Abstract
Craniofacial development is an intricate process of patterning, morphogenesis, and growth that involves many tissues within the developing embryo. Genetic misregulation of these processes leads to craniofacial malformations, which comprise over one-third of all congenital birth defects. Significant advances have been made in the clinical management of craniofacial disorders, but currently very few treatments specifically target the underlying molecular causes. Here, we review recent studies in which modeling of craniofacial disorders in primary patient cells, patient-derived induced pluripotent stem cells (iPSCs), and mice have enhanced our understanding of the etiology and pathophysiology of these disorders while also advancing therapeutic avenues for their prevention.
Introduction
The craniofacial complex is one of the most intricate and sophisticated parts of the human body. Its patterning and morphogenesis involve a dynamic interplay between the ectoderm, mesoderm, and endoderm, and a critical role is played by neural crest cells, which give rise to the majority of skeletal and connective tissues in the craniofacial region. These interactions are established and maintained by numerous genes, including those encoding a variety of transcription factors, growth factors, and receptors (1,2). Disruption of gene expression or function results in devastating craniofacial anomalies, which have a collective incidence rate of 1 in 600 births (3). Much of our current understanding of the etiology and pathophysiology of craniofacial disorders has been uncovered through the use of model systems. Mice are considered by many to be the gold standard for disease modeling, as they are anatomically and physiologically comparative to humans and can be genetically manipulated to mimic human phenotypes (4). Primary patient cells and patient-derived iPSCs have proven to be a valuable complement to the mouse model system by either highlighting species-specific differences or further validating the observations already made in mice (5). The modeling of craniofacial disorders has not only informed genetic risk assessment and patient prognosis but also identified potential targets for pharmaceutical intervention. In this review, we highlight recent efforts that have provided new information to advance the treatment of five general classes of human genetic disorders, with particular emphasis on the craniofacial region. In addition, we discuss how this information furthers our understanding of the molecular mechanisms regulating normal craniofacial development.
Craniosynostosis
The cranial sutures are fibrous joints that form between the five principal flat bones of the skull vault during embryogenesis. From the early fetal period through the first years of life, the cranial sutures are primary sites of bone growth and allow the skull vault to expand with the growing brain. Formation and maintenance of the suture, which include the osteogenic fronts of the jointed bones and their interposed mesenchyme, is critical to its function as a growth center. Dysfunction in genes that regulate the organization, proliferation, and/or differentiation within the suture can lead to its premature fusion, a relatively common birth defect known as craniosynostosis (6). Serious clinical problems associated with craniosynostosis include craniofacial deformities, increased intracranial pressure, and impaired brain development leading to learning difficulties or developmental delay. Treatment plans involve surgery to removes and reshapes large areas of the calvaria; however, for many patients suture re-fusion necessitates repeated surgeries (7). Thus, there is a clinical need to develop less invasive, more effective therapies for craniosynostosis. Recent studies that apply our current knowledge of the molecular players in normal and abnormal suture development have advanced the potential for these new therapies.
The pathogenesis of syndromic craniosynostosis is commonly associated with gain-of-function mutations in Fibroblast Growth Factor receptors (FGFRs) 1-3. FGF signaling promotes proliferation and differentiation in osteogenic cells, most notably in the cranial sutures (8,9). Apert syndrome is usually caused by dominant mutations in FGFR2 that increase ligand-dependent activation and subsequently enhance osteoblast differentiation (10). New findings show that expression or nanogel-mediated delivery of a soluble form of FGFR2 harboring the Apert mutation S252W blocks enhanced FGFR2 signaling and inhibits craniosynostosis in a mouse model for Apert syndrome (11,12).
The Bone Morphogenetic Protein (BMP) pathway plays a critical role in the development of the skull vault. Increased BMP signaling is associated with craniosynostosis (13–15), and antagonists of the pathway are being tested as a possible treatment to prevent post-operative re-fusion. In a recent study, delivery of the BMP antagonist GREMLIN1 via hydrogel that rapidly polymerized upon injection prevented bone re-growth in a mouse model for re-synostosis (16).
Hypophosphatasia, a metabolic disorder with craniosynostosis, is caused by loss-of-function mutations in ALPPL, the gene encoding Tissue-nonspecific Alkaline Phosphatase (TNAP). TNAP is an osteoblast surface protein that induces hydroxyapatite crystal growth by increasing inorganic phosphate (17,18). A recent report shows that craniosynostosis in Alppl knockout mice is rescued by subcutaneous injection of a mineral-targeted form of recombinant TNAP (19).
Craniofacial Dysmorphologies
Dysmorphic craniofacial features can often be quantified as anthropometric measurements outside the normal variance, and these can be isolated or occur in a syndrome. As such, current treatments are directed towards addressing the specific anomalies on a patient-by-patient basis. While craniofacial dysmorphologies can have phenotypic overlap, the underlying mechanism of disease is quite disparate. Due to their genetic heterogeneity, an exciting frontier in the treatment of craniofacial dysmorphologies is the possibility of targeted therapeutics such as genome editing (20–23).
Brachio-ocular-facial (BOF) syndrome is associated with missense mutations in the TFAP2A gene encoding AP-2α, a transcription factor with early roles in neural crest cell specification and survival (24–26). Generation of the first fully penetrant cleft lip and palate mouse model caused by mutations in Tfap2a revealed that one cause of clefting can be subtle changes in FGF pathway gene expression in the facial prominences. Manipulation of Fgf8 gene dosage partially rescued the phenotype, suggesting that FGF signaling and/or downstream effectors may be possible targets of pharmacological intervention in BOF syndrome and nonsyndromic cases of clefting associated with TFAP2A mutations (27).
Heterozygous mutations in BRAF are found in 50-75% of patients with cardio-facio-cutaneous (CFC) syndrome (21). BRAF is a serine threonine kinase that regulates the RAS-MAPK signaling pathway, and therefore CFC syndrome is classified as a RASopathy (28). BrafQ241R/+ mice exhibit embryonic/neonatal lethality with liver necrosis, edema, and craniofacial abnormalities, effectively mimicking the phenotypes of human patients. Interestingly, co-treatment with MEK inhibitors and histone demethylase inhibitors rescued the pathophysiology (29). This finding has implications not only for prospective therapies of CFC syndrome but for other RASopathies as well. It will be important to examine the epigenetic contributions to heart and skeletal defects in these disorders to inform upon the treatment potential of combined inhibition of HRAS signaling and histone demethylases.
Treacher Collins syndrome (TCS) is an autosomal dominant disorder which presents with hypoplasia of the facial bones, cleft palate, and low set, malformed ears (30). In a mouse model of TCS, haploinsufficiency of the Tcof1 gene encoding the nucleolar phosphoprotein treacle reduces ribosome biogenesis, causing deficient proliferation and extensive apoptosis of neuroepithelial cells via a nucleolar stress-induced, p53 pathway (31,32). The recent discovery that treacle also functions in DNA damage response/repair to limit oxidative stress-induced neuroepithelial cell death identified a novel underlying contributor to the pathogenesis of TCS. Excitingly, in utero treatment with antioxidants prevented DNA damage and minimized cell death in the neuroepithelium to substantially ameliorate the craniofacial anomalies in Tcof1+/-embryos (33). While previous work has shown that genetic and pharmacological inhibition of p53 can suppress the neuroepithelial apoptosis in Tcof+/- embryos, maternal antioxidant dietary supplementation may be a safer potential therapeutic for patients with TCS, given the risk of tumorigenesis associated with p53 manipulation (32,33).
Dental Anomalies
Developmental dental anomalies are defined as marked deviations from the normal color, contour, size, number, and degree of formation of teeth. These malformations can occur either as part of a syndrome or as an isolated finding (34). In Costello syndrome (CS), a RASopathy associated with craniofacial, cardiac, musculoskeletal, and neurodevelopmental abnormalities, characteristic dental phenotypes include class III malocclusion, enamel hypomineralization, and soft tissue hyperplasia (35). Nearly all individuals with CS have a heterozygous mutation in HRAS that results in constitutive activation of Ras signaling (36,37). A CS mouse model expressing HRasG12Vphenocopies many aspects of the syndrome and was used to understand the cellular mechanism underlying the hypomineralization of the enamel (38). In this model, enamel-forming ameloblasts lack polarity, and the ameloblast progenitor cells are hyperproliferative. Inhibition of MAPK led to complete rescue of the dental phenotype, whereas modulation of either MAPK or PI3K signaling corrected the defect in progenitor cell proliferation in CS mice (38). This work defined for the first time distinct roles of Ras signaling in tooth development and provided additional evidence for the use of Ras inhibitors in treating CS and other RASopathies.
Hereditary conditions involving nonsyndromic enamel conditions are referred to as amelogenesis imperfectas (AIs). The X-linked form of hypoplastic AI is associated with missense mutations in Amelogenin, an extracellular matrix protein secreted by ameloblasts (39,40). The murine Y62H Amelogenin mutation similarly results in the eruption of malformed tooth enamel with severely compromised mechanical properties (41). Recent work has demonstrated that this specific mutation disrupts proper intracellular trafficking of amelogenin and induces ER stress-related apoptosis in ameloblasts, classifying AI as a protein conformational disease for the first time (42). Treatment with 4-phenylbutyrate, which can act to relieve conformational abnormalities of the protein, rescued the enamel phenotype in affected female mice by promoting cell survival over apoptosis, offering a potential therapeutic option for patients with this form of AI (42,43).
Skeletal Dysplasias
Skeletal dysplasias represent one of the largest classes of birth defects, with over 450 recognizable conditions (44). The craniofacial defects in these disorders result from the combinatorial interactions of transcription factors, growth factors, and receptors responsible for the intricate genetic patterning and morphogenesis of craniofacial structures (45). With the advent of next-generation DNA sequencing, clinical phenotypes can be linked to key cellular processes of skeletal development, including proliferation, differentiation, and apoptosis. Dominant missense mutations in FGFR3 that reduce chondrocyte proliferation are associated with achondroplasia (ACH) and thanatophoric dysplasia (TD), the most common genetic forms of dwarfism (46–48). Craniofacial findings include macrocephaly, frontal bossing, and midface hypoplasia in ACH, and macrocrania, cloverleaf skull, and frontal bossing in TD. The severity of these chondrodysplasias is linked with the degree of constitutively activated FGFR3 signaling through MAPK or STAT1, and as such, therapeutic strategies have focused on decreasing excessive downstream signaling (49,50). Recent work in patient-specific iPSCs has identified statins as a potential drug to treat FGFR3-mediated chondrodysplasias. Treatment with statins rescued cartilage formation in chondrogenically differentiated TD1 and ACH iPSCs and led to significant recovery of bone growth in an ACH mouse model (51). While the precise mechanism of action remains to be determined, the success of statin treatment highlights a previously unappreciated role for anabolic activity during chondrogenesis (52–54).
Maintaining the proper balance between proliferation and differentiation is also critical for bone formation. Examination of the pathophysiology of Bent Bone Dysplasia Syndrome (BBDS) revealed an unexpected nuclear route for FGF signaling to regulate osteoprogenitor cell proliferation and differentiation via ribosome biogenesis (55). BBDS is a dominant disorder characterized by bent long bones in the lower extremities and craniofacial abnormalities including poorly mineralized calvaria, craniosynostosis, midface hypoplasia, micrognathia, low-set ears, and prenatal teeth. BBDS results from mutations in the transmembrane domain of FGFR2 that redistribute the receptor from the plasma membrane to the nucleolus, where it activates ribosomal DNA transcription by halting RUNX2-mediated repression (55,56). Inhibition of ribosomal RNA synthesis by small molecules has been shown to be effective in preclinical cancer models in mice and may be a potential therapeutic strategy to specifically target the pro-proliferative nucleolar role FGFR2 in BBDS and other FGFR2 gain-of-function disorders (57–60).
Cherubism is a condition caused by excessive osteoclast activity in the mandible and maxilla, which drives progressive proliferation of fibrous tissues and leads to severe facial deformities. Spontaneous regression of bone lesions is usually observed at puberty, and surgical intervention is only considered when functional or aesthetic concerns arise (61,62). Recently, two independent studies presented promising pharmacological therapeutic approaches to inhibit or delay the progression of cherubic lesions. Most patients with cherubism have gain-of-function mutations in the gene encoding SH3BP2, an adapter protein involved in the immune response. Sh3bp2 knock-in mice develop massive infiltration of macrophages into skeletal elements, including the jaw, which can be rescued by genetic inhibition of TNF-α expression (63). Consistent with the role of TNF-α, treatment with the anti-TNF-α inhibitor etanercept significantly reduced facial swelling and bone loss in neonatal mice. Furthermore, this phenotypic rescue was not recapitulated in adult mice, emphasizing the importance of early diagnosis and treatment of cheribusm (64). An effective therapy for patients with actively growing and established inflammatory lesions may be bone marrow (BM) transplants. Transplantation of wild type BM cells to Sh3pb2 knockin mice rescued the systemic inflammation and bone loss in adult cherubism that could not be ameliorated by etanercept treatment (65). Treatment with tacrolimus, an immunosuppressor that has been shown to inhibit activation of the calcineurin/NFATc pathway and osteoclastogenesis (66–68), led to significant clinical improvement in a 4-year old boy with an aggressive form of cherubism; specifically noted was stabilization of jaw size and intraosseous osteogenesis (69). Future studies are needed to determine the precise mechanism of action of tacrolimus and whether combined treatment with anti-inflammatories may further ameliorate the pathophysiology of cherubism.
Bone Mineral Density
Bone mineral density (BMD) is determined by the relative rates of bone deposition and resorption, which are carried out by osteoblasts and osteoclasts, respectively. Mutations in the genes controlling osteoblast and osteoclast function cause congenital disorders with abnormal BMD. While these conditions present with generalized skeletal abnormalities, the craniofacial findings have important clinical complications. In osteopenic and osteoporotic disorders, where bone resorption exceeds deposition, calvaria are undermineralized, malformed, and fractured. In osteopetrotic disorders, where bone deposition outpaces resorption, there is focal or widespread thickening of the calvaria, skull base, and facial bones. Recent studies have supported the use of biologics to restore the balance between bone anabolism and catabolism in congenital BMD disorders.
Genetic studies of congenital BMD disorders have demonstrated that the Wnt/LRP5 pathway increases bone density by promoting osteoblast production and function. Loss-of-function mutations in the Wnt co-receptor LRP5 cause the low bone mass disorder osteoporosis-pseudoglioma syndrome (OPPG), while LRP5 gain-of-function mutations cause higher bone mass disorders Van Buchem disease, osteosclerosis, and osteopetrosis (70–74). LRP5 mutations in higher bone mass disorders increase the co-receptor activity by disrupting the binding of the inhibitor sclerostin, which is inactivated in higher bone density disorder sclerosteosis (75–78). These studies laid the groundwork for development of an inhibitory antibody against sclerostin that is now in phase 3 clinical trials for the treatment of postmenopausal osteoporosis (79). New evidence supports repurposing anti-sclerostin to treat the very syndromes that advanced its discovery: depletion of sclerostin, either genetically or through the use of anti-sclerostin, increases the BMD of mouse models for OPGG (80,81). These findings also provide the rationale for use of a recombinant Wnt/LRP5 inhibitor or inhibitory antibody against LRP5 to block bone overgrowth in the osteopetrotic disorders.
There is strong evidence to suggest that anti-sclerostin will increase BMD in other skeletal fragility syndromes as well, such as osteogenesis imperfecta (OI) and hereditary hypophosphatemic rickets, despite differences in the molecular pathologies. While OI is largely caused by deficiencies in type I collagen production, modification, or secretion, mouse models for OI gain a significant increase in bone mass and strength when Wnt/LRP5 signaling is increased, through either expression of LRP5 gain-of-function mutation or treatment with anti-sclerostin (82–86). Additionally, anti-sclerostin significantly improved osteomalacia in DMP1 knockout mice, a model for hereditary hypophosphatemic rickets (82).
Mouse models with reduced BMD have enabled identification of promising new targets for protein-based therapies. Defective type I collagen biosynthesis in OI increases the bioavailability of TGFβ, leading to excessive TGFβ signaling (87). Promotion of osteoclast bone resorption by TGFβ signaling provides a rationale for the use of inhibitory antibodies against TGFβ. Indeed, anti-TGFβ treatment improved bone mass in mouse models for OI (87). Knockout of Nell1, which codes for a secreted bone-inducing factor, leads to age-related osteoporosis (88). Correspondingly, delivery of recombinant NELL1 was shown to increase bone formation via the Wnt pathway in both small and large animal models of osteoporosis (89).
Future Directions
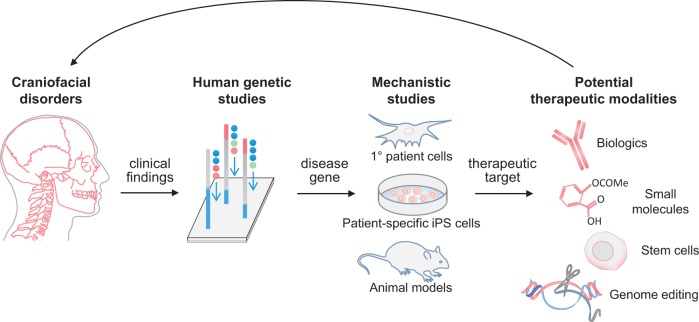
Studies that model congenital disorders in primary patient cells, iPSCs, and mice have advanced therapeutic opportunities for craniofacial disorders (Fig. 1). New technologies such as CRISPR/Cas9 that increase the speed, efficiency, and simplicity in genome editing will allow for rapid generation of cell lines and animal models that carry human disease-causing mutations (90–93). Specifically, genome-editing techniques offer a way to model Mendelian disorders in large animals, whose disease states may more closely resemble humans than the mouse models. Indeed, strategies for CRISPR/Cas9-modification in monkey, pig, and goat embryos have recently been reported (94–96). Genome editing will also aid in the study of congenital disorders associated with allelic or locus heterogeneity, which can complicate the diagnosis and treatment of these conditions (44,97). Introducing patient-specific mutations will help to identify genotype-phenotype correlations and subtle differences in the mechanistic effects of specific mutation. One of the most exciting clinical applications of genome editing is the possibility of correcting disease-causing genes. The therapeutic potential of CRISPR/Cas9 is currently being investigated in patient-derived iPSCs, organoid cultures, and mouse models (98–103). These studies raise high hopes for improving the clinical diagnosis, treatment, and outcome of patients with craniofacial and skeletal malformations.
Figure 1.
Pipeline for transforming insights from disease models into potential therapeutics for craniofacial disorders. Human genetic studies identify critical genes linked to craniofacial disease. Mechanistic studies, using primary patient cells, patient-specific iPS cells, and/or animal models, probe the disease gene’s role in craniofacial biology. Once the biological function of the gene is discovered, therapeutic targets can be identified. Having an in-depth view of the target’s biology aids in selecting therapeutic modalities, such as biologics, small molecules, stems cells, and possibly gene editing.
Acknowledgements
A.E.M. and O.D.K. co-supervised the writing of this review.
Conflict of Interest Statement. None declared.
Funding
This work was supported by the National Institutes of Health [R01DE025222 to A.E.M, U01-DE024440 and R01-DE024988 to O.D.K]; and March of Dimes [#6-FY15-233 to A.E.M.].
References
- 1.Thesleff I. (2006) The genetic basis of tooth development and dental defects. Am. J. Med. Genet. A, 140, 2530–2535. [DOI] [PubMed] [Google Scholar]
- 2.Minoux M., Rijli F.M. (2010) Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development, 137, 2605–2621. [DOI] [PubMed] [Google Scholar]
- 3.(2004) Global strategies to reduce the health care burden of craniofacial anomalies: report of WHO meetings on international collaborative research on craniofacial anomalies. Cleft Palate Craniofac. J, 41, 238–243. [DOI] [PubMed] [Google Scholar]
- 4.Rosenthal N., Brown S. (2007) The mouse ascending: perspectives for human-disease models. Nat. Cell Biol., 9, 993–999. [DOI] [PubMed] [Google Scholar]
- 5.Tiscornia G., Vivas E.L., Izpisua Belmonte, J.C. (2011) Diseases in a dish: modeling human genetic disorders using induced pluripotent cells. Nat. Med, 17, 1570–1576. [DOI] [PubMed] [Google Scholar]
- 6.Ishii M., Sun J., Ting M.C., Maxson R.E. (2015) The Development of the Calvarial Bones and Sutures and the Pathophysiology of Craniosynostosis. Curr. Top. Dev. Biol., 115, 131–156. [DOI] [PubMed] [Google Scholar]
- 7.Foster K.A., Frim D.M., McKinnon M. (2008) Recurrence of synostosis following surgical repair of craniosynostosis. Plast. Reconstr. Surg., 121, 70e–76e. [DOI] [PubMed] [Google Scholar]
- 8.Iseki S., Wilkie A.O., Morriss-Kay G.M. (1999) Fgfr1 and Fgfr2 have distinct differentiation- and proliferation-related roles in the developing mouse skull vault. Development, 126, 5611–5620. [DOI] [PubMed] [Google Scholar]
- 9.Rice D.P., Aberg T., Chan Y., Tang Z., Kettunen P.J., Pakarinen L., Maxson R.E., Thesleff I. (2000) Integration of FGF and TWIST in calvarial bone and suture development. Development, 127, 1845–1855. [DOI] [PubMed] [Google Scholar]
- 10.Yang F., Wang Y., Zhang Z., Hsu B., Jabs E.W., Elisseeff J.H. (2008) The study of abnormal bone development in the Apert syndrome Fgfr2+/S252W mouse using a 3D hydrogel culture model. Bone, 43, 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morita J., Nakamura M., Kobayashi Y., Deng C.X., Funato N., Moriyama K. (2014) Soluble form of FGFR2 with S252W partially prevents craniosynostosis of the apert mouse model. Dev. Dyn., 243, 560–567. [DOI] [PubMed] [Google Scholar]
- 12.Yokota M., Kobayashi Y., Morita J., Suzuki H., Hashimoto Y., Sasaki Y., Akiyoshi K., Moriyama K. (2014) Therapeutic effect of nanogel-based delivery of soluble FGFR2 with S252W mutation on craniosynostosis. PLoS One, 9, e101693.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong Y., Krakow D., Marcelino J., Wilkin D., Chitayat D., Babul-Hirji R., Hudgins L., Cremers C.W., Cremers F.P., Brunner H.G, et al. (1999) Heterozygous mutations in the gene encoding noggin affect human joint morphogenesis. Nat. Genet, 21, 302–304. [DOI] [PubMed] [Google Scholar]
- 14.Komatsu Y., Yu P.B., Kamiya N., Pan H., Fukuda T., Scott G.J., Ray M.K., Yamamura K., Mishina Y. (2013) Augmentation of Smad-dependent BMP signaling in neural crest cells causes craniosynostosis in mice. J. Bone Miner. Res., 28, 1422–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warren S.M., Brunet L.J., Harland R.M., Economides A.N., Longaker M.T. (2003) The BMP antagonist noggin regulates cranial suture fusion. Nature, 422, 625–629. [DOI] [PubMed] [Google Scholar]
- 16.Hermann C.D., Wilson D.S., Lawrence K.A., Ning X., Olivares-Navarrete R., Williams J.K., Guldberg R.E., Murthy N., Schwartz Z., Boyan B.D. (2014) Rapidly polymerizing injectable click hydrogel therapy to delay bone growth in a murine re-synostosis model. Biomaterials, 35, 9698–9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hessle L., Johnson K.A., Anderson H.C., Narisawa S., Sali A., Goding J.W., Terkeltaub R., Millan J.L. (2002) Tissue-nonspecific alkaline phosphatase and plasma cell membrane glycoprotein-1 are central antagonistic regulators of bone mineralization. Proc. Natl. Acad. Sci. U. S. A., 99, 9445–9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murshed M., Harmey D., Millan J.L., McKee M.D., Karsenty G. (2005) Unique coexpression in osteoblasts of broadly expressed genes accounts for the spatial restriction of ECM mineralization to bone. Genes Dev., 19, 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J., Campbell C., Nam H.K., Caron A., Yadav M.C., Millan J.L., Hatch N.E. (2015) Enzyme replacement for craniofacial skeletal defects and craniosynostosis in murine hypophosphatasia. Bone, 78, 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tekin M., Sirmaci A., Yuksel-Konuk B., Fitoz S., Sennaroglu L. (2009) A complex TFAP2A allele is associated with branchio-oculo-facial syndrome and inner ear malformation in a deaf child. Am. J. Med. Genet. A, 149A, 427–430. [DOI] [PubMed] [Google Scholar]
- 21.Niihori T., Aoki Y., Narumi Y., Neri G., Cave H., Verloes A., Okamoto N., Hennekam R.C., Gillessen-Kaesbach G., Wieczorek D., et al. (2006) Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat. Genet., 38, 294–296. [DOI] [PubMed] [Google Scholar]
- 22.Goodwin A.F., Oberoi S., Landan M., Charles C., Groth J., Martinez A., Fairley C., Weiss L.A., Tidyman W.E., Klein O.D., et al. (2013) Craniofacial and dental development in cardio-facio-cutaneous syndrome: the importance of Ras signaling homeostasis. Clin. Genet., 83, 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vincent M., Genevieve D., Ostertag A., Marlin S., Lacombe D., Martin-Coignard D., Coubes C., David A., Lyonnet S., Vilain C., et al. (2016) Treacher Collins syndrome: a clinical and molecular study based on a large series of patients. Genet. Med., 18, 49–56. [DOI] [PubMed] [Google Scholar]
- 24.Milunsky J.M., Maher T.A., Zhao G., Roberts A.E., Stalker H.J., Zori R.T., Burch M.N., Clemens M., Mulliken J.B., Smith R., et al. (2008) TFAP2A mutations result in branchio-oculo-facial syndrome. Am. J. Hum. Genet., 82, 1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milunsky J.M., Maher T.M., Zhao G., Wang Z., Mulliken J.B., Chitayat D., Clemens M., Stalker H.J., Bauer M., Burch M., et al. (2011) Genotype-phenotype analysis of the branchio-oculo-facial syndrome. Am. J. Med. Genet. A, 155A, 22–32. [DOI] [PubMed] [Google Scholar]
- 26.Knight R.D., Javidan Y., Zhang T., Nelson S., Schilling T.F. (2005) AP2-dependent signals from the ectoderm regulate craniofacial development in the zebrafish embryo. Development, 132, 3127–3138. [DOI] [PubMed] [Google Scholar]
- 27.Green R.M., Feng W., Phang T., Fish J.L., Li H., Spritz R.A., Marcucio R.S., Hooper J., Jamniczky H., Hallgrimsson B., et al. (2015) Tfap2a-dependent changes in mouse facial morphology result in clefting that can be ameliorated by a reduction in Fgf8 gene dosage. Dis. Model. Mech., 8, 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jindal G.A., Goyal Y., Burdine R.D., Rauen K.A., Shvartsman S.Y. (2015) RASopathies: unraveling mechanisms with animal models. Dis. Model. Mech., 8, 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoue S., Moriya M., Watanabe Y., Miyagawa-Tomita S., Niihori T., Oba D., Ono M., Kure S., Ogura T., Matsubara Y., et al. (2014) New BRAF knockin mice provide a pathogenetic mechanism of developmental defects and a therapeutic approach in cardio-facio-cutaneous syndrome. Hum. Mol. Genet., 23, 6553–6566. [DOI] [PubMed] [Google Scholar]
- 30.Trainor P.A., Dixon J., Dixon M.J. (2009) Treacher Collins syndrome: etiology, pathogenesis and prevention. Eur. J. Hum. Genet., 17, 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubbi C.P., Milner J. (2003) Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J., 22, 6068–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones N.C., Lynn M.L., Gaudenz K., Sakai D., Aoto K., Rey J.P., Glynn E.F., Ellington L., Du C., Dixon J., et al. (2008) Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med., 14, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakai D., Dixon J., Achilleos A., Dixon M., Trainor P.A. (2016) Prevention of Treacher Collins syndrome craniofacial anomalies in mouse models via maternal antioxidant supplementation. Nat. Commun., 7, 10328.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein O.D., Oberoi S., Huysseune A., Hovorakova M., Peterka M., Peterkova R. (2013) Developmental disorders of the dentition: an update. Am. J. Med. Genet. C Semin. Med. Genet., 163C, 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodwin A.F., Oberoi S., Landan M., Charles C., Massie J.C., Fairley C., Rauen K.A., Klein O.D. (2014) Craniofacial and dental development in Costello syndrome. Am. J. Med. Genet. A, 164A, 1425–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aoki Y., Niihori T., Kawame H., Kurosawa K., Ohashi H., Tanaka Y., Filocamo M., Kato K., Suzuki Y., Kure S., et al. (2005) Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet., 37, 1038–1040. [DOI] [PubMed] [Google Scholar]
- 37.Estep A.L., Tidyman W.E., Teitell M.A., Cotter P.D., Rauen K.A. (2006) HRAS mutations in Costello syndrome: detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. Am. J. Med. Genet. A, 140, 8–16. [DOI] [PubMed] [Google Scholar]
- 38.Goodwin A.F., Tidyman W.E., Jheon A.H., Sharir A., Zheng X., Charles C., Fagin J.A., McMahon M., Diekwisch T.G., Ganss B., et al. (2014) Abnormal Ras signaling in Costello syndrome (CS) negatively regulates enamel formation. Hum. Mol. Genet., 23, 682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lagerstrom M., Dahl N., Nakahori Y., Nakagome Y., Backman B., Landegren U., Pettersson U. (1991) A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1). Genomics, 10, 971–975. [DOI] [PubMed] [Google Scholar]
- 40.Kim S., Inoue S., Akisaka T. (1994) Ultrastructure of quick-frozen secretory ameloblasts of the rat molar tooth. Tissue Cell, 26, 29–41. [DOI] [PubMed] [Google Scholar]
- 41.Barron M.J., Brookes S.J., Kirkham J., Shore R.C., Hunt C., Mironov A., Kingswell N.J., Maycock J., Shuttleworth C.A., Dixon M.J. (2010) A mutation in the mouse Amelx tri-tyrosyl domain results in impaired secretion of amelogenin and phenocopies human X-linked amelogenesis imperfecta. Hum. Mol. Genet., 19, 1230–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brookes S.J., Barron M.J., Boot-Handford R., Kirkham J., Dixon M.J. (2014) Endoplasmic reticulum stress in amelogenesis imperfecta and phenotypic rescue using 4-phenylbutyrate. Hum. Mol. Genet., 23, 2468–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iannitti T., Palmieri B. (2011) Clinical and experimental applications of sodium phenylbutyrate. Drugs R D, 11, 227–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warman M.L., Cormier-Daire V., Hall C., Krakow D., Lachman R., LeMerrer M., Mortier G., Mundlos S., Nishimura G., Rimoin D.L., et al. (2011) Nosology and classification of genetic skeletal disorders: 2010 revision. Am. J. Med. Genet. A, 155A, 943–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neben C.L., Merrill A.E. (2015) Signaling Pathways in Craniofacial Development: Insights from Rare Skeletal Disorders. Curr. Top. Dev. Biol., 115, 493–542. [DOI] [PubMed] [Google Scholar]
- 46.Rousseau J., Sikorska H.M., Gervais A., Bisson G., Margaron P., Lamoureux G., Mirza S.A., van Lier J.E. (1994) Evaluation of a 99mTc-antimyosin kit for myocardial infarct imaging. J. Nucl. Biol. Med., 38, 43–53. [PubMed] [Google Scholar]
- 47.Shiang R., Thompson L.M., Zhu Y.Z., Church D.M., Fielder T.J., Bocian M., Winokur S.T., Wasmuth J.J. (1994) Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell, 78, 335–342. [DOI] [PubMed] [Google Scholar]
- 48.Henderson J.E., Naski M.C., Aarts M.M., Wang D., Cheng L., Goltzman D., Ornitz D.M. (2000) Expression of FGFR3 with the G380R achondroplasia mutation inhibits proliferation and maturation of CFK2 chondrocytic cells. J. Bone Miner. Res., 15, 155–165. [DOI] [PubMed] [Google Scholar]
- 49.Krejci P., Salazar L., Kashiwada T.A., Chlebova K., Salasova A., Thompson L.M., Bryja V., Kozubik A., Wilcox W.R. (2008) Analysis of STAT1 activation by six FGFR3 mutants associated with skeletal dysplasia undermines dominant role of STAT1 in FGFR3 signaling in cartilage. PLoS One, 3, e3961.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laederich M.B., Horton W.A. (2010) Achondroplasia: pathogenesis and implications for future treatment. Curr. Opin. Pediatr., 22, 516–523. [DOI] [PubMed] [Google Scholar]
- 51.Yamashita A., Morioka M., Kishi H., Kimura T., Yahara Y., Okada M., Fujita K., Sawai H., Ikegawa S., Tsumaki N. (2014) Statin treatment rescues FGFR3 skeletal dysplasia phenotypes. Nature, 513, 507–511. [DOI] [PubMed] [Google Scholar]
- 52.Yudoh K., Karasawa R. (2010) Statin prevents chondrocyte aging and degeneration of articular cartilage in osteoarthritis (OA). Aging (Albany NY), 2, 990–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simopoulou T., Malizos K.N., Poultsides L., Tsezou A. (2010) Protective effect of atorvastatin in cultured osteoarthritic chondrocytes. J. Orthop. Res., 28, 110–115. [DOI] [PubMed] [Google Scholar]
- 54.Baker J.F., Walsh P.M., Byrne D.P., Mulhall K.J. (2012) Pravastatin suppresses matrix metalloproteinase expression and activity in human articular chondrocytes stimulated by interleukin-1beta. J. Orthop. Traumatol., 13, 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neben C.L., Idoni B., Salva J.E., Tuzon C.T., Rice J.C., Krakow D., Merrill A.E. (2014) Bent bone dysplasia syndrome reveals nucleolar activity for FGFR2 in ribosomal DNA transcription. Hum. Mol. Genet., 23, 5659–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merrill A.E., Sarukhanov A., Krejci P., Idoni B., Camacho N., Estrada K.D., Lyons K.M., Deixler H., Robinson H., Chitayat D., et al. (2012) Bent bone dysplasia-FGFR2 type, a distinct skeletal disorder, has deficient canonical FGF signaling. Am. J. Hum. Genet., 90, 550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haddach M., Schwaebe M.K., Michaux J., Nagasawa J., O'Brien S.E., Whitten J.P., Pierre F., Kerdoncuff P., Darjania L., Stansfield R., et al. (2012) Discovery of CX-5461, the First Direct and Selective Inhibitor of RNA Polymerase I, for Cancer Therapeutics. ACS Med. Chem. Lett., 3, 602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bywater M.J., Poortinga G., Sanij E., Hein N., Peck A., Cullinane C., Wall M., Cluse L., Drygin D., Anderes K., et al. (2012) Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell, 22, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Devlin J.R., Hannan K.M., Hein N., Cullinane C., Kusnadi E., Ng P.Y., George A.J., Shortt J., Bywater M.J., Poortinga G., et al. (2016) Combination Therapy Targeting Ribosome Biogenesis and mRNA Translation Synergistically Extends Survival in MYC-Driven Lymphoma. Cancer Discov., 6, 59–70. [DOI] [PubMed] [Google Scholar]
- 60.Drygin D., Lin A., Bliesath J., Ho C.B., O'Brien S.E., Proffitt C., Omori M., Haddach M., Schwaebe M.K., Siddiqui-Jain A., et al. (2011) Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res., 71, 1418–1430. [DOI] [PubMed] [Google Scholar]
- 61.Papadaki M.E., Lietman S.A., Levine M.A., Olsen B.R., Kaban L.B., Reichenberger E.J. (2012) Cherubism: best clinical practice. Orphanet. J. Rare Dis., 7 Suppl 1, S6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reichenberger E.J., Levine M.A., Olsen B.R., Papadaki M.E., Lietman S.A. (2012) The role of SH3BP2 in the pathophysiology of cherubism. Orphanet. J. Rare Dis., 7 Suppl 1, S5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ueki Y., Lin C.Y., Senoo M., Ebihara T., Agata N., Onji M., Saheki Y., Kawai T., Mukherjee P.M., Reichenberger E., et al. (2007) Increased myeloid cell responses to M-CSF and RANKL cause bone loss and inflammation in SH3BP2 “cherubism” mice. Cell, 128, 71–83. [DOI] [PubMed] [Google Scholar]
- 64.Yoshitaka T., Ishida S., Mukai T., Kittaka M., Reichenberger E.J., Ueki Y. (2014) Etanercept administration to neonatal SH3BP2 knock-in cherubism mice prevents TNF-alpha-induced inflammation and bone loss. J. Bone Miner. Res., 29, 1170–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshitaka T., Kittaka M., Ishida S., Mizuno N., Mukai T., Ueki Y. (2015) Bone marrow transplantation improves autoinflammation and inflammatory bone loss in SH3BP2 knock-in cherubism mice. Bone, 71, 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lietman S.A., Yin L., Levine M.A. (2008) SH3BP2 is an activator of NFAT activity and osteoclastogenesis. Biochem. Biophys. Res. Commun., 371, 644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amaral F.R., Brito J.A., Perdigao P.F., Carvalho V.M., de Souza P.E., Gomez M.V., De Marco L., Gomez R.S. (2010) NFATc1 and TNFalpha expression in giant cell lesions of the jaws. J. Oral Pathol. Med., 39, 269–274. [DOI] [PubMed] [Google Scholar]
- 68.Duarte A.P., Gomes C.C., Gomez R.S., Amaral F.R. (2011) Increased expression of NFATc1 in giant cell lesions of the jaws, cherubism and brown tumor of hyperparathyroidism. Oncol. Lett., 2, 571–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kadlub N., Vazquez M.P., Galmiche L., L'Hermine A.C., Dainese L., Ulinski T., Fauroux B., Pavlov I., Badoual C., Marlin S., et al. (2015) The calcineurin inhibitor tacrolimus as a new therapy in severe cherubism. J. Bone Miner. Res., 30, 878–885. [DOI] [PubMed] [Google Scholar]
- 70.Gong Y., Slee R.B., Fukai N., Rawadi G., Roman-Roman S., Reginato A.M., Wang H., Cundy T., Glorieux F.H., Lev D., et al. (2001) LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell, 107, 513–523. [DOI] [PubMed] [Google Scholar]
- 71.Ai M., Holmen S.L., Van Hul W., Williams B.O., Warman M.L. (2005) Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Mol. Cell. Biol., 25, 4946–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Little R.D., Carulli J.P., Del Mastro R.G., Dupuis J., Osborne M., Folz C., Manning S.P., Swain P.M., Zhao S.C., Eustace B., et al. (2002) A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am. J. Hum. Genet., 70, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boyden L.M., Mao J., Belsky J., Mitzner L., Farhi A., Mitnick M.A., Wu D., Insogna K., Lifton R.P. (2002) High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med., 346, 1513–1521. [DOI] [PubMed] [Google Scholar]
- 74.Van Wesenbeeck L., Cleiren E., Gram J., Beals R.K., Benichou O., Scopelliti D., Key L., Renton T., Bartels C., Gong Y., et al. (2003) Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am. J. Hum. Genet., 72, 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Balemans W., Devogelaer J.P., Cleiren E., Piters E., Caussin E., Van Hul W. (2007) Novel LRP5 missense mutation in a patient with a high bone mass phenotype results in decreased DKK1-mediated inhibition of Wnt signaling. J. Bone Miner. Res., 22, 708–716. [DOI] [PubMed] [Google Scholar]
- 76.Brunkow M.E., Gardner J.C., Van Ness J., Paeper B.W., Kovacevich B.R., Proll S., Skonier J.E., Zhao L., Sabo P.J., Fu Y., et al. (2001) Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am. J. Hum. Genet., 68, 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balemans W., Ebeling M., Patel N., Van Hul E., Olson P., Dioszegi M., Lacza C., Wuyts W., Van Den Ende J., Willems P., et al. (2001) Increased bone density in sclerosteosis is due to the deficiency of a novel secreted protein (SOST). Hum. Mol. Genet., 10, 537–543. [DOI] [PubMed] [Google Scholar]
- 78.Semenov M., Tamai K., He X. (2005) SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J. Biol. Chem., 280, 26770–26775. [DOI] [PubMed] [Google Scholar]
- 79.McClung M.R., Grauer A., Boonen S., Bolognese M.A., Brown J.P., Diez-Perez A., Langdahl B.L., Reginster J.Y., Zanchetta J.R., Wasserman S.M., et al. (2014) Romosozumab in postmenopausal women with low bone mineral density. N. Engl. J. Med., 370, 412–420. [DOI] [PubMed] [Google Scholar]
- 80.Chang M.K., Kramer I., Keller H., Gooi J.H., Collett C., Jenkins D., Ettenberg S.A., Cong F., Halleux C., Kneissel M. (2014) Reversing LRP5-dependent osteoporosis and SOST deficiency-induced sclerosing bone disorders by altering WNT signaling activity. J. Bone Miner. Res., 29, 29–42. [DOI] [PubMed] [Google Scholar]
- 81.Kedlaya R., Veera S., Horan D.J., Moss R.E., Ayturk U.M., Jacobsen C.M., Bowen M.E., Paszty C., Warman M.L., Robling A.G. (2013) Sclerostin inhibition reverses skeletal fragility in an Lrp5-deficient mouse model of OPPG syndrome. Sci. Transl. Med., 5, 211ra158.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grafe I., Alexander S., Yang T., Lietman C., Homan E.P., Munivez E., Chen Y., Jiang M.M., Bertin T., Dawson B, et al. (2015) Sclerostin Antibody Treatment Improves the Bone Phenotype of Crtap Mice, a Model of Recessive Osteogenesis Imperfecta. J. Bone Miner. Res, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jacobsen C.M., Barber L.A., Ayturk U.M., Roberts H.J., Deal L.E., Schwartz M.A., Weis M., Eyre D., Zurakowski D., Robling A.G., et al. (2014) Targeting the LRP5 pathway improves bone properties in a mouse model of osteogenesis imperfecta. J. Bone Miner. Res., 29, 2297–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roschger A., Roschger P., Keplingter P., Klaushofer K., Abdullah S., Kneissel M., Rauch F. (2014) Effect of sclerostin antibody treatment in a mouse model of severe osteogenesis imperfecta. Bone, 66, 182–188. [DOI] [PubMed] [Google Scholar]
- 85.Sinder B.P., Eddy M.M., Ominsky M.S., Caird M.S., Marini J.C., Kozloff K.M. (2013) Sclerostin antibody improves skeletal parameters in a Brtl/+ mouse model of osteogenesis imperfecta. J. Bone Miner. Res., 28, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sinder B.P., White L.E., Salemi J.D., Ominsky M.S., Caird M.S., Marini J.C., Kozloff K.M. (2014) Adult Brtl/+ mouse model of osteogenesis imperfecta demonstrates anabolic response to sclerostin antibody treatment with increased bone mass and strength. Osteoporos. Int., 25, 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grafe I., Yang T., Alexander S., Homan E.P., Lietman C., Jiang M.M., Bertin T., Munivez E., Chen Y., Dawson B., et al. (2014) Excessive transforming growth factor-beta signaling is a common mechanism in osteogenesis imperfecta. Nat. Med., 20, 670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Desai J., Shannon M.E., Johnson M.D., Ruff D.W., Hughes L.A., Kerley M.K., Carpenter D.A., Johnson D.K., Rinchik E.M., Culiat C.T. (2006) Nell1-deficient mice have reduced expression of extracellular matrix proteins causing cranial and vertebral defects. Hum. Mol. Genet., 15, 1329–1341. [DOI] [PubMed] [Google Scholar]
- 89.James A.W., Shen J., Zhang X., Asatrian G., Goyal R., Kwak J.H., Jiang L., Bengs B., Culiat C.T., Turner A.S., et al. (2015) NELL-1 in the treatment of osteoporotic bone loss. Nat Commun., 6, 7362.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. (2013) RNA-programmed genome editing in human cells. Elife, 2, e00471.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. (2013) RNA-guided human genome engineering via Cas9. Science, 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harrison M.M., Jenkins B.V., O'Connor-Giles K.M., Wildonger J. (2014) A CRISPR view of development. Genes Dev., 28, 1859–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hai T., Teng F., Guo R., Li W., Zhou Q. (2014) One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res., 24, 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Li W., et al. (2014) Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell, 156, 836–843. [DOI] [PubMed] [Google Scholar]
- 96.Ni W., Qiao J., Hu S., Zhao X., Regouski M., Yang M., Polejaeva I.A., Chen C. (2014) Efficient gene knockout in goats using CRISPR/Cas9 system. PLoS One, 9, e106718.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu J.T., Campeau P.M., Lee B.H. (2014) Genotype-phenotype correlation–promiscuity in the era of next-generation sequencing. N. Engl. J. Med., 371, 593–596. [DOI] [PubMed] [Google Scholar]
- 98.Li H.L., Fujimoto N., Sasakawa N., Shirai S., Ohkame T., Sakuma T., Tanaka M., Amano N., Watanabe A., Sakurai H., et al. (2015) Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports, 4, 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Osborn M.J., Gabriel R., Webber B.R., DeFeo A.P., McElroy A.N., Jarjour J., Starker C.G., Wagner J.E., Joung J.K., Voytas D.F., et al. (2015) Fanconi anemia gene editing by the CRISPR/Cas9 system. Hum. Gene Ther., 26, 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schwank G., Koo B.K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K., et al. (2013) Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell, 13, 653–658. [DOI] [PubMed] [Google Scholar]
- 101.Xie F., Ye L., Chang J.C., Beyer A.I., Wang J., Muench M.O., Kan Y.W. (2014) Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res., 24, 1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu Y., Liang D., Wang Y., Bai M., Tang W., Bao S., Yan Z., Li D., Li J. (2013) Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell, 13, 659–662. [DOI] [PubMed] [Google Scholar]
- 103.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. (2014) Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol., 32, 551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]