in the progression of glioblastoma. Taking all this into account, the canonical pathways more significantly altered after SOX2 inhibition are those related with intracellular signaling cascades and amino-acid metabolism pathways associated with tumor propagation.
Consistent with what is known about SOX2 biological function, our data-set is enriched with genes involved in morphology determination, development and cellular proliferation and migration. Interestingly, “proliferation of tumor cells” (S2 Table), is one of the most repeated subcategories for which IPA analysis assigned an activation Z score close to -2, predicting it´s inhibition, which is in line with a putative role of SOX2 in cell proliferation.
One of the most exciting aspects of our study involved expanding our knowledge of the SOX2 transcriptome into the realm of lncRNAs. In this study we showed and classified the lncRNA landscape regulated by SOX2 in GSCs. Our microarray results showed a strong correlation with published reports, demonstrating the strength of our approach and providing confidence that we can use this data-set for de novo discovery of novel SOX2 targets, including lncRNAs. One previous study determined the differentially expressed lncRNAs between glioblastoma and brain tissues, showing 654 lncRNAs upregulated and 654 down-regulated [55]. To our knowledge, this is the first study that evaluates the differentially expression of lncRNAs in GSCs controlled by SOX2.
Among the transcripts regulated by SOX2 we found that SOX2OT was down-regulated in our data set, even though we did not validate it. SOX2OT is a lncRNA which harbors SOX2 gene in its intronic region and is transcribed in the same orientation as SOX2 [56]. Several studies have demonstrated a role of SOX2OT in the regulation of SOX2 gene in human stem cells [57,58] although little is known about the exact role of this non-coding RNA. SOX2OT has been associated with carcinogenesis and, for example in breast cancer is involved in the induction and/or maintenance of SOX2 expression [59], in esophageal squamous cell carcinoma has been shown to play a role in tumor initiation and/or progression as well as in regulation of the pluripotent state of stem cells [58], and proliferation in lung cancer cells [60]. Askarian-Amiri et al demonstrated that SOX2OT has a positive effect on SOX2 expression [59]. Published data suggest the mediation of lncRNA SOX2OT in pluripotency and tumorigenesis events, probably through regulation of SOX2 expression. These data together with our own results suggest a possible role of SOX2OT in the malignant phenotype of glioblastoma, however further functional and mechanistic studies will be necessary to elucidate the precise role of SOX2OT and other lncRNA candidates in the tumorigenicity of glioblastoma.
Although advance in managing and treating glioblastoma have been made, tumor recurrence and treatment resistance remains the major cause of glioblastoma mortality. Understanding how factors such as SOX2 drive the glioblastoma tumor phenotype will aid in the development of new therapeutic approaches based on targeting GSCs. Our study integrates for the first time the coding and non-coding transcriptome controlled by SOX2 in GSCs, gaining new insights about the molecular circuitries governing glioblastoma biology.
Conclusion
In conclusion we have performed a comprehensive analysis of differential expression of coding and non-coding transcripts controlled by SOX2 in GSCs. We performed gene set enrichment analysis to find the most relevant pathways and biological functions altered in our data set. This integrated analysis allows for a better understanding of the SOX2 transcriptome in GSCs.
Supporting Information
(XLS)
(XLS)
Acknowledgments
We are grateful to Dr. Yolanda Sánchez and Dr. John Laterra for critical reading of the manuscript.
Data Availability
The microarray data from this study are publicly available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE79302.
Funding Statement
This work was supported by the European Union (Marie Curie IRG270459 to MMA), the Instituto de Salud Carlos III y los Fondos Feder Europeos (PI13/125 to MMA), the Spanish Ministry of Economy and competitiveness (IEDI-2015-00638 to MMA), The L`OREAL-Unesco Foundation (to MMA), The Department of Health of the Government of Navarra 22/2015 (to MMA), The Basque Foundation for Health Research (BIOEF, BIO13/CI/005) and Fundación Caja Navarra (Convocatoria de Ayudas 2015 to MMA). AMAR is supported by a fellowship from the Friends of the University of Navarra Foundation.
References
- 1.Ostrom QT, Gittleman H, Stetson L, Virk SM, Barnholtz-Sloan JS. Epidemiology of gliomas. Cancer Treat Res. 2015;163: 1–14. 10.1007/978-3-319-12048-5_1 [DOI] [PubMed] [Google Scholar]
- 2.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14 Suppl 5: v1–49. 10.1093/neuonc/nos218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10: 459–466. 10.1016/S1470-2045(09)70025-7 [DOI] [PubMed] [Google Scholar]
- 4.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432: 396–401. [DOI] [PubMed] [Google Scholar]
- 5.Singh AK, Arya RK, Maheshwari S, Singh A, Meena S, Pandey P, et al. Tumor heterogeneity and cancer stem cell paradigm: updates in concept, controversies and clinical relevance. Int J Cancer. 2015;136: 1991–2000. 10.1002/ijc.28804 [DOI] [PubMed] [Google Scholar]
- 6.Wakimoto H, Mohapatra G, Kanai R, Curry WT Jr, Yip S, Nitta M, et al. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro Oncol. 2012;14: 132–144. 10.1093/neuonc/nor195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444: 756–760. [DOI] [PubMed] [Google Scholar]
- 8.Auffinger B, Spencer D, Pytel P, Ahmed AU, Lesniak MS. The role of glioma stem cells in chemotherapy resistance and glioblastoma multiforme recurrence. Expert Rev Neurother. 2015;15: 741–752. 10.1586/14737175.2015.1051968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17: 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussenet T, Dali S, Exinger J, Monga B, Jost B, Dembele D, et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLOS ONE. 2010;5: e8960 10.1371/journal.pone.0008960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41: 1238–1242. 10.1038/ng.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40: 499–507. 10.1038/ng.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alonso MM, Diez-Valle R, Manterola L, Rubio A, Liu D, Cortes-Santiago N, et al. Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PLOS ONE. 2011;6: e26740 10.1371/journal.pone.0026740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27: 40–48. 10.1634/stemcells.2008-0493 [DOI] [PubMed] [Google Scholar]
- 15.Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, et al. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics. 2011;12: 11-2164-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10: 155–159. 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- 17.Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y, et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst. 2013;9: 407–411. 10.1039/c2mb25386k [DOI] [PubMed] [Google Scholar]
- 18.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464: 1071–1076. 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48: R45–53. 10.1530/JME-12-0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14: 4655–4669. 10.3390/ijms14034655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, et al. Human cancer long non-coding RNA transcriptomes. PLOS ONE. 2011;6: e25915 10.1371/journal.pone.0025915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang XQ, Sun S, Lam KF, Kiang KM, Pu JK, Ho AS, et al. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol Dis. 2013;58: 123–131. 10.1016/j.nbd.2013.05.011 [DOI] [PubMed] [Google Scholar]
- 23.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3: Article3. [DOI] [PubMed] [Google Scholar]
- 24.Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W. Bioinformatics and computational biology solutions using R and Bioconductor Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S. ed. New York, NY: Springer; 2005. [Google Scholar]
- 25.Nuutinen S, Panula P. Histamine in neurotransmission and brain diseases. Adv Exp Med Biol. 2010;709: 95–107. [DOI] [PubMed] [Google Scholar]
- 26.Chen L, Li X, Liu L, Yu B, Xue Y, Liu Y. Erastin sensitizes glioblastoma cells to temozolomide by restraining xCT and cystathionine-gamma-lyase function. Oncol Rep. 2015;33: 1465–1474. 10.3892/or.2015.3712 [DOI] [PubMed] [Google Scholar]
- 27.Dinan TG. Serotonin: current understanding and the way forward. Int Clin Psychopharmacol. 1996;11 Suppl 1: 19–21. [PubMed] [Google Scholar]
- 28.Yang S, Li WS, Dong F, Sun HM, Wu B, Tan J, et al. KITLG is a novel target of miR-34c that is associated with the inhibition of growth and invasion in colorectal cancer cells. J Cell Mol Med. 2014;18: 2092–2102. 10.1111/jcmm.12368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carson WE, Haldar S, Baiocchi RA, Croce CM, Caligiuri MA. The c-kit ligand suppresses apoptosis of human natural killer cells through the upregulation of bcl-2. Proc Natl Acad Sci U S A. 1994;91: 7553–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flanagan JG, Chan DC, Leder P. Transmembrane form of the kit ligand growth factor is determined by alternative splicing and is missing in the Sld mutant. Cell. 1991;64: 1025–1035. [DOI] [PubMed] [Google Scholar]
- 31.Lea RW, Dawson T, Martinez-Moreno CG, El-Abry N, Harvey S. Growth hormone and cancer: GH production and action in glioma? Gen Comp Endocrinol. 2015;220: 119–123. 10.1016/j.ygcen.2015.06.011 [DOI] [PubMed] [Google Scholar]
- 32.Minchenko DO, Kharkova AP, Hubenia OV, Minchenko OH. Insulin receptor, IRS1, IRS2, INSIG1, INSIG2, RRAD, and BAIAP2 gene expressions in glioma U87 cells with ERN1 loss of function: effect of hypoxia and glutamine or glucose deprivation. Endocr Regul. 2013;47: 15–26. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Wakeman TP, Lathia JD, Hjelmeland AB, Wang XF, White RR, et al. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28: 17–28. 10.1002/stem.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28: 5–16. 10.1002/stem.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert CA, Daou MC, Moser RP, Ross AH. Gamma-secretase inhibitors enhance temozolomide treatment of human gliomas by inhibiting neurosphere repopulation and xenograft recurrence. Cancer Res. 2010;70: 6870–6879. 10.1158/0008-5472.CAN-10-1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou HY, Katsman Y, Dhaliwal NK, Davidson S, Macpherson NN, Sakthidevi M, et al. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev. 2014;28: 2699–2711. 10.1101/gad.248526.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferri A, Favaro R, Beccari L, Bertolini J, Mercurio S, Nieto-Lopez F, et al. Sox2 is required for embryonic development of the ventral telencephalon through the activation of the ventral determinants Nkx2.1 and Shh. Development. 2013;140: 1250–1261. 10.1242/dev.073411 [DOI] [PubMed] [Google Scholar]
- 38.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18: 1433–1445. 10.1101/gr.078378.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5: e944014 10.4161/21541272.2014.944014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu LJ, Gazin C, Lawson ND, Pages H, Lin SM, Lapointe DS, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11: 237-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22: 1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lathia JD, Li M, Sinyuk M, Alvarado AG, Flavahan WA, Stoltz K, et al. High-throughput flow cytometry screening reveals a role for junctional adhesion molecule a as a cancer stem cell maintenance factor. Cell Rep. 2014;6: 117–129. 10.1016/j.celrep.2013.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alvarado AG, Turaga SM, Sathyan P, Mulkearns-Hubert EE, Otvos B, Silver DJ, et al. Coordination of self-renewal in glioblastoma by integration of adhesion and microRNA signaling. Neuro Oncol. 2016;18: 656–666. 10.1093/neuonc/nov196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arber C, Precious SV, Cambray S, Risner-Janiczek JR, Kelly C, Noakes Z, et al. Activin A directs striatal projection neuron differentiation of human pluripotent stem cells. Development. 2015;142: 1375–1386. 10.1242/dev.117093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inoue A, Takahashi H, Harada H, Kohno S, Ohue S, Kobayashi K, et al. Cancer stem-like cells of glioblastoma characteristically express MMP-13 and display highly invasive activity. Int J Oncol. 2010;37: 1121–1131. [DOI] [PubMed] [Google Scholar]
- 46.Reddy EM, Chettiar ST, Kaur N, Ganeshkumar R, Shepal V, Shanbhag NC, et al. Dlxin-1, a member of MAGE family, inhibits cell proliferation, invasion and tumorigenicity of glioma stem cells. Cancer Gene Ther. 2011;18: 206–218. 10.1038/cgt.2010.71 [DOI] [PubMed] [Google Scholar]
- 47.Dietrich J, Diamond EL, Kesari S. Glioma stem cell signaling: therapeutic opportunities and challenges. Expert Rev Anticancer Ther. 2010;10: 709–722. 10.1586/era.09.190 [DOI] [PubMed] [Google Scholar]
- 48.Liebelt BD, Shingu T, Zhou X, Ren J, Shin SA, Hu J. Glioma Stem Cells: Signaling, Microenvironment, and Therapy. Stem Cells Int. 2016;2016: 7849890 10.1155/2016/7849890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kucharzewska P, Christianson HC, Belting M. Global profiling of metabolic adaptation to hypoxic stress in human glioblastoma cells. PLOS ONE. 2015;10: e0116740 10.1371/journal.pone.0116740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basak O, Taylor V. Identification of self-replicating multipotent progenitors in the embryonic nervous system by high Notch activity and Hes5 expression. Eur J Neurosci. 2007;25: 1006–1022. [DOI] [PubMed] [Google Scholar]
- 51.Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39: 13–25. [DOI] [PubMed] [Google Scholar]
- 52.Irshad K, Mohapatra SK, Srivastava C, Garg H, Mishra S, Dikshit B, et al. A combined gene signature of hypoxia and notch pathway in human glioblastoma and its prognostic relevance. PLOS ONE. 2015;10: e0118201 10.1371/journal.pone.0118201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaetani P, Hulleman E, Levi D, Quarto M, Scorsetti M, Helins K, et al. Expression of the transcription factor HEY1 in glioblastoma: a preliminary clinical study. Tumori. 2010;96: 97–102. [DOI] [PubMed] [Google Scholar]
- 54.Hulleman E, Quarto M, Vernell R, Masserdotti G, Colli E, Kros JM, et al. A role for the transcription factor HEY1 in glioblastoma. J Cell Mol Med. 2009;13: 136–146. 10.1111/j.1582-4934.2008.00307.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu S, et al. LncRNA pro fi le of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol. 2012;40: 2004–2012. 10.3892/ijo.2012.1413 [DOI] [PubMed] [Google Scholar]
- 56.Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, et al. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33: 461–463. [DOI] [PubMed] [Google Scholar]
- 57.Amaral PP, Neyt C, Wilkins SJ, Askarian-Amiri ME, Sunkin SM, Perkins AC, et al. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 2009;15: 2013–2027. 10.1261/rna.1705309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shahryari A, Rafiee MR, Fouani Y, Oliae NA, Samaei NM, Shafiee M, et al. Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells. 2014;32: 126–134. 10.1002/stem.1542 [DOI] [PubMed] [Google Scholar]
- 59.Askarian-Amiri ME, Seyfoddin V, Smart CE, Wang J, Kim JE, Hansji H, et al. Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in breast cancer. PLOS ONE. 2014;9: e102140 10.1371/journal.pone.0102140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou Z, Zhao W, Zhou J, Shen L, Zhan P, Xu C, et al. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol. 2014;53: 380–388. 10.1016/j.biocel.2014.06.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
Data Availability Statement
The microarray data from this study are publicly available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE79302.