Abstract
The diamondback moth, Plutella xylostella, is one of the most important pests of cruciferous crops. We have earlier shown that N6-(2-hydroxyethyl)-adenosine (HEA) exhibits insecticidal activity against P. xylostella. In the present study we investigated the possible mechanism of insecticidal action of HEA on P. xylostella. HEA is a derivative of adenosine, therefore, we speculated whether it acts via P. xylostella adenosine receptor (PxAdoR). We used RNAi approach to silence PxAdoR gene and used antagonist of denosine receptor (AdoR) to study the insecticidal effect of HEA. We cloned the whole sequence of PxAdoR gene. A BLAST search using NCBI protein database showed a 61% identity with the Drosophila adenosine receptor (DmAdoR) and a 32–35% identity with human AdoR. Though the amino acids sequence of PxAdoR was different compared to other adenosine receptors, most of the amino acids that are known to be important for adenosine receptor ligand binding and signaling were present. However, only 30% binding sites key residues was similar between PxAdoR and A1R. HEA, at a dose of 1 mg/mL, was found to be lethal to the second-instar larvae of P. xylostella, and a significant reduction of mortality and growth inhibition ratio were obtained when HEA was administered to the larvae along with PxAdoR-dsRNA or antagonist of AdoR (SCH58261) for 36, 48, or 60 h. Especially at 48 h, the rate of growth inhibition of the PxAdoR knockdown group was 3.5-fold less than that of the HEA group, and the corrected mortality of SCH58261 group was reduced almost 2-fold compared with the HEA group. Our findings show that HEA may exert its insecticidal activity against P. xylostella larvae via acting on PxAdoR.
Introduction
Adenosine is an endogenous nucleoside that modulates numerous physiological processes, including oxygen and metabolic balance in tissues. Intracellular adenosine is an important intermediate in the biosynthetic pathway that generates adenosine triphosphate (ATP). However, the nucleoside can also be released from cells or formed extracellularly from ATP, adenosine diphosphate (ADP), and adenosine monophosphate (AMP). The extracellular adenosine acts as a stress hormone, causing neuromodulation, vasoconstriction [1–2]. Adenosine regulates cellular functions by binding to G protein coupled receptors (GPCRs). Four mammalian subtypes of the AdoR, namely, A1, A2A, A2B, and A3, have been identified, and their genes have been cloned. The subtypes have been shown to modulate intracellular levels of adenosine 3’,5’-cyclic monophosphate (cAMP) in different ways [3]. In certain cells, the adenosine receptor (AR) is also coupled to the calcium-mobilizing G protein subunit, Gαq [4]. The adenosine derivative N6-(2-hydroxyethyl)-adenosine (HEA) was first isolated from the Cordyceps pruinosa and was identified as a calcium antagonist that inhibits muscle contraction [5].
The diamondback moth, Plutella xylostella, is one of the most important pests of cruciferous crops. The use of chemical insecticides is the preferred method for its control, however, it leads to the development of insecticide-resistance among pests and poses concern over possible environmental and human health hazards. Therefore, a search for new insect-control agents is highly warranted. Entomopathogenic fungi may be an alternative source of insect-control agents because these fungi constitute a rich source of bioactive chemicals [6].
Our earlier study has shown that the Paecilomyces cicadae, an entomopathogenic fungi, exerts an insecticidal effect on diamondback moth, Plutella xylostella [7]. The insecticidal active compound was isolated from the fruiting body of P. cicadae which is often used as traditional Chinese medicine, and was identified to be HEA [8]. Most of the published articles of HEA focus on humans, no published studies reporting insecticidal activity of HEA apart from ours [9]. It has been shown that overexpression of DmAdoR (increasing adenosine) in vivo causes lethality or severe developmental anomalies on Drosophila melanogaster [10–12]. We are interested in whether HEA, an adenosine derivative, has insecticidal activity to P. xylostella via PxAdoR. Therefore, we examined the PxAdoR gene expression, growth and mortality of P. xylostella by down regulating PxAdoR gene function using RNAi technology and AR inhibitor.
Materials and Methods
Insect strains
Susceptible strains of P. xylostella 2 instar larvae were provided by Nanjing Agricultural University. The moths were reared on Chinese cabbage in the laboratory in the Institute of Plant Protection, Zhejiang Institute of Subtropical Crops, at 25 ± 1°C, 16 h/8 h light/dark photoperiod and 65 ± 5% relative humidity.
RNA extraction and cDNA synthesis
Total RNA was extracted from the whole bodies of P. xylostella second instar larvae using Trizol reagent (Invitrogen/Life Technologies, Paisley, UK) as per manufacturer's instructions and then quantitated by calculating the absorbance ratio and by agarose gel electrophoresis. First strand cDNA was synthesized from 2 μg of RNA with the RevertAid First Strand cDNA Synthesis Kit K1622 (Thermo Scientific, US) by following the manufacturer's protocol.
Cloning of Full-length PxAdoR cDNA
The mRNA was reverse transcribed to cDNA with the common primer named Oligo (dT), and the Gene Specific Primers (GSPs) were generated for the amplifications. A second set of primers that extend from the unknown end of the message back to the known region of the 3’end was provided by the poly (A) tail, while an appended homopolymer tail was used for the 5’end [13]. Degenerate primers PxTA-F and PxTA-R (Table 1) were designed for the amplification of a partial PxAdoR cDNA. A set of specific primers was synthesized based on the sequence of putative insert for 5'- and 3'-rapid amplification of cDNA ends (RACE). rCai-R1 and rCai-R2 were used for 5'-RACE, and rCai-F1 and rCai-F2 (Table 1) were used for 3'-RACE. The Reverse Transcriptase-Ploymerase Chain Reaction (RT-PCR) and PCR were uesd to amplify the ends of transcripts. RACE was performed by using the 5'-Full RACE Kit and 3'-Full RACE Core Set Version 2.0 (TakaRa, Dalian, China) according to the manufacturer's protocol. RACE products were gel purified and sequenced by Sangon Biotech Company (Shanghai, China).
Table 1. List of primers used in the study.
| Primer name | Primer sequence 5’-3’ |
|---|---|
| PxTA-F | GAGCGAGGTGATGACGGTGGA |
| PxTA-R | GACGATGATGGCGAGGTTCTGC |
| rCai-F1 | GCAGATGAGCGAGGTGATGACGGTG |
| rCai-F2 | CCTCGCCATCATCGTCTTCTTCTTCATCA |
| rCai-R1 | GGCGAGGTTCTGCGTGGCCTTGAC |
| rCai-R2 | CACCGTCATCACCTCGCTCATCTGCTT |
| Pxds-F | CCCGAAGGACGAGACAA |
| Pxds-R | CAGCGAAACAACATTACCAC |
| Xiaocaiye-18F | GACTCAACACGGGAAATCTCACCA |
| Xiaocaiye-18R | CCAGACAAATCGCTCCACCAACTA |
Bioinformatic analysis
Comparative and bioinformatic analyses of nucleotide sequences and deduced amino acids sequences were carried out at http://www.ncbi.nlm.nih.gov and http://cn.expasy.org. The nucleotide sequence, deduced amino acids sequence, and open reading frame (ORF) were analyzed, and sequence comparison was conducted through database searches using BLAST programs (http://www.ncbi.nlm.nih.gov/BLAST/). The phylogenetic analysis of PxAdoR from other species was done by Clusta X program version 1.83 using default parameters [14] and manual adjustment where necessary. A phylogenetic tree was constructed using MEGA (molecular evolutionary genetics analysis) program, version 4.0 [15] from Clustal W1.6 alignment.
PxAdoR Gene Silencing
A double-stranded RNA (348 bp) corresponding to a portion of the PxAdoR was synthesized by using a method that eliminates the cloning step [16]. Here, templates for in vitro transcription were produced by adding T7 promoter sequences to each 5’ end of cDNA template prepared by SMART-RACE cDNA Amplification Kit (Clontech) through polymerase chain reaction (PCR). The primers Pxds-F and Pxds-R (Table 1) used to generate a cDNA with T7 promoter sequences, were designed based on the PxAdoR sequence at positions 1314–1661. The PCR products were examined on agarose gel prior to in vitro transcription to verify that the products show a single band and the expected sizes. All dsRNA preparations were quantified spectrophotometrically and stored at −20°C until use.
Second instar larvae of P. xylostella were starved for 24 h and fed for 48 h with cabbage dishes which were cut into 10 cm diameter to fit the size of a petri dish [17], and drenched in 500 ng/mL [17]of Pxds or green fluorescent protein (GFP) -dsRNA 5 mins, and aired. Being a nontoxic reporter gene, GFP-dsRNA (dsGFP) was used as a control.
Analysis of silencing specificity
To assess the off-target effects of the silencing method used in this study, we checked the expression levels [17] of PxAdoR along with dsGFP control by Quantitative RT-PCR 48 h after RNAi. qRT-PCR assays were performed in a 25 μL reaction volume by using SYBR premix Ex-Taq Perfect Real-time Kit (TakaRa, Dalian, China), with 10 pmol of primers and 20 ng total RNA-equivalent of each cDNA generated by reverse transcription in a Thermal Cycler Dice Real-Time System ABI7500 (Applied Biosystems, USA) with Primers for PxAdoR target sequences, as well as for endogenous 18S ribosomal RNA control, were designed by prime 5.0 (Table 1). Serial (10×) dilutions of cDNA samples were prepared to generate the relative standard curves for each of the genes tested for each qRT-PCR run. The thermal cycling conditions are as follows: 95°C for 10 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. A dissociation step at 95°C for 15 s and 60°C for 30 s was added as a final step for melt curve analysis. The cycle threshold (Ct) value was used to calculate the transcript quantity of target genes on the basis of the standard curve. All calculations were performed by the software that accompanies the employed qRT-PCR machine. Each qPCR measurement was carried out independently for a minimum of three times, and the mean value was used for quantification. The 2−△△Ct method was used to analyze the relative changes in gene expression. The expression of the 18S gene was used as a control and the larvae fed the dsGFP were used as the negative control.
Statistical analysis
The results are presented as means ± standard deviation (SD). Statistical analysis was performed using analysis of variance (ANOVA) followed by LSD test or Student’s t-test using SPSS ver. 20.0 software. A significant difference was defined as P < 0.05 and very significant difference was defined as P < 0.01.
Effect of RNAi and receptor inhibitor on the insecticidal activity of HEA
Cabbages were cut into 10 cm diameter to fit the size of a petri dish, drenched in 1 mg/mL [9] HEA for 30 s, and aired. The second instar larvae were first allowed to feed on dsRNA for 48 hours and then ten of these larvae were transferred to cabbage treated with 1mg/ml HEA in a petri dish.
In the same method, the AR inhibitor SCH58261 was used as a positive control and the untreated larvae were used as a negative control. Each group consisted of five replicates. The mortalities and rate of growth inhibition were recorded after 24, 36, 48, and 60 h. Corrected mortality (%) = (mortality of treatment−mortality of control)/(1−mortality of control)×100. Rate of growth inhibition (%) = [(weight of control−weight of treatment)/ weight of control]×100.
Results
Cloning and Characterization of the PxAdoR Gene from P. xylostella
Transcriptome sequencing as part of our earlier study provided a 297 bp fragment. BLAST analysis showed that the nucleotide sequence of this fragment shared about 82% and 77% identities with Papilio machaon AR A2b-like and Nasonia vitripennis AR A2b.
Based on the sequence information, specific primers were designed for 5'-and 3'- RACE of the related gene. 5'-RACE generated a 784 bp fragment, and 3'-RACE produced a 960 bp fragment. Splicing the sequences generated a 1828 bp fragment, which was identified as the full-length PxAdoR gene (GenBank accession NO.KR258794). The ORF of PxAdoR cDNA encoded a protein of 440 amino acids residues with a calculated molecular mass of 64.74243 kDa and an isoelectric point of 11.495 (Fig 1).
Fig 1. cDNA sequence of Plutella xylostella Adenosine Receptor.
The line box area indicates the open reading frame (ORF), which encodes protein of 440 amino acids. The start codon (ATG) and the stop codon (TGA) are highlighted in black. The sequence was deposited in the GenBank (Assession No.KR258794).
Alignment of P. xylostella AdoR with other known AdoRs
After the sequences of the PxAdoR gene were determined, the evolutionary position of the gene among the various AdoR genes was investigated. The sequence of PxAdoR was submitted to NCBI for BLAST searches, and results showed that PxAdoR presents some homology with insect AdoR genes from other species, with 78% identity with Papilio machaon AdoR, 77% and 78% identity with Amyelois transitella and Papilio xuthus AdoR, and 61% identity with Drosophila melanogaster AdoR. The gene exhibited low similarity with human AdoR, with 33% identity with human A1R, 35% identity with A2aR, 32% identity with A2bR, and 34% identity with A3R (Table 2).
Table 2. The similarity of protein sequences between PxAdoR and other insect adenosine receptors.
| Description | Query cover | Identities |
|---|---|---|
| PREDICTED: adenosine receptor A2b-like [Papilio xuthus] | 100% | 78% |
| PREDICTED: adenosine receptor A2b-like [Papilio machaon] | 100% | 78% |
| PREDICTED: adenosine receptor A3 [Amyelois transitella] | 99% | 77% |
| PREDICTED: adenosine receptor A2b [Bombyx mori] | 99% | 73% |
| Adenosine receptor A2a [Papilio xuthus] | 68% | 74% |
| Adenosine receptor A2a [Papilio machaon] | 68% | 74% |
| GH18390 [Drosophila grimshawi] | 80% | 60% |
| hypothetical protein KGM_02847 [Danaus plexippus] | 68% | 72% |
| hypothetical protein FF38_04892 [Lucilia cuprina] | 79% | 62% |
| PREDICTED: uncharacterized protein LOC106087647 [Stomoxys calcitrans] | 78% | 62% |
| PREDICTED: uncharacterized protein LOC105233440 [Bactrocera dorsalis] | 82% | 58% |
| adenosine receptor, isoform B [Drosophila melanogaster] | 82% | 61% |
| PREDICTED: LOW QUALITY PROTEIN: adenosine receptor A2b [Plutella xylostella] | 47% | 99% |
| PREDICTED: adenosine receptor A2b [Apis mellifera] | 68% | 68% |
| PREDICTED: adenosine receptor A2b-like [Megachile rotundata] | 70% | 68% |
| PREDICTED: adenosine receptor A2b [Nasonia vitripennis] | 68% | 69% |
| hypothetical protein L798_01328 [Zootermopsis nevadensis] | 83% | 60% |
| AAEL002894-PA [Aedes aegypti]) | 67% | 65% |
| PREDICTED: adenosine receptor A2b-like isoform X1 [Wasmannia auropunctata] | 66% | 64% |
| PREDICTED: adenosine receptor A2b-like isoform X1 [Vollenhovia emeryi] | 66% | 65% |
| Adenosine receptor A2a [Operophtera brumata] | 60% | 72% |
| PREDICTED: adenosine receptor A2a [Tribolium castaneum] | 83% | 57% |
| adenosine receptor A2a [Homo sapiens] | 65% | 35% |
| adenosine receptor A2b [Homo sapiens] | 65% | 32% |
| adenosine receptor A3 isoform A [Homo sapiens] | 65% | 34% |
| adenosine receptor A1 [Homo sapiens] | 68% | 33% |
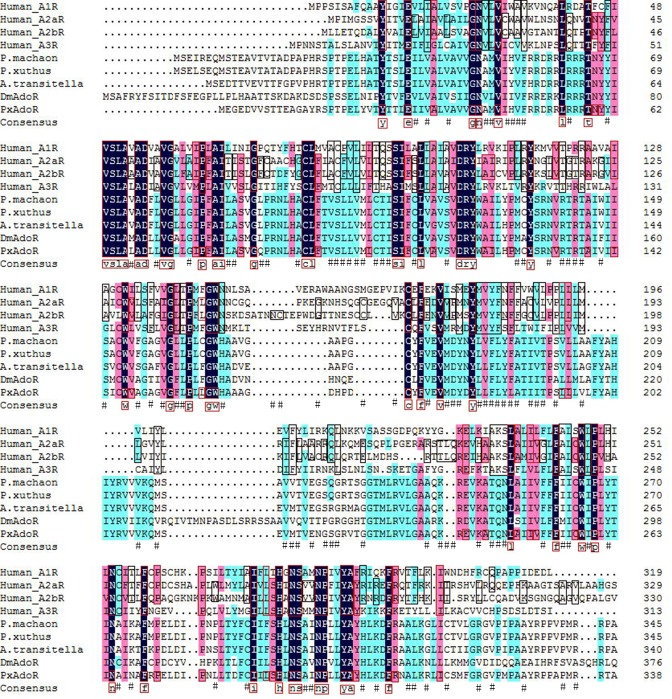
The predicted ORF of the PxAdoR gene encodes a protein of 440 amino acids. Using the method of Clustal W, a comparison of amino acids sequence showed that the N-terminal sequence of PxAdoR had a low homology with human A2b adenosine receptor, but was more similar with Drosophila AdoR and other insects AdoRs. The human adenosine receptor have 193 binding sites key residues [18], only 71 binding sites key residues of A1R, 84 of A2aR, 85 of A2bR, and 64 of A3R are identical compared to that of PxAdoR (Fig 2).
Fig 2. Alignment of the human A2b adenosine receptor and N-terminal part of AdoR amino acids sequences of other insects and Plutella xylostella.
Different color shows different homology levels. Black means 100% homology level, pink shows 75% homology level, blue stands for 50% homology level, and yellow marks 33% homology level. The PxAdoR and human adenosine receptors share the same key residues marked by red solid border and the mismatched key residues between PxAdoR and human adenosine receptors are marked by black solid border. “#” sign indicates the special residues of PxAdoR compared with human adenosine receptors.
The PxAdoR belongs to the G protein coupled receptors, has seven transmembrane helices [19] and contains the most important amino acids for adenosine receptor ligand binding [20–21].
PxAdoR silencing and specificity
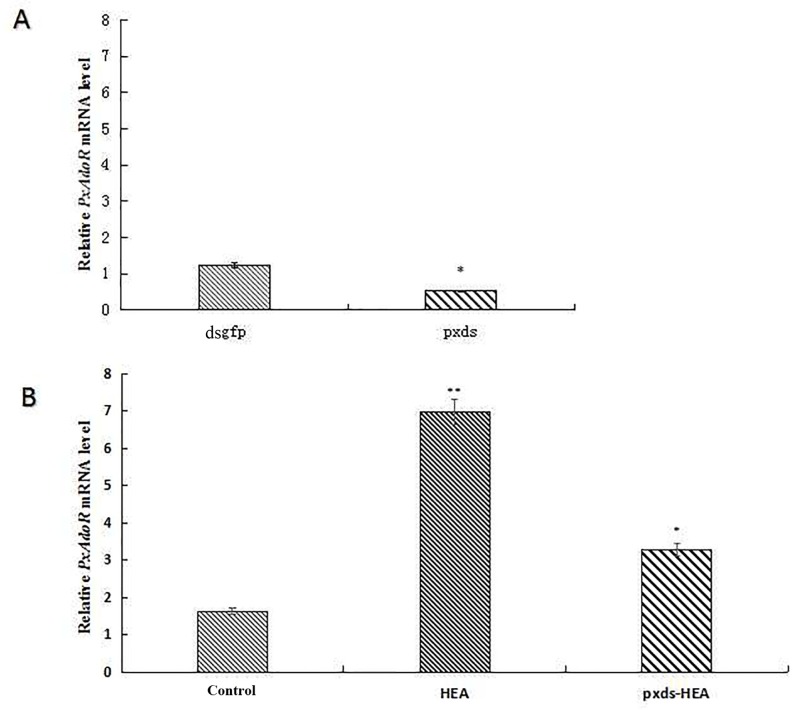
Quantitative RT-PCR analyzes the RNAi experiments showed that PxAdoR gene expression in the second instar larvae after RNAi was reduced by 2.36-fold compared to dsGFP-fed control (Table 3). The findings show that dsRNA feeding resulted in a significantly reduced the expression of PxAdoR gene (Fig 3A).
Table 3. The effect on relative PxAdoR mRNA level by qRT-PCR.
| Sample Name | △Ct (n = 3) | Relative PxAdoR mRNA level (2-△△Ct) (n = 3) |
|---|---|---|
| dsGFP1 | 22.153±0.144 | 1.239±0.664 |
| dsGFP2 | 22.612±0.357 | |
| dsGFP3 | 21.161±0.087 | |
| Pxds1 | 22.818±0.133 | 0.525±0.111 |
| Pxds2 | 23.057±0.113 | |
| Pxds3 | 23.443±0.267 | |
| Control1 | 17.893±0.069 | 1.611±0.543 |
| Control2 | 16.868±0.132 | |
| Control3 | 17.047±0.422 | |
| HEA1 | 14.996±0.018 | 6.850±0.584 |
| HEA2 | 15.125±0.064 | |
| HEA3 | 15.241±0.556 | |
| Pxds-HEA1 | 16.207±0.096 | 3.652±1.099 |
| Pxds-HEA2 | 15.600±0.412 | |
| Pxds-HEA3 | 16.390±0.026 |
Fig 3. Relative PxAdoR mRNA level.
(A) Effect of PxAdoR silencing by 500 ng/mL dsRNA feeding. (B) Effect of HEA feeding after silencing PxAdoR. Analyzed sample were cDNAs that were reverse-transcribed from pooled RNA samples of 10 larvae. Bars represent the means ± standard deviations of technical replicates. *P < 0.05; **P < 0.001 (A) compared with the dsgfp group. (B) compared with the control.
After treatment with 1 mg/mL HEA, the relative PxAdoR mRNA level showed a highly significant, almost 4.3-fold (P < 0.001) increase compared to the control (Table 3). The relative PxAdoR mRNA level of Pxds-HEA group exhibited a significantly reduced expression in comparison with the HEA group but was higher than the control. This data shows that HEA affects PxAdoR gene expression and suggests that it could induce insecticidal activity via P. xylostella AdoRs (Fig 3B).
Effect of RNAi and receptor inhibitor on the insecticidal activity of HEA on P. xylostella larvae
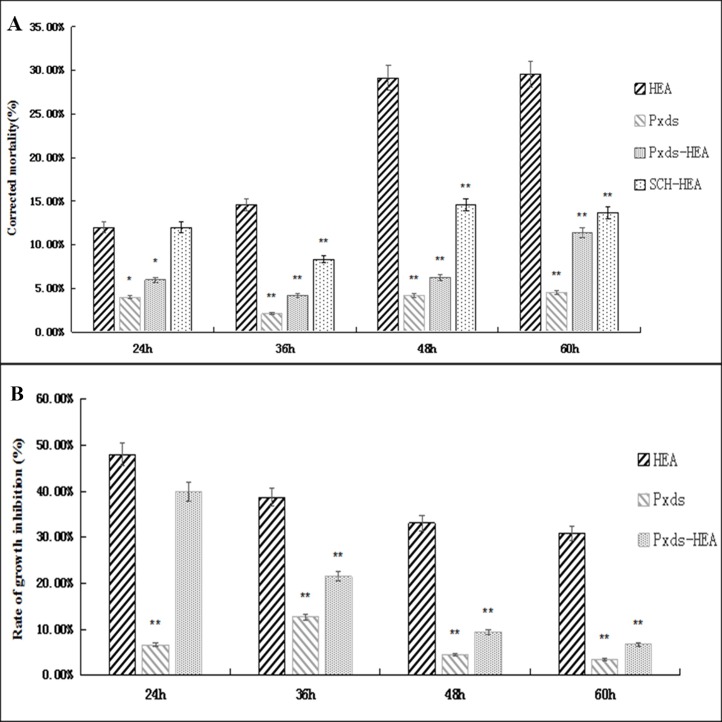
The mortality and rate of growth inhibition of the group fed with Pxds were the lowest compared to control group and the corrected mortality rates were found to be below 5% at 24, 36, 48, and 60 h time points. This data shows that Pxds feeding affected the insect body minimally. The corrected mortality of the Pxds–HEA group showed a significant reduction at all time points (24 h: from 12% to 6%; P < 0.05; 36 h: 14% to 4%; P < 0.001; 48 h: 29% to 6%; P < 0.001; 60 h: 29% to 11%; P < 0.001) compared with the HEA group at each time point. These results show that silencing PxAdoR gene can reduce the lethal effect of HEA on P. xylostella larvae. The corrected mortality of the SCH-HEA group at 24 h did not show a significant difference compared to the HEA group, however, a significant reduction in mortality was observed from 36 h to 60 h time points (P < 0.001). In particular, at 48 and 60 h, almost 2-fold reduction in mortality was observed (Fig 4A).
Fig 4. Impact of RNAi and Receptor Inhibitor on the insecticidal effect of HEA.
Larvae were fed with dsRNA or receptor inhibitor (SCH58261) with or without HEA for a period of 24 h to 60 h. *P < 0.05; **P < 0.001 compared with the HEA group. (A) Effect of PxAdoR silencing and SCH58261 on the mortality induced by HEA. Corrected mortality (%) = (mortality of treatment−mortality of control)/(1−mortality of control)×100. (B) Effect of PxAdoR silencing on the rate of larvae growth. Rate of growth inhibition (%) = [(weight of control−weight of treatment)/ weight of control]×100.
The rate of growth inhibition of the Pxds–HEA group at 24 h was not significantly (P > 0.05) different compared to the HEA group. However, a statistically significant (P < 0.001) reduction was observed at 36 h to 60 h (36 h: 38% to 21%; 48 h: 33% to 9%; 60 h: 30% to 6%) (Fig 4B). This shows that the insecticidal activity of HEA on P. xylostella larvae is via PxAdoR.
Discussion
In this study we investigated the possible underlying mechanism of insecticidal activity of HEA on P. xylostella. qRT-PCR and RACE was used to clone the gene of P. xylostella adenosine receptor (PxAdoR). According to the Franchetti’s study that A1R had 10 key binding sites residues (M180, L88, T91, H278, T277, W247, N254, L258, F185, V181) interacting with N6 derivatives CPA which was the agonist of A1R [22]. Only 3 residues of PxAdoR (H278, W247, N254) were same as that of A1R, also the similarity was 30%. We speculated that HEA which was the N6 derivatives adenosine has the same binding sites as CPA. Activation of A1R of human and mammals has effect of sedative, hypnotic, anticonvulsant [23], antiinflammatory [24]. HEA has the same effect for human through activating A1R.[25,26]. Thus, HEA could be a possible environmentally safe biological pesticide that is nontoxic to the human body.
Since PxAdoR exhibited considerable homology with AdoRs of other insects, this provides a possibility of selecting adenosine-like insecticides through other insect AdoRs e.g. adenosine receptor agonists 5’-N-Ethylcarboxamidoadenosine (NECA), N6-Cyclopentyladenosine (CPA), 2-Chloro-N6-cyclopentyladenosine (CCPA), and non-purine adenosine receptor agonist Paeoniflorin. This can further broaden the adenosine-AdoR insecticide spectrum.
Based on the sequence of the receptor gene, we designed specific primers for a double-stranded RNA synthesis. After dsRNA feeding to reduce the expression of PxAdoR, P. xylostella were fed with HEA and receptor antagonist SCH58261. The corrected mortality and the rate of growth inhibition were analyzed to find whether HEA acts through PxAdoR. Our data showed that Pxds–HEA group, with larvae that were fed with dsRNA and HEA, had a significantly lower mortality and rate of growth inhibition compared to the group treated with only HEA, thus showing that HEA acts through PxAdoR.
HEA was first isolated from the entomopathogenic fungi C. pruinosa and it was identified to be a calcium antagonist. That instance marked the first time that a synthetically known compound has been isolated biologically [5]. As part of our previous study we showed that P. cicadae exerts an insecticidal effect on diamondback moth, P. xylostella and isolated the insecticidal active compound, HEA, from the fruiting body of P. cicadae, which is used in ancient traditional Chinese medicine [9].
Based on our previous observations, we assessed the possibility of using HEA as a pesticide against the diamondback moth, P. xylostella. Being an adenosine derivative, HEA will be contacted with AdoRs. In a previous study, Tomas Dolezal et al. have shown that varying adenosine levels in Drosophila in vivo results in metamorphic changes such as developmental delay or fat body disintegration [27]. Another study by Monika Zuberova et al. showed that increasing extracellular adenosine levels in Drosophila affects the energy stores leading to wasting and death [10].
To address the question whether HEA, an adenosine derivative, induces insecticidal effects through its interaction with AdoRs, we used RNAi silencing of PxAdoR gene by feeding P. xylostella with specific dsRNA [28,17]. Our findings show that feeding dsRNA reduces expression of PxAdoR gene. We found that RNAi exerts evident effect from 24 h to 60 h after feeding dsRNA. Silencing of PxAdoR gene through RNAi could inhibit the lethal effect of HEA and the growth of P. xylostella larvae at 36–60 h. Further, treatment with an antagonist of A2A receptor could also weaken the effect of HEA on P. xylostella. These findings show that the HEA acts against P. xylostella via adenosine receptors.
Magazanik et al. studied the effects of adenosine and ATP on blowfly larvae, Calliphora vicina, and found that presynaptic AdoRs could regulate transmitter release at insect motor nerve terminals [29]. This finding showed that adenosine modulates neuronal activity via AdoRs. To determine if adenosine modulates neuronal activity in invertebrate neurons, Malik et al. conducted whole-cell recordings and found that adenosine can depress neuronal activity via AdoRs [30]. It could also be due to elevated Ca2+ due to the inactivation of N-methyl-D-aspartate receptors [31], which are Ca2+ permeable glutamate-gated ion channels that are regulated by [Ca2+] and are necessary for fast excitatory neurotransmission in the central nervous system [32]. It is known that adenosine activates the AR through the second messenger cAMP for Ca2+ control. However, there are no similar studies on PxAdoR. Therefore, as part of our future studies, we intend to investigate how the HEA effect on second messenger cAMP and Ca2+ by activation of AR of P. xylostella. This approach is expected to provide an in-depth understanding of the insecticidal mechanism of HEA.
In conclusion, our findings show that HEA acts on P. xylostella and induces insecticidal activity through PxAdoR, P. xylostella adenosine receptors. Since PxAdoR has a very low homology with human AdoRs, HEA can potentially be used as an environmentally safe pesticide against the diamondback moth, P. xylostella, further more, may screening pesticide with PxAdoR.
Acknowledgments
We thank Dr. Yidong Wu, Dr. Yihua Yang (College of Plant Protection, Nanjing Agricultural University) for providing the experimental equipment and material.
Data Availability
Data are available from Figshare (URL: https://dx.doi.org/10.6084/m9.figshare.3494744.v4; DOI:10.6084/m9.figshare.3494744).
Funding Statement
This work was supported by the Major priority projects of Zhejiang Province 2008C12004-3, Zhejiang Provincial Natural Science Foundation of China LY12C14004, Wenzhou Science and Technology Program Projects (CN) N20150010 and Agricultural science and Technology Achievements Transformation Fund Project of Zhejiang Province 2010D70088. The four fundings was received by Chai YQ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dunwiddie TV and Masino SA. The role and regulation of adenosine in the central nervous system. Annu Rev Neurosci. 2001; 24: 31–55. [DOI] [PubMed] [Google Scholar]
- 2.Nyce J W. Insight into adenosine receptor function using antisense and gene-knockout approaches. Trends Pharmacol Sci.1999; 20: 79–83. [DOI] [PubMed] [Google Scholar]
- 3.Newell EA, Exo JL, Verrier JD, Jackson TC, Gillespie DG, Janesko-Feldman K et al. 2′,3′-cAMP, 3′-AMP, 2′-AMP and adenosine inhibit TNF-α and CXCL10 production from activated primary murine microglia via A 2A receptors. Brain Res, 2014; 1594:27–35. 10.1016/j.brainres.2014.10.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cascio M G, Valeri D, Tucker S J, Marini P. A1-adenosine acute withdrawal response and cholecystokinin-8 induced contractures are regulated by Ca 2+—and ATP-activated K + channels. Pharmacol Res, 2015; s 95–96:82–91. 10.1016/j.phrs.2015.03.014 [DOI] [PubMed] [Google Scholar]
- 5.Furuya T, Hirotani M, Matsuzawa M. N6-(2-hydroxyethyl)-adenosine, a biologically active compound from cultured mycelia of Cordyceps and Isaria species. Phytochem, 1983; 22(22):2509–2512. [Google Scholar]
- 6.Kim J R, Yeon S H, Kim H S, Ahn YJl. Larvicidal activity against Plutella xylostella of cordycepin from the fruiting body of Cordyceps militaris. Pest Manag Sci, 2002; 58(7):713–7. [DOI] [PubMed] [Google Scholar]
- 7.Chen G J. Experiment on the Indoor Toxity of Paecilomyces cicadae APC20 to Plutella xylostella. J Anhui Agric Sci, 2008; 36(6):2380–2381. [Google Scholar]
- 8.Chai YQ, Jin YW, Chen GJ, Liu YG, Li XL, Wang GE. Separation of Insecticidal Ingredient of Paecilomyces cicadae (Miquel) Samson. Agric Sci China, 2007; 6(11):1352–1358. [Google Scholar]
- 9.Chai YQ, Chen GJ, Jin YW, Liu YG, Li XL, Wang GE. Use of N6-(2-hydroxyethyl)-adenosine in preparation of crop pesticide.United States patent US 20140256669. 2014.
- 10.Zuberova M, Fenckova M, Simek P, Janeckova L, Dolezal T. Increased Extracellular Adenosine in Drosophila That Are Deficient in Adenosine Deaminase Activates a Release of Energy Stores Leading to Wasting and Death. Dis Model Mech. 2010; 3(11–12): 773–784. 10.1242/dmm.005389 [DOI] [PubMed] [Google Scholar]
- 11.Dolezelova E, Nothacker HP, Civelli O, Bryant PJ, Zurovec M. A Drosophila adenosine receptor activates cAMP and calcium signaling. Insect Biochem Mol Biol, 2007;37(4):318–29. [DOI] [PubMed] [Google Scholar]
- 12.Kucerova L, Broz V, Fleischmannova J, Santruckova E, Sidorow R, Dolezal V, et al. Characterization of the Drosophila adenosine receptor: the effect of adenosine analogs on cAMP signaling in Drosophila cells and their utility for in vivo experiments. J Neuro, 2012; 121(3):383–95. [DOI] [PubMed] [Google Scholar]
- 13.Okamura Y, Takeyama H, Shirai M, Kajiyama T, Kambara H, Matsunaga T. Rapid amplification of cDNA ends (RACE). Methods Mol Biol, 2011; 703(5):107–22. [DOI] [PubMed] [Google Scholar]
- 14.Thompson JD, Gibson TJ, Plewniak F Jeanmougin F Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. Nucleic Acids Res, 1997; 25(24): 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol, 2007;24(8):1596–1599. [DOI] [PubMed] [Google Scholar]
- 16.Schepers. RNA Interference in Practice. Comp Funct Genomics, 2005; 6(5–6):320–322. [Google Scholar]
- 17.Bautista MAM, Miyata T, Miura K, Tananka T. RNA interference-mediated knockdown of a cytochrome P450, CYP6BG1, from the diamondback moth, Plutella xylostella, reduces larval resistance to permethrin. Insect Biochem Mol Biol, 2009; 39(1):38–46. 10.1016/j.ibmb.2008.09.005 [DOI] [PubMed] [Google Scholar]
- 18.Cheng F, Xu Z, Liu G, Tang Y. Insights into binding modes of adenosine A2B, antagonists with ligand-based and receptor-based methods. Eur J Med Chem, 2010; 45(8):3459–71. 10.1016/j.ejmech.2010.04.039 [DOI] [PubMed] [Google Scholar]
- 19.Tuccinardi T, Ortore G, Manera C, Saccomanni G, Martinelli A. Adenosine receptor modelling. A1/A2a selectivity. Eur J Med Chem, 2006; 41(3):321–9. [DOI] [PubMed] [Google Scholar]
- 20.Gao ZG, Chen A, Barak D, Kim SK, Muller CE, Jacobson Ka. Identification by site-directed mutagenesis of residues involved in ligand recognition and activation of the human A3 adenosine receptor. J Biol Chem, 2002; 277(21):19056–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SK, Gao ZG, Van Rompaey P, Gross AS, Chen A, Van Calenbergh S, et al. Modeling the adenosine receptors: comparison of the binding domains of A2A agonists and antagonists. J Med Chem, 2003; 46(23):4847–59. [DOI] [PubMed] [Google Scholar]
- 22.Franchetti P, Cappellacci L, Vita P, Petrelli R, Lavecchia A, Kachler S, et al. N6-Cycloalkyl and N6-bicycloalkyl-C5'(C2')-modified adenosine derivatives as high-affinity and selective agonists at the human A1 adenosine receptor with antinociceptive effects in mice. J Med Chem. 2009, 52(8):2393–406. 10.1021/jm801456g [DOI] [PubMed] [Google Scholar]
- 23.Mare P. Anticonvulsant action of 2-chloroadenosine against pentetrazol-induced seizures in immature rats is due to activation of A1 adenosine receptors. J Neural Transm, 2010, 117(11):1269–77. 10.1007/s00702-010-0465-9 [DOI] [PubMed] [Google Scholar]
- 24.Siddaramappa U N, Kaczmarek E, Fatteh N, Burns N, Lucas R,Kurt R,et al. Adenosine A1 Receptors Promote, Vasa Vasorum, Endothelial Cell Barrier Integrity via Gi,and Akt-Dependent Actin Cytoskeleton Remodeling. Plos One, 2013, 8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng X.X, Chai Y.Q, Zhu B.C, Jin Y.W, Li X.L, Yu L.S. The protective effects of N6-(2-hydroxyethyl)-adenosine extracted from Ophiocordyceps sobolifera on renal ischemia reperfusion injury (IRI) in mice. Jun Wu Xue Bao. 2015; 34(2): 311‐320. (in chinese). [Google Scholar]
- 26.Zhu B.C, Chai Y.Q, Zhan S.S, Peng X.X, Jin Y.W, Chen G.J. Effects of active components from Ophiocordyceps sobolifera on Anticonvulsions. J. Mycosystema. 2016; 35: 619‐627. (in chinese) [Google Scholar]
- 27.Dolezal T, Dolezelova E, Zurovec M, Bryant PJ. A role for adenosine deaminase in Drosophila larval development. PLoS Biol, 2005; 3(7):e201–e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim ZX, Robinson KE, Jain RG, Sharath Chandra G, Asokan R, Asgari S, et al. Diet-delivered RNAi in Helicoverpa armigera—Progresses and challenges. J Insect Physio, 2015; 85:86–93. [DOI] [PubMed] [Google Scholar]
- 29.Magazanik L G, Fedorova I M. Modulatory Role of Adenosine Receptors in Insect Motor Nerve Terminals. Neurochem Res, 2003; 28(3–4):617–24. [DOI] [PubMed] [Google Scholar]
- 30.Malik A, Buck LT. Adenosinergic modulation of neuronal activity in the pond snail Lymnaea stagnalis. J Exp Biol, 2010; 213(213):1126–32. [DOI] [PubMed] [Google Scholar]
- 31.Ehlers MD, Zhang S, Bernhadt JP, Huganir R. Inactivation of NMDA receptors by direct interaction of calmodulin with the NR1 subunit. Cell, 1996; 84(5):745–55. [DOI] [PubMed] [Google Scholar]
- 32.Albensi BC. The NMDA receptor/ion channel complex: a drug target for modulating synaptic plasticity and excitotoxicity. Curr Pharm Des, 2007; 13(31):3185–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available from Figshare (URL: https://dx.doi.org/10.6084/m9.figshare.3494744.v4; DOI:10.6084/m9.figshare.3494744).