Abstract
Various genetic mutations associated with cancer are known to alter cell signaling, but it is not clear whether they dysregulate signaling pathways by altering the abundance of pathway proteins. Using a combination of RNA sequencing and ultrasensitive targeted proteomics, we defined the primary components—16 core proteins and 10 feedback regulators—of the epidermal growth factor receptor (EGFR)–mitogen-activated protein kinase (MAPK) pathway in normal human mammary epithelial cells and then quantified their absolute abundance across a panel of normal and breast cancer cell lines as well as fibroblasts. We found that core pathway proteins were present at very similar concentrations across all cell types, with a variance similar to that of proteins previously shown to display conserved abundances across species. In contrast, EGFR and transcriptionally controlled feedback regulators were present at highly variable concentrations. The absolute abundance of most core proteins was between 50,000 and 70,000 copies per cell, but the adaptors SOS1, SOS2, and GAB1 were found at far lower amounts (2000 to 5000 copies per cell). MAPK signaling showed saturation in all cells between 3000 and 10,000 occupied EGFRs, consistent with the idea that adaptors limit signaling. Our results suggest that the relative stoichiometry of core MAPK pathway proteins is very similar across different cell types, with cell-specific differences mostly restricted to variable amounts of feedback regulators and receptors. The low abundance of adaptors relative to EGFR could be responsible for previous observations that only a fraction of total cell surface EGFR is capable of rapid endocytosis, high-affinity binding, and mitogenic signaling.
INTRODUCTION
Cancer is a genetic disease frequently associated with alterations in signaling pathways. Dysregulated signaling can promote sustained cell proliferation and reduced apoptosis, which are both hallmarks of cancer (1). An important question is how specific genetic mutations can alter signaling pathways to produce the regulatory changes associated with cancer. Mutations can modify quantitative parameters associated with protein-protein interactions, for example, by changing the affinity between interacting proteins (2) or the loss of specific nodes within a signaling network (3). Alternatively, genetic alterations can lead to an increase in protein abundance, such as that observed with amplification of the HER2 gene, which can result in a loss of target specificity and modified kinetic parameters during signaling (4, 5). Previous modeling studies have also suggested that quantitative differences in the abundance of multiple signaling pathway proteins can produce significant differences in signaling outcomes (6–9). Thus, both qualitative and quantitative changes in signaling protein abundance can alter the functional topology of signaling networks.
Although numerous specific mutations are known to alter cell signaling, the extent to which they dysregulate signaling networks through altered protein abundance is largely unstudied. The overexpression of several proteins is known to be important in cancer, but these were initially identified because of their association with oncogenic viruses rather than because they displayed altered abundances (10, 11). Alterations in gene expression are commonly observed in cancers, but it is increasingly recognized that correlations between mRNA and protein abundances are often low (12,13), and thus, altered gene expression cannot be assumed to lead to changes in protein abundance. Proteomics studies providing “deep” coverage have started to address the issue of quantitative protein differences between cancer subtypes, but it is not clear whether quantitative differences in specific protein abundances between normal and cancer cells are important in disease progression or simply reflect cell type–specific variations (14, 15).
One of the most important signaling pathways in cancer is the mitogen-activated protein kinase (MAPK) [also known as extracellular signal–regulated kinase (ERK)] pathway, which has a critical role in both stimulating proliferation and suppressing apoptosis. Understanding MAPK regulation is central to efforts to rationally design new antiproliferative drugs and other therapies (16). Members of the epidermal growth factor receptor (EGFR) family are potent regulators of MAPK in both normal and transformed epithelial cells (17). Both mutations and amplified expression of the gene encoding this receptor are associated with poor prognosis in cancer (18, 19), and drugs targeting it are effective in subsets of cancers (20). Unfortunately, resistance to drugs targeting the EGFR family is frequent. How frequently this is due to secondary mutations or to altered abundance of other EGFR-MAPK pathway proteins is not yet clear.
Investigating quantitative differences in signaling proteins is challenging. First, the complete set of proteins that constitute a specific signaling pathway is rarely known. In the case of the EGFR-MAPK pathway, the core (or canonical) pathway components have been well described for a number of cell types. However, often there are multiple isotypes of these proteins, such as SOS1 and SOS2, which appear to have overlapping but distinct functional properties (21, 22). There are also numerous regulators that can modify the activity of specific pathway proteins by either forming complexes (23) or altering posttranslational modifications (24). Differences in the abundance of pathway regulators could have a marked effect on signaling, but identifying which are functionally active in a specific cell type is rarely done. There are also technological problems in reliably quantifying protein abundance in cells. Antibody-based approaches require specific antibodies as well as purified protein standards, which are usually not available (25). Current detection technologies for antibodies also have a limited dynamic range (26). Mass spectrometry–based proteomics, including label-free and isobaric labeling strategies, provide broad coverage of relatively abundant, nonmodified proteins, but they have limited sensitivity and are therefore often unable to detect low-abundance signaling proteins. Moreover, the quantification accuracy for global shotgun proteomics is often questionable because of several common issues such as ratio suppression of isobaric reagents and missing data (27, 28). Targeted approaches using selected reaction monitoring (SRM) and heavy isotope–labeled standard peptides can greatly improve the quantification accuracy of low-abundance proteins but require previous knowledge of which pathway components are important (29). Thus, it is important to first establish the essential components of a signaling pathway before conducting a comparative analysis of their abundance in different cell types.
To investigate whether there are quantitative alterations in the EGFR-MAPK signaling pathways in cancer cells, we first characterized this pathway in 184A1 human mammary epithelial cells (HMECs), which is a well-studied, nontransformed model system. We identified both the core pathway proteins and the feedback regulators that modulate their activity by using a selective perturbation strategy together with transcriptional profiling and shotgun proteomics. By using a combination of RNA sequencing (RNA-Seq) and targeted proteomics, we then quantified mRNA and protein abundance of these components across a panel of breast cancer cell lines as well as normal human fibroblasts. In agreement with previous studies, the correlation between mRNA and protein abundances was relatively low (12). However, we found that all pathway proteins identified in normal cells were also found in cancer cells at very similar amounts, with the exception of EGFR itself and transcriptionally controlled feedback regulators. Surprisingly, we found that in most cell lines, EGFR was present at far greater concentrations than were the adaptor proteins that couple it to the MAPK pathway, indicating that adaptor abundance is generally limiting for EGFR-MAPK signaling. Our results suggest that the relative abundance and stoichiometry of most core EGFR-MAPK pathway proteins are highly conserved, indicating that relative abundance and stoichiometry are important for pathway function. Variability in receptor abundance and the feedback regulators that modulate the activity of core pathway proteins, rather than differences in expression of the core proteins themselves, is most likely responsible for cell type–specific responses to EGF
RESULTS
Identification of core pathway proteins and the feedback regulators that modulate EGF-stimulated MAPK phosphorylation
To define the abundance of EGFR-MAPK pathway proteins in nontransformed cells, we initially used a mammary epithelial cell line that displays constitutive autocrine signaling through EGFR [184A1 HMECs (30, 31)]. We started with the core “compendium” pathway described by Kirouac et al. (32) because it contains most of the proteins experimentally associated with the EGFR-MAPK pathway in these cells (33–35). We added the adaptors SHC1 and GAB1 (GRB2-associated binding protein 1) because of multiple studies indicating their importance in EGFR signaling (36, 37) and because of their pronounced phosphorylation in response to EGF addition (discussed further below). We added RASA1 (RasGap) because of its important role in negatively regulating RAS activation and the ability of EGFR activation to control its localization and activity (37, 38). We also included the known isotypes of the different pathway proteins, for example, MAP2K1 and MAP2K2. However, several proteins were omitted from the analysis because of their multiple roles in cells, such as CAV1 and PEBP1 (also known as RKIP). In addition, the very high abundance of those proteins (>106 copies per cell) indicates that their abundance is unlikely to be limiting to MAPK signaling (39).
Stimulation of EGFR can also result in activation of many non-MAPK signaling pathways (such as that of the kinase AKT) that can affect the overall amount of phosphorylation of MAPK (40). In the case of HMECs, however, inhibition of the AKT pathway has little, if any, effect on EGFR-induced MAPK activation; thus, we did not examine the amounts of those pathway proteins (41). Similarly, inhibitors of the kinases PKC, PKA, JAK2, JNK, and p38-MAPK have no effect on EGFR-MAPK signaling in HMECs (42); hence, proteins comprising those pathways were not included in our analysis. SRC family kinase inhibitors affect ligand shedding in HMECs, but not EGF-induced MAPK signaling (42), indicating that SRC family kinases are indirect rather than direct modulators of MAPK signaling in HMECs.
Overall EGFR-MAPK pathway activity also depends on feedback regulators that either enhance or inhibit the activity of the core pathway proteins (42). To identify which of these regulatory proteins are active in HMECs, we used a perturbation strategy, assuming that proteins whose abundance or state of phosphorylation responded to modulations in EGFR-MAPK pathway activity were most likely to be involved in pathway regulation (Fig. 1A). Because HMECs are autocrine cells that depend on constitutive signaling through the EGFR-MAPK pathway (31), we reasoned that important regulators should be present or phosphorylated in the basal state. Thus, inhibiting normal autocrine signaling in HMECs by blocking EGFR with the monoclonal antibody 225 mAb (42) should show a reciprocal effect to adding exogenous EGF. Because of the relatively low sensitivity of global proteomics measurements in detecting EGF-induced protein changes (34), we used transcriptome assays (microarray and RNA-Seq) as a first-pass surrogate for relative protein abundance. To determine which EGF-induced changes resulted from activation of the MAPK pathway, we identified genes whose expression was modulated by the addition of the MEK inhibitor U0126. Finally, we looked at the effect of EGFR-MAPK pathway perturbation on both protein Tyr phosphorylation and Ser/Thr phosphorylation (table S1).
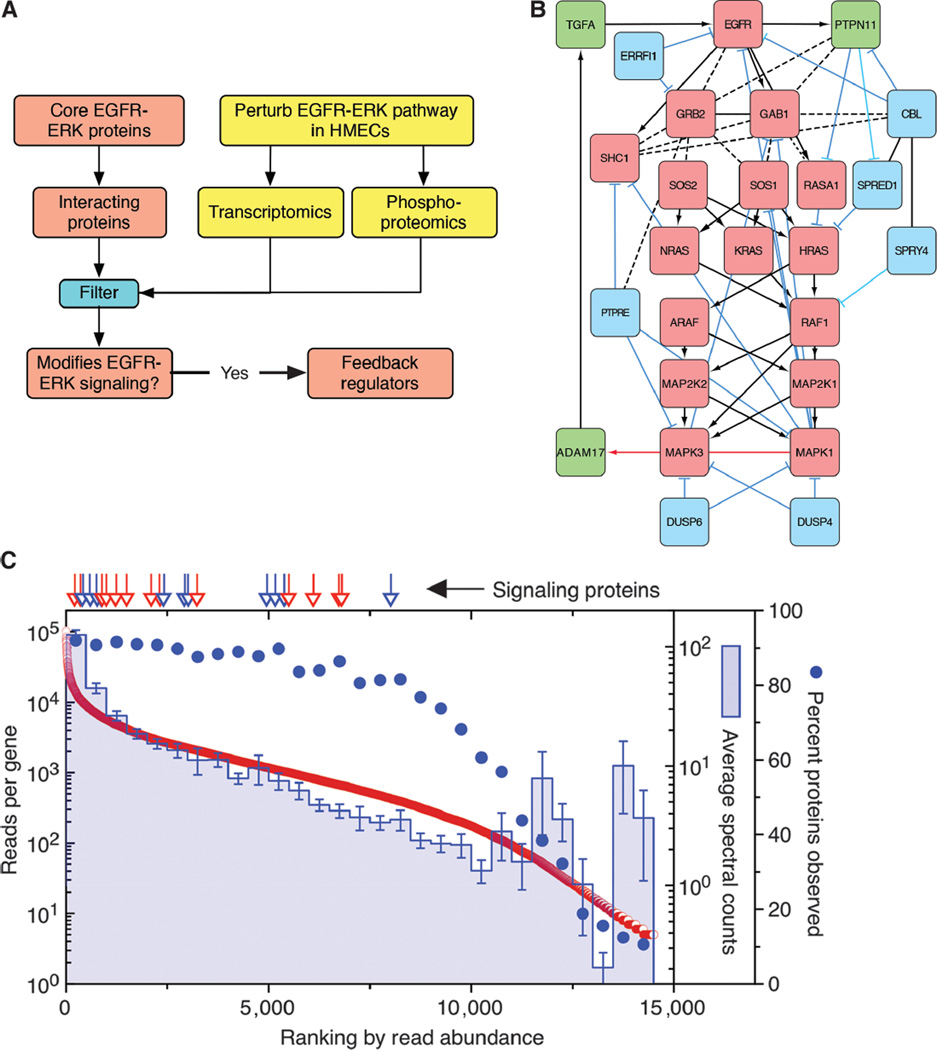
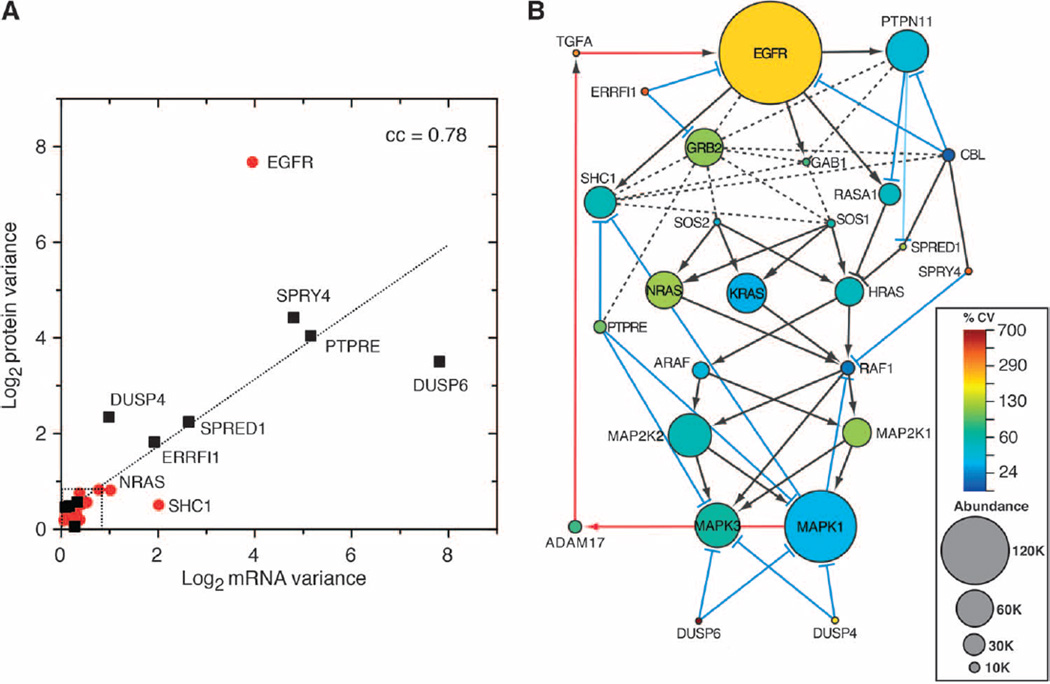
Fig. 1. Identification of core and primary feedback regulators of the EGFR-MAPK pathway.
(A) Schematic of the “omics” assays (yellow) and analysis (red and blue). HMECs were perturbed with EGF (10 ng/ml), 10 µM U0126, or 225 mAb (10 µg/ml) overnight to identify genes whose expression was significantly altered or with EGF or 225 mAb to assess changes in protein phosphorylation. From the results (see table S1), significantly altered genes or proteins that interact with core MAPK pathway proteins and altered pathway activity were classified as feedback regulators. (B) Map of the EGFR-MAPK interaction network. Core proteins are in red, positive feedback regulators are in green, and negative feedback regulators are in blue. Activating interactions are shown as arrows, inhibiting interactions are shown as blue “T” lines, and protein-protein interactions are shown as dotted lines. Red arrow indicates unknown biochemical mechanisms. (C) HMEC 184A1 cells analyzed by global RNA-Seq and shotgun proteomics. Genes were then ranked by the sum of their mapped reads. Spectral counts of the corresponding genes were then averaged in bins of N = 500. The percent of genes in each bin for which spectral counts were recorded is indicated with filled circles. Arrows indicate ranking of gene expression of either core EGFR-MAPK pathway proteins (red) or feedback regulators (blue). Error bars are the SD of the mean of the spectral counts per bin. Data are listed in table S2.
Proteins whose mRNA expression changed by at least twofold or displayed significant changes in either Tyr or Ser/Thr phosphorylation were then restricted to those experimentally shown to directly interact with canonical EGFR-MAPK pathway proteins. The list was then further restricted to proteins documented to have either a positive or a negative effect on MAPK pathway activity. For example, addition of EGF induced a substantial (~5-fold) increase in the Tyr phosphorylation of PTPN18. However, this phosphatase has been shown to inhibit the ability of HER2 to activate MAPK but has no effect on EGFR-MAPK activation (43). Thus, it was excluded from the list. The result of these restrictions was a set of 17 regulatory feedback proteins (table S1). Several were positive pathway regulators, such as the EGFR ligands (TGFA, AREG, HBEGF, and EREG) and their releasing protease (ADAM17) as well as the phosphatase PTPN11 (also known as SHP2). However, most were negative regulators that have been reported to attenuate the activity of the core signaling components (44), with three being members of the dual-specificity protein phosphatase family (DUSP4, DUSP5, and DUSP6) and four being members of the sprouty family (SPRED1, SPRED2, SPRY2, and SPRY4). For our initial study, we selected the two members of the DUSP and sprouty families that showed the most robust response to EGF addition or inhibition (DUSP4, DUSP6, SPRED1, and SPRY4). We also selected the ligand TGFΑ because it has been shown to be critical for autocrine signaling in HMECs (30). This yielded a total of 26 proteins for this study (Fig. 1B).
Detection of feedback regulators by global proteomics measurements
To determine the abundance of the EGFR-MAPK pathway proteins in HMECs, we initially used global shotgun proteomics. We were able to detect unique peptides for all of the 15 core proteins of the EGFR-MAPK pathway (table S2), with the exception of KRAS, most likely because it has few unique peptides due to its strong homology with other members of the RAS family. Several pathway proteins, such as SOS1, SOS2, and TGFA, were represented by only a single peptide, indicating low abundance. Of the 11 feedback regulators, 4 (DUSP4, DUSP6, SPRED1, and SPRY4) were not detected.
To determine whether the abundance of EGFR-MAPK core proteins and feedback regulators was correlated with their mRNA expression, we performed deep transcriptome profiling using RNA-Seq, identifying about 14,500 expressed genes. We then compared the abundance of the transcripts with the abundance of corresponding proteins, estimated by spectral counts (45). We found that there was a general correspondence between transcript abundance and spectral counts (Fig. 1C), in agreement with previous studies (46). The probability of observing any given protein remained between 80 and 90% for the top 8000 transcripts, but fell sharply thereafter. Thus, there should be a very high probability of detecting a protein encoded by abundant transcripts. Surprisingly, we found that mRNA expression for feedback regulators was similar to the core proteins despite their lack of representation in the proteomics data. This suggests that the EGFR-MAPK feedback regulators are either inefficiently translated or rapidly degraded.
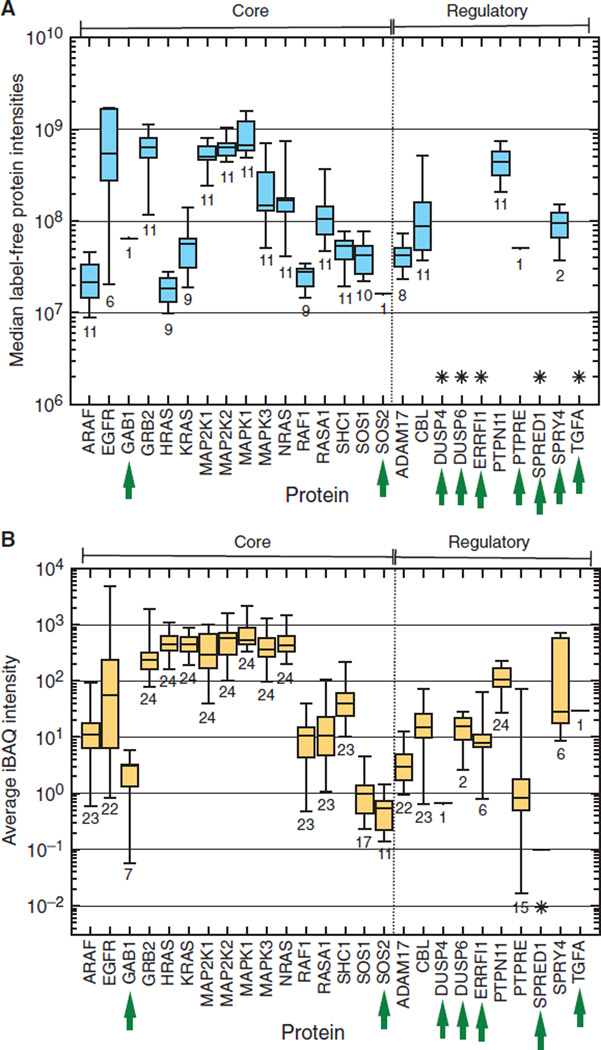
To determine whether feedback regulators generally displayed low abundance in cancer cells, we examined the data from two studies that attempted to comprehensively quantify protein expression in multiple cancer cell lines. The study of Geiger et al. (47) identified an average of about 10,000 distinct proteins in 11 cancer cell lines. Most of the core proteins of the MAPK pathway were detected in these lines (Fig. 2A), although EGFR was detected in only about half of them, and GAB1 and SOS2 were detected in only a single line (fig. S1A). Only half of the feedback regulators were detected in any of the cell lines, suggesting that either there is cell specificity in the pattern of expression of feedback regulators or their abundance is near the limit of detection of current global proteomics technologies. Relative protein abundance across all of the lines was highly variable, especially in the case of EGFR.
Fig. 2. Relative abundance of proteins of the EGFR-MAPK signaling pathway as assessed by deep proteomics surveys.
(A) Reported abundances of proteins in the EGFR-MAPK pathway from the study of Geiger et al. (47) with n = 11 different cell lines. Proteins are grouped into either core components or feedback regulators as described in the text. Within groups, proteins are listed alphabetically. Data from the label-free quantification intensity values for both core and regulated components of the EGFR-MAPK pathway are plotted. The box encloses the upper and lower quartiles, the midline is the median value, and the whiskers show the data range. Numbers below each protein group indicates the number of cell types in which that protein was detected. Green arrows indicate proteins that were observed in less than half of the surveyed cell types. Asterisks (*) indicate proteins that were not detected or reported. (B) Same as in (A), except the study was that of Lawrence et al. (14). In this survey, the iBAQ (intensity-based absolute quantification) label-free method was used for protein quantification, using n = 20 different cell types (n = 2 replicates each) and n = 4 tumors.
A more recent study by Lawrence et al. (2015) compared protein abundance across 20 breast cancer lines and 4 tumors, yielding peptides to almost 13,000 distinct proteins, with at least 9000 proteins found in each cell line (14). Here, most of the feedback regulators were detected in at least some of the cell lines, except for SPRED1 (Fig. 2B). The general protein abundance pattern was similar to that observed by Geiger et al. (47), with the exception of relatively higher amounts of RAS proteins. RAS, MEK, and ERK (encoded by RAS, MAP2K, and MAPK, respectively) were present at very similar amounts in all cell lines and tumors, but lower-abundance proteins and feedback regulators were present at highly variable amounts (fig. S1B). These data suggest that there could be differences in either the presence or abundance of multiple EGFR-MAPK pathway proteins in cancer cells.
Estimation of the real versus observed variability in cellular protein abundance
The high degree of observed variability in the presence and/or abundance of EGFR-MAPK pathway proteins in different cell types could be due to either real biological differences or limitations in the particular proteomics technologies used. To estimate the relative contribution of real versus methodological variability to the observed protein abundance variance between cell types, we calibrated our data against a “gold standard” set of highly conserved proteins. These proteins were identified in the study by Khan et al. (48), who investigated proteins that display relatively constant expression across multiple animal species despite significant variations in their mRNA expression. Although these proteins were identified in hematopoietic cells, we postulate that the same proteins in other cell lines will also be conserved and thus can be used as internal calibration proteins.
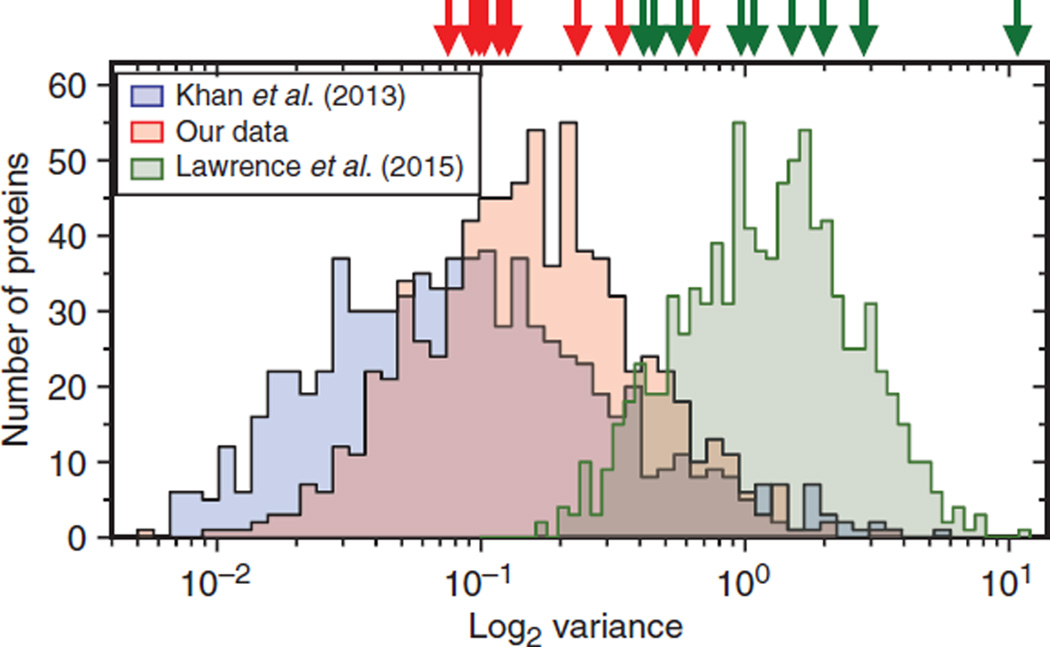
To test this idea, we compared the relative expression of “conserved” proteins and EGFR-MAPK pathway proteins in a subset of the cell lines used in the Lawrence et al. study (14) that showed highly variable detection (between 16 and 20 of 26 EGFR-MAPK proteins). These lines included nontumorigenic MCF10A cells and HER2-overexpressing (SKBR3), hormone receptor–positive (MCF7), and triple-negative (BT20 and HS578T) breast cancer subtypes. We also included normal human dermal fibroblasts (NHDFs) to serve as a nonepithelial cell control as well as our original 184A1 HMEC line. We first performed transcriptional profiling of the cell lines using RNA-Seq, yielding 12,261 genes expressed in common. Global comparative proteome analysis of all of our cell lines using isobaric tags for relative and absolute quantitation (iTRAQ)–based quantification (49) yielded a total of 2862 high-confidence proteins across all of our cell lines, including 11 proteins in the EGFR-MAPK pathway. There were 781 proteins (~27%) corresponding to those with conserved protein abundances across species (table S3). The abundance variance distribution of these conserved proteins across our cell lines was similar to that reported by Khan et al. (48) (median log2 variance of 0.15 versus 0.08, respectively; Fig. 3), whereas nonconserved proteins showed significantly greater variance (log2 variance of 0.34, P < 0.0001; fig. S2). The log2 variance of the RNA-Seq data on conserved proteins in our cells yielded a higher value of 0.29 (fig. S2), despite the generally higher precision of RNA-Seq measurements (50, 51), consistent with protein rather than mRNA abundances being under selective pressure (48). The core EGFR-MAPK pathway proteins displayed a median log2 variance of 0.12 (table S6), essentially the same as the highly conserved protein set (Fig. 3). Signaling protein abundances were also less variable than the corresponding mRNAs (median value of 0.12 versus 0.30).
Fig. 3. Abundance variance of highly conserved proteins and MAPK pathway proteins appears different depending on the approach used for protein quantification.
Blue distribution is log2 sample variance SILAC (stable isotope labeling with amino acids in cell culture) data from Khan et al. (48), using n = 5 biological replicates per species and n = 3 species (N = 15 total samples). Red distribution is iTRAQ data from the current study (n = 7 cell types), and green curve is data from the study of Lawrence et al. (14) (n = 24 samples). Data were sorted into 50 equal bins of between 15 and 40 protein variance values each. Red arrows represent the variance values of MAPK pathway proteins found in our data set for comparison, whereas the green arrows are data on the same proteins in the Lawrence et al. (14) data set.
When we extracted the abundance values of the highly conserved protein set from the study of Lawrence et al. (14) and compared their variance with our values and those of Khan et al. (48), we found them to be substantially greater (median log2 variance of 1.23 versus 0.15 and 0.08; Fig. 3). The abundance variance of the EGFR-MAPK pathway protein was similarly shifted (median log2 variance of 1.1). These results support the idea that the high variability in the measurement of signaling proteins observed by previous investigators (Fig. 2) was likely due to the use of proteomics methods with relatively low precision.
Measuring low-abundance proteins of the EGFR-MAPK pathway using targeted proteomics
RNA-Seq analysis of our panel of cell lines showed that mRNA transcripts of all of the core proteins and feedback regulators of the EGFR-MAPK pathway could be detected at some level, although some displayed highly variable mRNA expression, especially those encoding proteins that function in feedback regulation (Fig. 4A, top, and table S4). This shows that, at least at the mRNA level, all the components of the EGFR-MAPK pathway in HMECs are also found in the other cell types. To increase our ability to detect potentially low-abundance proteins with high precision, we used our ultrasensitive targeted proteomics approach (PRISM-SRM) together with isotopically labeled peptides as internal standards. PRISM-SRM can quantify very low concentrations of proteins (50 to 100 pg/ml in human serum), providing the sensitivity needed to quantify even low amounts of signaling protein (52). For each targeted protein in the EGFR-MAPK pathway, we first selected two highly detectable unique surrogate peptides that had no potential posttranslational modification sites. For each surrogate peptide, three transitions [specific pairs of mass/charge ratio (m/z) values associated with the precursor and fragment ions of the peptide] were selected on the basis of their abundances, the intensity of the SRM signal, and the absence of coeluting interference.
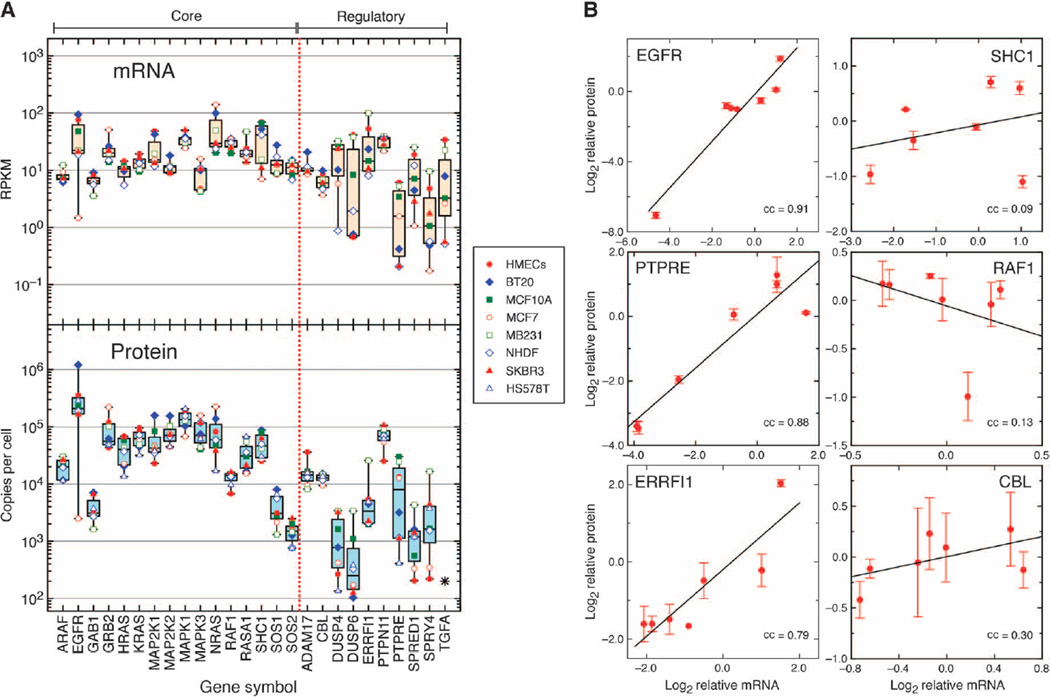
Fig. 4. Variability of mRNA and protein abundance of EGFR-MAPK pathway components across cell lines using RNA-Seq and targeted proteomics.
(A) Top: the expression of mRNA for the species indicated on the x axis was determined by RNA-Seq and normalized to reads per kilobase per million mapped reads (RPKM). Symbols correspond to values from the indicated cell lines (n = 8). Boxes represent the statistics of each species as described in the legend of Fig. 2A. Bottom: absolute quantification of the indicated proteins by targeted SRM (n = 8 cell types, each representing n = 4 samples), corrected for cell number and normalized to measured EGFR abundance as described in Materials and Methods. (B) Relationship between relative mRNA versus protein abundances of selected EGFR-MAPK pathway components across all cell lines shown in (A). The log2 value of the mRNA of each cell line (pooled from n = 4 samples) divided by the average of all lines was plotted against the comparable protein value. Error bars are SD from n = 4 samples. The lines are linear regression of the values with Pearson’s correlation coefficient (cc).
We found that SRM-based targeted quantification allowed the detection and quantification of all signaling pathway proteins across all our cell lines, with the exception of DUSP4 in the fibroblasts and TGFA in several cell lines. Transcriptomics analysis showed very low expression of DUSP4 mRNA in fibroblasts but high expression of DUSP1 mRNA (table S4), which probably serves an analogous role (53). TGFΑ is one of seven secreted EGFR ligands that can activate the receptor in an autocrine fashion. Transcriptome analysis showed that the expression pattern of genes encoding different EGFR autocrine ligands was highly variable across all of the epithelial cells, with TGFA and AREG being the most commonly expressed. However, all ligands appeared to be absent in fibroblasts and the HS578T breast cancer cell line (Table 1). Although peptides from TGFΑ were sometimes detected, they were usually below the concentration needed for reliable quantification.
Table 1. Expression of mRNA encoding autocrine ligands in multiple cell lines.
Libraries from each cell line were prepared as described in Materials and Methods, sequenced, and mapped against the reference human genome. Reads mapping to the indicated genes were converted to RPKM using Avadis NGS. Values in the top 10,000 ranking of gene expression are in boldface.
| Gene symbol |
Cell type |
|||||||
|---|---|---|---|---|---|---|---|---|
| BT20 | HMECs | MCF10A | MCF7 | MB231 | NHDF | SKBR3 | HS578T | |
| AREG | 1.0 | 142.4 | 84.6 | 26.4 | 31.7 | 1.6 | 1.4 | 0.1 |
| BTC | 0.7 | 1.1 | 1.1 | 0.8 | 1.1 | 0.2 | 1.7 | 0.2 |
| EGF | 0.5 | 0.2 | 0.4 | 0.6 | 1.4 | 0.4 | 3.6 | 0.5 |
| EPGN | 0.2 | 11.7 | 28.5 | 0.2 | 0.5 | 0.3 | 2.0 | 0.0 |
| EREG | 0.5 | 53.2 | 1.7 | 0.4 | 2.7 | 0.5 | 0.3 | 0.0 |
| HBEGF | 0.2 | 4.2 | 1.2 | 0.6 | 11.1 | 0.4 | 0.3 | 1.8 |
| TGFA | 7.9 | 34.0 | 3.3 | 2.6 | 22.5 | 0.5 | 0.6 | 0.1 |
Abundances of core MAPK pathway proteins across different cell lines
Using the measured peak area ratio of endogenous peptides relative to isotope-labeled internal standards, we calculated absolute concentrations of all proteins in terms of protein copies per cell (table S5). We found that the abundance pattern of the signaling pathway proteins was similar to that observed in the study of Lawrence et al. (14) (Fig. 2B) but with much less variability between different cell types (Fig. 4A, bottom). In general, the core signaling pathway proteins were more abundant than the feedback regulators, with the exception of GAB1, SOS1, and SOS2, which were present at low concentrations in all cell types.
The availability of absolute protein abundance values for components of the EGFR-MAPK pathway together with RNA-Seq data from the same samples allowed us to quantify the relationship between mRNA expression and signaling protein abundance in the different cell types (fig. S3). We found that mRNA expression generally displayed a lower dynamic range than protein abundance (Fig. 4A, top), in agreement with previous studies (13). At the level of individual proteins, we found that the correlation between mRNA expression and protein abundance was strong for some and weak or nonexistent for others. For example, EGFR abundance strongly correlated with mRNA expression [Pearson’s correlation coefficient (cc) of 0.91; Fig. 4B]. The negative feedback regulators PTPRE and ERRFI1 also showed high correlations between mRNA and protein expression (Fig. 4B). Conversely, SHC1 showed essentially no correlation (cc = 0.09), whereas RAF1 displayed a negative correlation (cc = −0.13). Overall, the median Pearson’s correlation coefficient between mRNA expression and protein abundance was 0.42, indicating that mRNA expression is a poor surrogate for estimating the relative abundance of most signaling proteins.
The low variance of some MAPK pathway proteins across different cell lines (Fig. 3) suggests the presence of evolutionary pressure to maintain their absolute concentrations (48). To extend this analysis to all pathway proteins measured by SRM, we first calibrated the expected variance values. By comparing the abundance variance of a common set of proteins measured by multiple techniques (table S6), we estimate that the observed log2 variance of highly conserved proteins measured by SRM should be <0.7 (see Materials and Methods). When we compared the variability of mRNA and protein abundance for all 26 of the measurable proteins of the EGFR-MAPK pathway, we found a strong correlation between mRNA and protein variance (cc = 0.78; Fig. 5A). All of the core members of the EGFR-MAPK signaling pathway, with the exception of EGFR, displayed relatively low protein abundance variance (median, 0.38; Fig. 5A and table S6). In contrast, transcriptionally controlled feedback regulators were highly variable in both their protein and mRNA abundances (median variance, 2.03 and 1.45, respectively). Proteins regulated by phosphorylation (ADAM17, CBL, GAB1, and PTPN11) displayed an average variance of 0.52, essentially the same as the core components (table S6).
Fig. 5. Median abundance and variability of proteins in the EGFR-MAPK pathway in a panel of cell lines.
(A) Plot of log2 variance of mean mRNA and protein abundance of EGFR-MAPK pathway proteins across a panel of cell lines. Red symbols are core components. Black squares are feedback regulators. Line is linear regression of all values. Dotted box is the median variance of highly conserved proteins +1 SD, derived as described in Materials and Methods. Proteins falling outside of the dotted box are individually labeled. (B) Size of each node is directly proportional to median protein abundance with a minimum node size of 7 and a maximum node size of 390. Node color reflects the calculated percent coefficient of variation of the protein (n = 7 cell lines, each value being the average of n = 4 samples). Edges are as described in the legend of Fig. 1B.
Stoichiometric bottlenecks in the EGFR-MAPK signaling pathway
Although the abundances of most core proteins in the EGFR-MAPK pathway were similar across cell types, their stoichiometry relative to each other was quite distinct (Fig. 4A, bottom). To better understand the relationship between network topology and protein stoichiometry, we added information on both median protein abundance and protein variance to our reconstructed network (Fig. 5B). EGFR and several of its directly interacting proteins (PTPN11, SHC1, and GRB2) were relatively abundant as were the isoforms of RAS (NRAS, KRAS, and HRAS), MEK (MAP2K1 and MAP2K2), and ERK (MAPK1 and MAPK3). In contrast, abundances of the adaptor GAB1 as well as SOS1 and SOS2 were relatively low. The abundance of both ARAF and RAF1 was low compared to that of either upstream RAS species or downstream MAP2Ks. The abundance of most feedback regulators was very low, especially those that showed the greatest variability in their expression, such as DUSP4 and DUSP6.
The abundance of EGFR was generally much greater than its downstream adaptor proteins. For example, the median ratio of EGFR/GRB2 and EGFR/SHC1 was ~4:1. The exception was MCF7 cells in which the abundance of EGFR was much less than its downstream adaptors (EGFR/GRB2 and EGFR/SHC1 ratios of 1:90 and 1:10, respectively). The abundance of SOS1 and SOS2 was also lower than that of the upstream adaptor GRB2 (GRB2/SOS ~10:1) and the downstream RAS isoforms with which they interact (SOS/RAS ~1:35). GAB1 was also present at stoichiometries far below its primary interaction partners GRB2 and PTPN11, with mean ratios of GRB2/GAB1 and PTPN11/GAB1 of ~18:1.
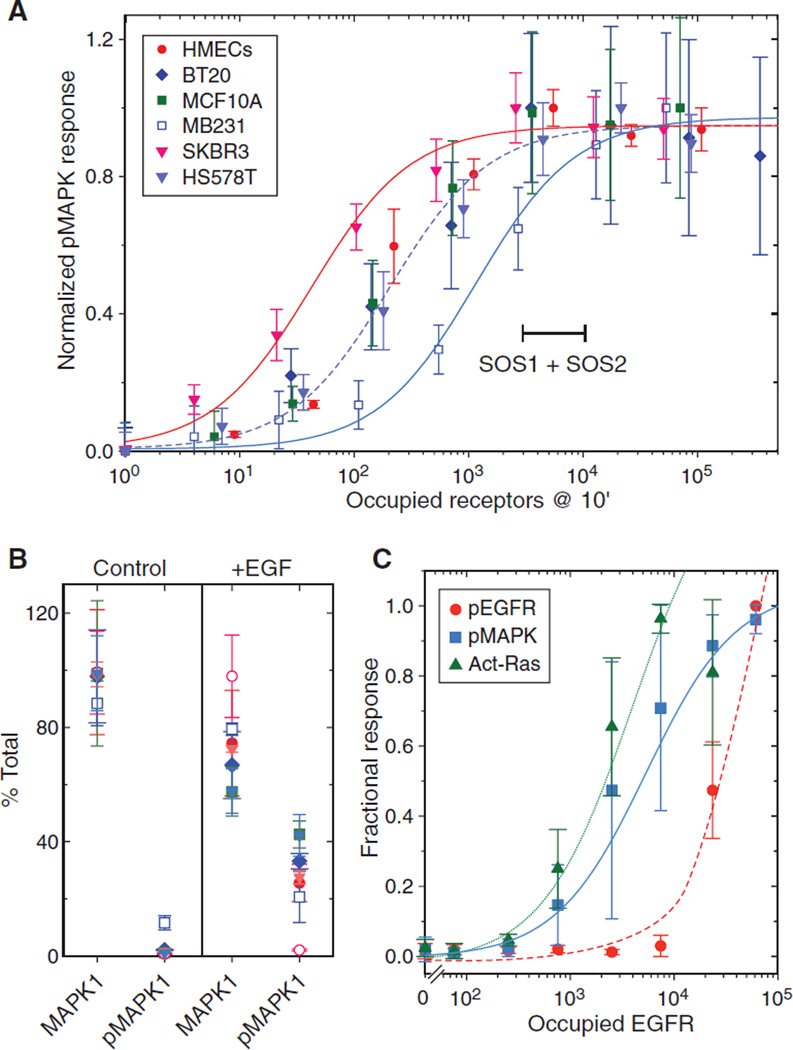
The much greater abundance of EGFR relative to its downstream adaptor proteins could explain previous reports that many of the cellular responses to EGF saturate at low receptor occupancy. To explore the idea that adaptors might be limiting in the activation of the MAPK pathway, we examined the relationship between EGFR occupancy and maximum MAPK activation. Different cell lines were treated with a range of EGF concentrations for 10 min, at which time MAPK phosphorylation was maximal (fig. S4). As outlined in Materials and Methods, we then converted EGF dose to absolute EGFR occupancy using the measured receptor abundance and EGF concentrations as input parameters (54). This approach yields very accurate estimates of total receptor occupancy (fig. S5). Our analysis showed that amounts of phosphorylated MAPK at 10 min for all the cell lines were saturated between 3000 and 10,000 occupied receptors (Fig. 6A). This roughly corresponds to the abundance of SOS1 + SOS2 (2000 to 10,000 per cell). Half-maximum activation of the MAPK pathway of most of the cell lines was ~250 occupied EGFR, with the exception of SKBR3 cells at ~50 receptors and MDA-MB231 cells at ~1200.
Fig. 6. Maximal phosphorylation of MAPK in a panel of responsive cell lines occurs well below maximal receptor occupancy.
(A) Plot of the amounts of occupied EGFR and activated (phosphorylated, “p”) MAPK in the indicated cell lines. Data are the mean response of n = 5 independent experiments normalized to a scale of 0 to 1 ± SEM as a function of occupied receptors at 10 min. Sigmoidal curves were fit to data from SKBR3 (red), HS578T (dashed), and MDA-MB231 (blue) cell lines. Range marker corresponds to the abundance range of SOS1 + SOS2 in evaluated cell lines. Results from MCF7 cells were not included because of their lack of significant response. (B) Abundance of MAPK1 or doubly phosphorylated MAPK1 in cells treated with and without EGF (10 ng/ml) for 10 min assayed by quantitative SRM-based proteomics (55). Results are from n = 3 samples with technical replicates expressed as percent of total MAPK1 ± SD. Open circles are from MCF7 cells, whereas other symbols are the same as in (A). (C) HMEC 184A1 treated with EGF (10 ng/ml) for 5 min and occupied EGFR calculated as described in Materials and Methods. The amounts of phosphorylated MAPK (blue squares) and phosphorylated EGFR (red circles) were measured using an enzyme-linked immunosorbent assay; RAS activity was measured by a pull-down assay (42). Data are the mean response of n = 4 independent experiments normalized to a scale of 0 to 1 ± SEM and fit to a sigmoid function.
To ensure that the cellular abundance of MAPK was not limiting for signaling, we treated cells with a saturating dose of EGF for 10 min and then quantified the absolute abundance of total and phosphorylated MAPK1 by targeted proteomics (55). We found very little phosphorylated MAPK1 in cells in the absence of EGF (Fig. 6B), except in the case of MDA-MB231 cells, which have an activating RAS mutation (56). After EGF addition, however, only about 30% of the pool of MAPK1 was converted into the doubly phosphorylated form, confirming that MAPK abundance was not limiting (Fig. 6B). We also examined the relationship between EGFR occupancy and its phosphorylation to downstream RAS activation and MAPK phosphorylation. Maximal activation of both RAS and MAPK was observed when only a small percentage of total EGFR was phosphorylated (Fig. 6C), showing that receptor activation is not limiting. Activation of RAS and MAPK displayed very similar dose responses, with half-maximal responses at <3000 occupied receptors, indicating that any limiting pathway components are likely to be between the EGFR and RAS. Together, these data support the idea that adaptor abundance limits the extent of MAPK signaling.
Because increased signaling through the EGFR-MAPK pathway is frequently associated with cancer, we sought to determine whether gene amplification of adaptors showed a similar association. To address this, we used the copy number variation (CNV) data from the COSMIC database and determined whether amplification of genes in the EGFR-MAPK pathway occurred at a higher frequency than random. We found that SHC1 and GRB2, as well as EGFR and KRAS, were amplified at a significantly higher frequency than random (P < 0.05; table S7). This was true for both breast cancer and other cancers. However, SOS1 and SOS2 as well as GAB1 displayed amplification frequencies similar to the bulk of the cellular genes (fig. S6), suggesting that the amplification of only a subset of adaptor proteins is associated with cancer.
DISCUSSION
We initiated this study to determine whether cancer cells displayed quantitative differences in the abundance of proteins that could dysregulate the EGFR-MAPK pathway. Previous studies have clearly established an association between the overexpression of select signaling proteins, such as receptors, and cell transformation. Thus, we sought to systematically evaluate the abundance of proteins that comprise the core components and primary regulators of the EGFR-MAPK pathway and determine whether there were any alterations associated with the cancer phenotype. By using deep transcriptional profiling and targeted proteomics, however, we were able to show that all of the core proteins and the great majority of feedback regulators were found in both normal epithelial cells and fibroblasts as well as all of the surveyed cancer cell lines. Furthermore, most pathway proteins were found at very similar concentrations across all cell lines.
We were surprised that several proteins important in the EGFR-MAPK pathway were absent in previous proteomics surveys, despite the reported detection sensitivities of those studies of 10,000 to 12,000 proteins per cell type, which is between 85 and 95% of the estimated number of expressed genes (13, 14, 47). Our analysis of previous data sets suggests that this was likely a result of the very low abundance of the missing proteins, which prevented them from being reliably detected. For example, in the data-driven approach used by Geiger et al. (47), lower-abundance peptides are less frequently selected and are more difficult to match across instrument runs (57). The iBAQ protein quantification approach used by Lawrence et al. (14) uses both peptide intensity and detection frequency to extend its dynamic range (58), thus introducing additional noise into abundance measurements, especially for proteins at the limits of detectability. Because of the inherent limitations of previously used proteomics technologies, both highly variable and low-abundance proteins in the EGFR-MAPK pathway frequently appeared to be absent.
In contrast to global, label-free approaches for estimating protein abundance, we used highly purified, specific labeled peptides together with multistage separations to detect proteins that were sometimes present at only hundreds of copies per cell. The precision of the PRISM-SRM proteomics approach allowed us to rigorously evaluate the relative abundance of signaling proteins across different cell lines. By using a gold standard set of proteins that has previously been shown to be under selective pressure for constant protein expression, we found comparably low variance of most EGFR-MAPK pathway proteins. With the exception of the transcriptionally controlled feedback regulators and EGFR itself, all pathway proteins were found at remarkably similar concentrations in all cell types. This strongly suggests that both the presence and absolute abundance of these proteins are under selective pressure (48), and, thus, the relative abundance and stoichiometry of pathway proteins are likely important for effective signaling or regulation.
Despite the very similar concentrations of EGFR-MAPK pathway proteins in all the cell types we examined, there were still distinct differences in their response to EGF. Sensitivity differences were likely due to the variable expression of EGFR family members. For example, the greater sensitivity of SKBR3 cells to EGF (Fig. 6A) is likely caused by their over-expression of HER2, which is known to increase the affinity and activity of EGFR (5). However, the number of occupied EGFRs needed to elicit a maximal MAPK response was very low and similar for all cell types, typically corresponding to <5% receptor occupancy. We found that RAS activation showed a similar EGF dose response as did MAPK activation, consistent with our previous results that showed that initial MAPK phosphorylation is proportional to RAS activity (42). Thus, any limiting pathway component(s) between EGFR and RAS would also limit the extent of MAPK signaling. The most likely candidates are SOS1 and SOS2, which form a molecular complex with GRB2 to couple EGFR to RAS activation (59). Unlike the major core components of the EGFR-MAPK pathway, such as RAS, MAP2K, and MAPK that are present at between 60,000 and 120,000 copies per cell, SOS1 and SOS2 are only present at between 1000 and 6000 copies. Other adaptors that are associated with MAPK signaling, such as GRB2, SHC1, and GAB1, are also much less abundant than EGFR, ranging from 3000 to 55,000 as compared with the median EGFR abundance of 210,000 per cell. Thus, at full occupancy, most of the EGFRs will probably not be able to form complexes with the adaptor proteins we examined here.
If adaptors are limiting for signaling, then an increase in their expression could facilitate cancer development. We found a significantly greater frequency of gene amplification for both SHC1 and GRB2 relative to the average in cancer. Amplification of GRB2 and SHC1 in cancers has been reported previously (60, 61) and is probably responsible for the relatively high GRB2 abundance and GRB2/EGFR ratios that we observed in MCF7 cells (Fig. 4A). The core proteins with the lowest abundance in the EGFR pathway (SOS1, SOS2, GAB1, and RAF1) showed no significant amplification, although functional mutations in some of these proteins are associated with diseases, such as Noonan syndrome (62, 63). This suggests that the relative stoichiometry of these proteins in the EGFR-MAPK pathway is important to their function.
The low stoichiometry of adaptors to EGFR suggests a ready explanation for the classic observation that EGFR signaling occurs through a relatively small class of “high-affinity” receptors. Formation of a stable receptor-adaptor complex would be expected to increase the affinity of the receptor for EGF (64), which would only be seen below adaptor saturation. The measured number of high-affinity receptors on HMECs [~40,000 (65)] is similar to the measured number of GRB2 adaptors (table S5). Similarly, the required GRB2 binding for occupancy-induced EGFR endocytosis would explain why it saturates below full receptor occupancy (66, 67). We only observed high amounts of EGFR self-phosphorylation at receptor occupancies much higher than needed to obtain maximal MAPK signaling, which presumably corresponds to maximal adaptor binding. Similar observations have been made previously (68). The reason for this is not clear, but it is tempting to speculate that adaptor protein binding could sterically inhibit self-phosphorylation of the EGFR on multiple residues. Regardless, the substantial difference between the dose of EGF necessary to produce a maximal biological response and that required to produce EGFR phosphorylation detectable in standard assays suggests that phosphoproteomics studies using high EGF concentrations should be interpreted with caution.
Our observations on the high similarity in both the types and abundance of core EGFR-MAPK pathway proteins in both normal and cancer cells have important implications with respect to efforts to build predictive models of cell signaling. Previous models lacked information on relative protein abundance, which can limit their ability to predict outcomes (8). For example, most previous models assumed that adaptors were in excess relative to the EGFR, thus making receptor occupancy limiting to MAPK activation (38, 69). That is clearly not the case in most circumstances. Instead, EGFR and other receptor tyrosine kinases likely compete for the less abundant adaptor proteins controlling downstream signal transduction (70). Competition for adaptors has been shown to be an important mechanism for regulating differential cellular responses in other receptor systems (71, 72), and such a mechanism could be important in the EGFR pathway as well.
The protein abundance measurements that we have established for the EGFR-MAPK pathway can provide the foundation for “universal” signaling models that include cell-specific feedback regulators. However, all of the environmental inputs and mechanisms that contribute to the steady-state concentrations of positive and negative feedback regulators are not known. Our results show that the expression level of positive and negative regulators of the MAPK pathway varies widely between different cell types and thus likely constitutes the most important source of cell type specificity. The expression level of these feedback regulators is dictated by the basal activation state of the EGFR-MAPK signaling pathway itself and, thus, is likely to be influenced by the mutational status of pathway components, such as BRAF, and crosstalk with other pathways. Thus, the mechanisms that control the amounts of the different MAPK pathway feedback regulators will need to be understood to predict how they change in response to the activation of any given receptor system and the role each plays in shaping cell-specific responses to EGF.
MATERIALS AND METHODS
Cell culture
Breast cell lines BT20, MCF10A, MCF7, MDA-MB231, HS578T, and SKBR3 were obtained from the American Type Culture Collection and were grown as previously described (73). HMEC line 184A1 was obtained from M. Stampfer (Lawrence Berkeley National Laboratory) and maintained in DFCI-1 medium as previously described (74). Cells were plated in 15-cm dishes or 96-well plates, grown for 24 hours, starved in serum-free medium for 18 hours, and starved again for an additional hour before treatment. Primary NHDF cells were obtained from Lonza and cultured to confluence in Fibroblast Growth Media-2 (Lonza).
Sample preparation
For protein samples, cells were washed twice with ice-cold phosphate-buffered saline (PBS), detached from the plate with 1.5 ml of trypsin, harvested in 8.5 ml of PBS with 10% fetal bovine serum, and counted. Cells were spun down in 50-ml Falcon-type tubes at 200g for 5 min at 25°C, resuspended in 1 ml of cold PBS, and transferred to a low-retention microcentrifuge tube. Cells were spun for 1 min at 500g, supernatant was removed, and the protein pellets were snap-frozen in liquid nitrogen. For RNA extraction, cells were washed twice with ice-cold PBS, lysed in 1 ml of RLT buffer (RNeasy Mini Kit, Qiagen) supplemented with 1:100 β-mercaptoethanol, transferred to a microcentrifuge tube, and snap-frozen in liquid nitrogen.
EGF response assays
Cells were treated with 10× stock of an EGF (PeproTech) dilution series for 10 min. Cells were treated with varying concentrations of EGF for 10 min before the evaluation of phosphorylated MAPK amounts by immunofluorescence as previously described (73) using an Operetta high-content imaging system (PerkinElmer). Data are the average of replicate wells generated using the Columbus image data storage and analysis system (PerkinElmer).
Transcriptomics
Genomic DNA was removed from RNA samples using a Qiagen RNase-Free DNase Set kit. RNA integrity was ascertained with a Bioanalyzer, and all samples had an RNA integrity number between 9 and 10. A Ribo-Zero Gold rRNA Removal Kit was used to enrich transcripts, and a SOLiD Total RNA-Seq Kit was used to construct template complementary DNA (cDNA) for RNA-Seq. The ribosomal depleted mRNA was fragmented using hydrolysis, followed by ligation with strand-specific adapters and reverse transcript to generate cDNA. Fragments greater than 150 base pairs were subsequently selected using Agencourt AMPure XP beads. The isolated cDNA went through 15 cycles of amplification to produce enough templates for the SOLiD EZ Bead system to generate a templated bead library for ligation-based sequencing on the SOLiD 3 platform using barcoding, with a minimum of 2.2 × 107 mapped reads per cell line. Reads were normalized to the average of all lines after sequencing, yielding 12,261 common expressed genes.
Proteomics
Cell pellets from different cell lines were lysed in 100 µl of lysis buffer containing 8 M urea in 100 mM NH4HCO3 (pH 7.8). Proteins were reduced by 5 mM dithiothreitol for 1 hour at 37°C and alkylated using 20 mM iodoacetamide for 1 hour at room temperature in the dark. Samples were diluted eightfold with 50 mM NH4HCO3 and digested by sequencing grade modified trypsin at a 1:50 enzyme-to-protein ratio (w/w) at 37°C for 3 hours. Each sample was then desalted by C18 solid phase extraction and concentrated to a volume of ~50 µl. The final peptide concentration was measured using bicinchoninic acid assay.
Global proteomics
For mass spectrometry–based shotgun proteome analysis, we used the accurate mass and time (AMT) tag approach (75). An existing AMT tag database encompassing the monoisotopic mass and normalized chromatographic elution times of peptides identified from previous liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses of HMEC proteins under a range of experimental conditions (33, 76–78) was used as a base reference database for the LC–Fourier transform ion cyclotron resonance measurements in this study. Details of LC-MS/MS analysis and data filtering involved in peptide identification have been described elsewhere (78). Criteria that would yield an overall confidence of greater than 95% at the unique peptide level were established for filtering raw peptide identifications.
For comparative proteomics, peptides were labeled with 8-plex iTRAQ reagents according to the manufacturer’s instructions (AB Sciex). The iTRAQ-labeled peptide mixtures were analyzed on a high-resolution, reversed-phase capillary LC system coupled with a Thermo Fisher Scientific LTQ-Orbitrap Velos mass spectrometer. Mobile phases consisted of 0.1% formic acid in water and 0.1% formic acid acetonitrile operated at a constant flow of 300 nl/min, with a gradient profile over the course of 100 min. The 10 most abundant parent ions, excluding single-charge states, were selected for MS/MS using high-energy collisional dissociation with a normalized collision energy setting of 40%.
Peptides were identified on the basis of tandem MS/MS spectra using the SEQUEST search algorithm against a human protein database (UniProtKB, released May 2010), and the abundance information across 8-plex samples was extracted from the reporter ion intensities within a given spectra. All peptides were identified with <0.1% false discovery rate by using an MS-generating function score (MS-GF) <1 × 10−10 and a decoy database searching strategy. The reporter ion intensities for each peptide were summed for all identified spectra for each channel in each biological condition. Relative abundances at peptide level were rolled up to the protein level using the software tool DAnTE (79), with the abundances being log2-transformed and normalized by the central tendency approach.
SRM assay configuration and LC-SRM measurements
To detect pathway proteins by an SRM assay, 10 tryptic peptides without miscleavage (except those peptides containing inhibitory motifs for trypsin) were initially chosen for representing each target protein based on existing LC-MS/MS results from our own laboratory and public data repositories such as PeptideAtlas, GPM, and PRIDE. For the core pathway proteins without existing LC-MS/MS data, in silico digestion was performed for peptide selection. All selected peptides were unique to the given proteins with no predicted posttranslational modifications. The selected peptides were further evaluated by two prediction tools: the ESP predictor and CONSeQuence software. Five peptides per protein with moderate hydrophobicity, high spectral counts, and high score from the prediction tools were selected for peptide synthesis. The synthesized crude heavy isotope–labeled peptides were further evaluated for peptide response and fragmentation pattern. For each peptide, three transitions were selected on the basis of their abundances and optimal collision energy (CE) values, which is achieved by direct infusion of the individual peptides and/or multiple LC-SRM runs with CE ramping. Two peptides with the best response were selected to configure final SRM assays for each target protein, and the best transition (the one with the most intense SRM signal and without clear evidence of coeluting interference) was used to quantify the target protein. The potential interference for given transitions was assessed on the basis of the relative intensity ratios between the three transitions for both light and heavy peptides using a similar approach as previously reported (52).
With the crude heavy isotope–labeled internal standards spiked in, all cell line samples were initially measured by regular LC-SRM using the scheduled SRM algorithm (80). For the core pathway proteins that cannot be reproducibly detected and quantified by regular LC-SRM, highly sensitive PRISM-SRM assays (52) were used to measure their relative abundances. All LC-SRM measurements were performed using the nanoACQUITY UPLC system coupled online to a TSQ Vantage triple quadrupole mass spectrometer (Thermo Scientific), and SRM data were analyzed using Skyline software (81).
Except for the secreted proteins TGFA and DUSP4, all the targeted proteins were confidently quantified by PRISM-SRM across all the eight cell lines. To obtain absolute protein concentration values of those core pathway proteins, high-purity light peptides (>95%) were purchased and used to determine their corresponding crude heavy peptide purity and the spiked-in concentrations in cell line samples. On the basis of the peak area ratio of endogenous light peptides over heavy-labeled internal standards, known concentrations of heavy internal standards, and cell density, the relative copy number of each protein per cell was estimated. This was then corrected for extraction and digestion efficiency by normalizing copy numbers to the number of EGFRs directly measured by steady-state 125I-EGF binding, which includes estimates of internal receptor pools (65).
Estimating EGFR occupancy
The level of binding of EGF to its receptor at any given time point was calculated by numeric integration of the rate equations describing the forward and reverse rate constants as well as receptor internalization (54). These equations accurately describe the dynamics of cells interacting with the EGFR at short time intervals (<15 min) (82). Rate constants used were as follows: ka = 1.2 × 106 M−1s−1; kd = 3.67 × 10−2 s−1; ke = 4.0 × 10−3 s−1; kt = 1.17 × 10−3 s−1. The value of Vr was adjusted to yield the initial number of EGFR measured for each cell type in our SRM proteomics measurements. The simulated volume was 2 ml, and cell number was 1.2 × 106. Calculated occupied receptors included both receptors at the cell surface and those that were internalized.
To validate the accuracy of the estimates, we treated 184A1 cells with concentrations of 125I-labeled EGF ranging from 0.1 to 120 ng/ml for 5 min and then quantified the total amount of labeled EGF associated with the cells. We then compared the amount to that predicted by our calculations (fig. S5).
Data analysis and normalization
Proteomics data from iTRAQ proteomics were normalized to an internal standard that was created by mixing equal amounts of protein from all seven cell lines being analyzed. iTRAQ data were then expressed as the log2 ratio to the standard. Sample variance was calculated for the ratios for each protein detected across all seven cell lines. RNA-Seq data were rolled up to the gene level and then filtered to remove non–protein-coding reads. Total reads were normalized to the average across all seven cell lines and then converted to RPKM using Avadis NGS. RPKM values for each gene were then converted to the log2 ratio of the average value across all seven cell lines. Sample variance of the log2 ratios was then calculated for each gene. Significance of differences of the distribution of gene expression or protein abundance variances between groups was calculated using an unpaired sample z test.
To assess amplification frequency of genes in the EGFR-MAPK pathway that could be associated with cancer, we downloaded CNV data from COSMIC database. The CNV data were converted into frequencies of copy number gain, and the frequency distribution was modeled as a two-component mixture of beta distributions. One component represents the bulk of the genes (~80%) that are amplified at random. The other minor component with a higher frequency of amplification represents a high-amplification subset of genes. The parameters of the two beta distributions were inferred using expectation-maximization algorithm. The null hypothesis is that the frequency is the same as the bulk of the genes. The null hypothesis was rejected at P < 0.05. All network maps were generated with Cytoscape 3.0.2 (83).
Comparing precision estimates across different proteomics measurements
The median log2 variance of our iTRAQ data was 0.15 with an SD of 0.3, which corresponds to the median value of 0.08 for the Khan et al. data (48), with an SD of 0.43. Because these proteins are under selective pressure to conserve their relative abundance across cell lines, we consider these “best case” variance values limited by the stochastic nature of protein expression and the precision of our proteomics measurements.
Proteins measured by SRM should have the same biological variance as those measured by iTRAQ. We had 11 proteins with corresponding iTRAQ and SRM data. However, because iTRAQ data tend to be compressed in their dynamic range (84), this will tend to reduce the observed variance. We excluded the EGFR data because their extreme variability and dynamic range skew the data. For the remaining 10 proteins, the median variance for the iTRAQ data was 0.12 with an SD of 0.15, approximately the same as the full data set, indicating that they are a representative set of proteins. The SRM data for the same 10 proteins displayed a median variance value of 0.48, with an SD of 0.18. Thus, the observed variance of SRM data was about four times greater than that of the iTRAQ data. To estimate the limits of low-variance proteins from SRM data, we used the median value +1 SD or a log2 variance of <0.7. For the corresponding mRNA data, the median variance was 0.28 with an SD of 0.57, or <0.85. These are the limits shown in Fig. 4B.
Supplementary Material
Acknowledgments
Some of the experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, a national scientific user facility sponsored by the U.S. Department of Energy under contract DE-AC05-76RL0 1830.
Funding: Portions of the research were supported by NIH grants DP2OD006668, P41GM103493, U24-CA-16001901, UC4-DK104167, and U54-HL127365.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/9/436/rs6/DC1
Fig. S1. Relative abundance of proteins of the EGFR-MAPK signaling pathway as assessed by deep proteomics surveys.
Fig. S2. Comparison of log2 variance distribution of conserved versus nonconserved proteins and mRNA.
Fig. S3. Relationship between mRNA expression and protein abundances of EGFR-MAPK pathway components across all cell lines.
Fig. S4. Kinetic response of cells to EGF as a function of ligand dose.
Fig. S5. Comparison between simulated and measured EGF binding to cells.
Fig. S6. Two-component mixture model of the copy number gain frequencies for breast and all cancers.
Table S1. EGFR-MAPK pathway–associated genes regulated by pathway perturbation.
Table S2. RNA expression and protein abundance of HMECs ranked by RNA-Seq reads.
Table S3. iTRAQ and transcriptome analysis of proteins with conserved abundances.
Table S4. Transcriptomics analysis of EGFR-MAPK pathway components across seven cell lines.
Table S5. SRM analysis of all of the EGFR-MAPK pathway proteins.
Table S6. Variance analysis of signaling proteins in the EGFR-MAPK pathway.
Table S7. Copy number gain or loss frequency for genes in the EGFR pathway.
Author contributions: T.S., M.N., W.-J.Q., R.D.S., P.K.S., and H.S.W. designed the experiments; M.N., Y.G., C.D.N., W.B.C., and L.M.M. conducted the experiments; T.S., M.N., J.E.M., K.D.R., V.A.P., and H.S.W analyzed the data; M.N. and H.S.W. wrote the paper and generated the figures.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The SRM proteomics data were deposited in the Panorama public database at https://panoramaweb.org/labkey/yUpp0c.url. The RNA-Seq data were deposited in the Gene Expression Omnibus repository with accession number GSE81032.
REFERENCES AND NOTES
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Zhao L, Sun T, Pei J, Ouyang Q. Mutation-induced protein interaction kinetics changes affect apoptotic network dynamic properties and facilitate oncogenesis. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E4046–E4054. doi: 10.1073/pnas.1502126112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogervorst FBL, Nederlof PM, Gille JJP, McElgunn CJ, Grippeling M, Pruntel R, Regnerus R, van Welsem T, van Spaendonk R, Menko FH, Kluijt I, Dommering C, Verhoef S, Schouten JP, van’t Veer LJ, Pals G. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003;63:1449–1453. [PubMed] [Google Scholar]
- 4.Nobeli I, Favia AD, Thornton JM. Protein promiscuity and its implications for biotechnology. Nat. Biotechnol. 2009;27:157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- 5.Worthylake R, Opresko LK, Wiley HS. ErbB-2 amplification inhibits down-regulation and induces constitutive activation of both ErbB-2 and epidermal growth factor receptors. J. Biol. Chem. 1999;274:8865–8874. doi: 10.1074/jbc.274.13.8865. [DOI] [PubMed] [Google Scholar]
- 6.Klinke II DJ. Signal transduction networks in cancer: Quantitative parameters influence network topology. Cancer Res. 2010;70:1773–1782. doi: 10.1158/0008-5472.CAN-09-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer RL, D’Alessandro A, Kar S, Kramer B, She B, Kaschek D, Hahn B, Wrangborg D, Karlsson J, Kvarnström M, Jirstrand M, Lehmann W-D, Timmer J, Höfer T, Klingmüller U. Heterogeneous kinetics of AKT signaling in individual cells are accounted for by variable protein concentration. Front Physiol. 2012;3:451. doi: 10.3389/fphys.2012.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stites EC, Aziz M, Creamer MS, Von Hoff DD, Posner RG, Hlavacek WS. Use of mechanistic models to integrate and analyze multiple proteomic datasets. Biophys. J. 2015;108:1819–1829. doi: 10.1016/j.bpj.2015.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkes EH, Terfve C, Gribben JG, Saez-Rodriguez J, Cutillas PR. Empirical inference of circuitry and plasticity in a kinase signaling network. Proc. Natl. Acad. Sci. U.S.A. 2015;112:7719–7724. doi: 10.1073/pnas.1423344112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraus MH, Popescu NC, Amsbaugh SC, King CR. Overexpression of the EGF receptor-related proto-oncogene erbB-2 in human mammary tumor cell lines by different molecular mechanisms. EMBO J. 1987;6:605–610. doi: 10.1002/j.1460-2075.1987.tb04797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Libermann TA, Nusbaum HR, Razon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield MD, Ullrich A, Schlessinger J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 12.Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 13.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Pääbo S, Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence RT, Perez EM, Hernández D, Miller CP, Haas KM, Irie HY, Lee S-I, Blau CA, Villén J. The proteomic landscape of triple-negative breast cancer. Cell Rep. 2015;11:630–644. doi: 10.1016/j.celrep.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, Yang JY, Broom BM, Verhaak RGW, Kane DW, Wakefield C, Weinstein JN, Mills GB, Liang H. TCPA: A resource for cancer functional proteomics data. Nat Methods. 2013;10:1046–1047. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 17.Citri A, Yarden Y. EGF-ERBB signalling: Towards the systems level. Nat. Rev. Mol. Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 18.Nicholson RI, Gee JMW, Harper ME. EGFR and cancer prognosis. Eur. J. Cancer. 2001;37(Suppl. 4):S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 19.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 20.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat. Rev. Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian X, Esteban L, Vass WC, Upadhyaya C, Papageorge AG, Yienger K, Ward JM, Lowy DR, Santos E. The Sos1 and Sos2 Ras-specific exchange factors: Differences in placental expression and signaling properties. EMBO J. 2000;19:642–654. doi: 10.1093/emboj/19.4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang S-s, Van Aelst L, Bar-Sagi D. Differential interactions of human Sos1 and Sos2 with Grb2. J. Biol. Chem. 1995;270:18212–18215. doi: 10.1074/jbc.270.31.18212. [DOI] [PubMed] [Google Scholar]
- 23.Anastasi S, Baietti MF, Frosi Y, Alemà S, Segatto O. The evolutionary conserved EBR module of RALT/MIG6 mediates suppression of the EGFR catalytic activity. Oncogene. 2007;26:7833–7846. doi: 10.1038/sj.onc.1210590. [DOI] [PubMed] [Google Scholar]
- 24.Mohi MG, Neel BG. The role of Shp2 (PTPN11) in cancer. Curr. Opin. Genet. Dev. 2007;17:23–30. doi: 10.1016/j.gde.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Aebersold R, Burlingame AL, Bradshaw RA. Western blots versus selected reaction monitoring assays: Time to turn the tables? Mol. Cell. Proteomics. 2013;12:2381–2382. doi: 10.1074/mcp.E113.031658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gassmann M, Grenacher B, Rohde B, Vogel J. Quantifying Western blots: Pitfalls of densitometry. Electrophoresis. 2009;30:1845–1855. doi: 10.1002/elps.200800720. [DOI] [PubMed] [Google Scholar]
- 27.Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC. iTRAQ underestimation in simple and complex mixtures: “The good, the bad and the ugly”. J. Proteome Res. 2009;8:5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- 28.Xie F, Liu T, Qian W-J, Petyuk VA, Smith RD. Liquid chromatography-mass spectrometry-based quantitative proteomics. J. Biol. Chem. 2011;286:25443–25449. doi: 10.1074/jbc.R110.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi T, Su D, Liu T, Tang K, Camp II DG, Qian W-J, Smith RD. Advancing the sensitivity of selected reaction monitoring-based targeted quantitative proteomics. Proteomics. 2012;12:1074–1092. doi: 10.1002/pmic.201100436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong J, Opresko LK, Dempsey PJ, Lauffenburger DA, Coffey RJ, Wiley HS. Metalloprotease-mediated ligand release regulates autocrine signaling through the epidermal growth factor receptor. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6235–6240. doi: 10.1073/pnas.96.11.6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stampfer MR, Pan CH, Hosoda J, Bartholomew J, Mendelsohn J, Yaswen P. Blockage of EGF receptor signal transduction causes reversible arrest of normal and immortal human mammary epithelial cells with synchronous reentry into the cell cycle. Exp. Cell Res. 1993;208:175–188. doi: 10.1006/excr.1993.1236. [DOI] [PubMed] [Google Scholar]
- 32.Kirouac DC, Saez-Rodriguez J, Swantek J, Burke JM, Lauffenburger DA, Sorger PK. Creating and analyzing pathway and protein interaction compendia for modelling signal transduction networks. BMC Syst. Biol. 2012;6:29. doi: 10.1186/1752-0509-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs JM, Mottaz HML, Yu R, Anderson DJ, Moore RJ, Chen WNU, Auberry KJ, Strittmatter EF, Monroe ME, Thrall BD, Camp II DG, Smith RD. Multidimensional proteome analysis of human mammary epithelial cells. J. Proteome Res. 2004;3:68–75. doi: 10.1021/pr034062a. [DOI] [PubMed] [Google Scholar]
- 34.Waters KM, Liu T, Quesenberry RD, Willse AR, Bandyopadhyay SL, Kathmann E, Weber TJ, Smith RD, Wiley HS, Thrall BD. Network analysis of epidermal growth factor signaling using integrated genomic, proteomic and phosphorylation data. PLOS One. 2012;7:e34515. doi: 10.1371/journal.pone.0034515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Y, Zhang C, Croucher DR, Soliman MA, St-Denis N, Pasculescu A, Taylor L, Tate SA, Hardy WR, Colwill K, Dai AY, Bagshaw R, Dennis JW, Gingras A-C, Daly RJ, Pawson T. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature. 2013;499:166–171. doi: 10.1038/nature12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montagner A, Yart A, Dance M, Perret B, Salles J-P, Raynal P. A novel role for Gab1 and SHP2 in epidermal growth factor-induced Ras activation. J. Biol. Chem. 2005;280:5350–5360. doi: 10.1074/jbc.M410012200. [DOI] [PubMed] [Google Scholar]
- 38.Borisov N, Aksamitiene E, Kiyatkin A, Legewie S, Berkhout J, Maiwald T, Kaimachnikov NP, Timmer J, Hoek JB, Kholodenko BN. Systems-level interactions between insulin–EGF networks amplify mitogenic signaling. Mol. Syst. Biol. 2009;5:256. doi: 10.1038/msb.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiel C, Ebhardt HA, Burnier J, Portugal C, Sabidó E, Zimmermann T, Aebersold R, Serrano L. Quantification of ErbB network proteins in three cell types using complementary approaches identifies cell-general and cell-type-specific signaling proteins. J. Proteome Res. 2014;13:300–313. doi: 10.1021/pr400878x. [DOI] [PubMed] [Google Scholar]
- 40.Wang C-C, Cirit M, Haugh JM. PI3K–dependent cross-talk interactions converge with Ras as quantifiable inputs integrated by Erk. Mol. Syst. Biol. 2009;5:246. doi: 10.1038/msb.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodland KD, Bollinger N, Ippolito DL, Opresko K, Coffey RJ, Zangar R, Wiley HS. Multiple mechanisms are responsible for transactivation of the epidermal growth factor receptor in mammary epithelial cells. J. Biol. Chem. 2008;283:31477–31487. doi: 10.1074/jbc.M800456200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joslin EJ, Shankaran HL, Opresko K, Bollinger N, Lauffenburger DA, Wiley HS. Structure of the EGF receptor transactivation circuit integrates multiple signals with cell context. Mol. Biosyst. 2010;6:1293–1306. doi: 10.1039/c003921g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gensler M, Buschbeck M, Ullrich A. Negative regulation of HER2 signaling by the PEST-type protein-tyrosine phosphatase BDP1. J. Biol. Chem. 2004;279:12110–12116. doi: 10.1074/jbc.M309527200. [DOI] [PubMed] [Google Scholar]
- 44.Legewie S, Herzel H, Westerhoff HV, Blüthgen N. Recurrent design patterns in the feedback regulation of the mammalian signalling network. Mol. Syst. Biol. 2008;4:190. doi: 10.1038/msb.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H, Sadygov RG, Yates JR., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 46.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geiger T, Wehner A, Schaab C, Cox J, Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics. 2012;11:M111 014050. doi: 10.1074/mcp.M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan Z, Ford MJ, Cusanovich DA, Mitrano A, Pritchard JK, Gilad Y. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science. 2013;342:1100–1104. doi: 10.1126/science.1242379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.L Ross P, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Łabaj PP, Leparc GG, Linggi BE, Markillie LM, Wiley HS, Kreil DP. Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling. Bioinformatics. 2011;27:i383–i391. doi: 10.1093/bioinformatics/btr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor RC, Webb Robertson B-J, Markillie M, Serres MH, Linggi BE, Aldrich JT, Hill EA, Romine MF, Lipton MS, Wiley HS. Changes in translational efficiency is a dominant regulatory mechanism in the environmental response of bacteria. Integr. Biol. 2013;5:1393–1406. doi: 10.1039/c3ib40120k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi T, Fillmore TL, Sun X, Zhao R, Schepmoes AA, Hossain M, Xie F, Wu S, Kim J-S, Jones N, Moore RJ, Paša-Tolić L, Kagan J, Rodland KD, Liu T, Tang K, Camp II DG, Smith RD, Qian W-J. Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum. Proc. Natl. Acad. Sci. U.S.A. 2012;109:15395–15400. doi: 10.1073/pnas.1204366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang C-Y, Tan T-H. DUSPs, to MAP kinases and beyond. Cell Biosci. 2012;2:24. doi: 10.1186/2045-3701-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knauer DJ, Wiley HS, Cunningham DD. Relationship between epidermal growth factor receptor occupancy and mitogenic response. J. Biol. Chem. 1984;259:5623–5631. [PubMed] [Google Scholar]
- 55.Shi T, Gao Y, Gaffrey MJ, Nicora CD, Fillmore TL, Chrisler WB, Gritsenko MA, Wu C, He J, Bloodsworth KJ, Zhao R, Camp II DG, Liu T, Rodland KD, Smith RD, Wiley HS, Qian W-J. Sensitive targeted quantification of ERK phosphorylation dynamics and stoichiometry in human cells without affinity enrichment. Anal. Chem. 2015;87:1103–1110. doi: 10.1021/ac503797x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozma SC, Bogaard ME, Buser K, Saurer SM, Bos JL, Groner B, Hynes NE. The human c-Kirsten ras gene is activated by a novel mutation in codon 13 in the breast carcinoma cell line MDA-MB231. Nucleic Acids Res. 1987;15:5963–5971. doi: 10.1093/nar/15.15.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim YJ, Zhan P, Feild B, Ruben SM, He T. Reproducibility assessment of relative quantitation strategies for LC-MS based proteomics. Anal. Chem. 2007;79:5651–5658. doi: 10.1021/ac070200u. [DOI] [PubMed] [Google Scholar]
- 58.Schwänhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 59.Buday L, Downward J. Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell. 1993;73:611–620. doi: 10.1016/0092-8674(93)90146-h. [DOI] [PubMed] [Google Scholar]
- 60.Lockwood WW, Chari R, Coe BP, Girard L, MacAulay C, Lam S, Gazdar AF, Minna JD, Lam WL. DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene. 2008;27:4615–4624. doi: 10.1038/onc.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daly RJ, Binder MD, Sutherland RL. Overexpression of the Grb2 gene in human breast cancer cell lines. Oncogene. 1994;9:2723–2727. [PubMed] [Google Scholar]
- 62.Kobayashi T, Aoki Y, Niihori T, Cavé H, Verloes A, Okamoto N, Kawame H, Fujiwara I, Takada F, Ohata T, Sakazume S, Ando T, Nakagawa N, Lapunzina P, Meneses AG, Gillessen-Kaesbach G, Wieczorek D, Kurosawa K, Mizuno S, Ohashi H, David A, Philip N, Guliyeva A, Narumi Y, Kure S, Tsuchiya S, Matsubara Y. Molecular and clinical analysis of RAF1 in Noonan syndrome and related disorders: Dephosphorylation of serine 259 as the essential mechanism for mutant activation. Hum. Mutat. 2010;31:284–294. doi: 10.1002/humu.21187. [DOI] [PubMed] [Google Scholar]
- 63.Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, Li L, Yassin Y, Tamburino AM, Neel BG, Kucherlapati RS. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat. Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 64.Gilson MK, Given JA, Bush BL, McCammon JA. The statistical-thermodynamic basis for computation of binding affinities: A critical review. Biophys. J. 1997;72:1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burke PM, Wiley HS. Human mammary epithelial cells rapidly exchange empty EGFR between surface and intracellular pools. J. Cell. Physiol. 1999;180:448–460. doi: 10.1002/(SICI)1097-4652(199909)180:3<448::AID-JCP16>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 66.Goh LK, Huang F, Kim W, Gygi S, Sorkin A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J. Cell Biol. 2010;189:871–883. doi: 10.1083/jcb.201001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lund KA, Opresko LK, Starbuck C, Walsh BJ, Wiley HS. Quantitative analysis of the endocytic system involved in hormone-induced receptor internalization. J. Biol. Chem. 1990;265:15713–15723. [PubMed] [Google Scholar]
- 68.Krall JA, Beyer EM, MacBeath G. High- and low-affinity epidermal growth factor receptor-ligand interactions activate distinct signaling pathways. PLOS One. 2011;6:e15945. doi: 10.1371/journal.pone.0015945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schoeberl B, Eichler-Jonsson C, Gilles ED, Müller G. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat. Biotechnol. 2002;20:370–375. doi: 10.1038/nbt0402-370. [DOI] [PubMed] [Google Scholar]
- 70.Gordus A, Krall JA, Beyer EM, Kaushansky A, Wolf-Yadlin A, Sevecka M, Chang BH, Rush J, MacBeath G. Linear combinations of docking affinities explain quantitative differences in RTK signaling. Mol. Syst. Biol. 2009;5:235. doi: 10.1038/msb.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meakin SO, MacDonald JIS, Gryz EA, Kubu CJ, Verdi JM. The signaling adapter FRS-2 competes with Shc for binding to the nerve growth factor receptor TrkA. A model for discriminating proliferation and differentiation. J. Biol. Chem. 1999;274:9861–9870. doi: 10.1074/jbc.274.14.9861. [DOI] [PubMed] [Google Scholar]
- 72.Wilson TJ, Garner LI, Metcalfe C, King E, Margraf S, Brown MH. Fine specificity and molecular competition in SLAM family receptor signalling. PLOS One. 2014;9:e92184. doi: 10.1371/journal.pone.0092184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niepel M, Hafner M, Pace EA, Chung M, Chai DH, Zhou L, Schoeberl B, Sorger PK. Profiles of basal and stimulated receptor signaling networks predict drug response in breast cancer lines. Sci. Signal. 2013;6:ra84. doi: 10.1126/scisignal.2004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Band V, Sager R. Distinctive traits of normal and tumor-derived human mammary epithelial cells expressed in a medium that supports long-term growth of both cell types. Proc. Natl. Acad. Sci. U.S.A. 1989;86:1249–1253. doi: 10.1073/pnas.86.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen Y, Conrads TP, Veenstra TD, Udseth HR. An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 76.Chen W-NU, Yu L-R, Strittmater EF, Thrall BD, Camp II DG, Smith RD. Detection of in situ labeled cell surface proteins by mass spectrometry: Application to the membrane subproteome of human mammary epithelial cells. Proteomics. 2003;3:1647–1651. doi: 10.1002/pmic.200300468. [DOI] [PubMed] [Google Scholar]
- 77.Liu T, Qian W-J, Strittmater EF, Camp II DG, Anderson GA, Thrall BD, Smith RD. High-throughput comparative proteome analysis using a quantitative cysteinyl-peptide enrichment technology. Anal. Chem. 2004;76:5345–5353. doi: 10.1021/ac049485q. [DOI] [PubMed] [Google Scholar]
- 78.Liu T, Qian W-J, Chen W-NU, Jacobs JM, Moore RJ, Anderson DJ, Gritsenko MA, Monroe ME, Thrall BD, Camp DGI, Smith RD. Improved proteome coverage by using high efficiency cysteinyl peptide enrichment: The human mammary epithelial cell proteome. Proteomics. 2005;5:1263–1273. doi: 10.1002/pmic.200401055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Polpitiya AD, Qian W-J, Jaitly N, Petyuk VA, Adkins JN, Camp II DG, Anderson GA, Smith RD. DAnTE: A statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24:1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi T, Fillmore TL, Gao Y, Zhao R, He J, Schepmoes AA, Nicora CD, Wu C, Chambers JL, Moore RJ, Kagan J, Srivastava S, Liu AY, Rodland KD, Liu T, Camp II DG, Smith RD, Qian W-J. Long-gradient separations coupled with selected reaction monitoring for highly sensitive, large scale targeted protein quantification in a single analysis. Anal. Chem. 2013;85:9196–9203. doi: 10.1021/ac402105s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.DeWitt AE, Dong JY, Wiley HS, Lauffenburger DA. Quantitative analysis of the EGF receptor autocrine system reveals cryptic regulation of cell response by ligand capture. J. Cell Sci. 2001;114:2301–2313. doi: 10.1242/jcs.114.12.2301. [DOI] [PubMed] [Google Scholar]
- 83.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rauniyar N, Yates JR., III Isobaric labeling-based relative quantification in shotgun proteomics. J. Proteome Res. 2014;13:5293–5309. doi: 10.1021/pr500880b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.