Abstract
High-resolution mass spectrometry (MS)-based proteomics is a powerful method for the identification of soluble protein complexes and large-scale affinity purification screens can decode entire protein interaction networks. In contrast, protein complexes residing on chromatin have been much more challenging, because they are difficult to purify and often of very low abundance. However, this is changing due to recent methodological and technological advances in proteomics. Proteins interacting with chromatin marks can directly be identified by pulldowns with synthesized histone tails containing posttranslational modifications (PTMs). Similarly, pulldowns with DNA baits harbouring single nucleotide polymorphisms or DNA modifications reveal the impact of those DNA alterations on the recruitment of transcription factors. Accurate quantitation – either isotope-based or label free – unambiguously pinpoints proteins that are significantly enriched over control pulldowns. In addition, protocols that combine classical chromatin immunoprecipitation (ChIP) methods with mass spectrometry (ChIP-MS) target gene regulatory complexes in their in-vivo context. Similar to classical ChIP, cells are crosslinked with formaldehyde and chromatin sheared by sonication or nuclease digested. ChIP-MS baits can be proteins in tagged or endogenous form, histone PTMs, or lncRNAs. Locus-specific ChIP-MS methods would allow direct purification of a single genomic locus and the proteins associated with it. There, loci can be targeted either by artificial DNA-binding sites and corresponding binding proteins or via proteins with sequence specificity such as TAL or nuclease deficient Cas9 in combination with a specific guide RNA. We predict that advances in MS technology will soon make such approaches generally applicable tools in epigenetics.
Introduction
Gene expression starts with regulatory proteins binding to DNA at the promoter and enhancer regions, which have the potential to alter the local chromatin environment or recruit polymerases and other proteins of the core transcriptional machinery. These proteins bind directly to specific DNA sequences or interact with other chromatin components such as specific posttranslational modifications (PTMs) on histone tails. The composition of complexes in the regulatory region of a gene decides whether a gene is actively transcribed, repressed, or held in an intermediated state. A true understanding of how a gene is controlled in normal function or disease requires identification of the complete inventory of regulatory proteins and complexes that reside in its regulatory regions, as well as their interactions and modifications.
High resolution, quantitative mass spectrometry (MS)-based proteomics has turned into a powerful tool to study diverse aspects of proteins in a global and unbiased manner (1), and in particular protein-protein interactions (2). Initial strategies involved stringent purification of protein complexes followed by SDS-PAGE separation and MS-based analysis of visually selected gel bands. Today, dramatic improvements in shot-gun proteomics allow direct in-solution digestion of immuno-precipitates, which defines significantly enriched proteins over background binders (3). This requires accurate quantitation, which can be achieved either in isotope labelled formats such as SILAC (4), or by the increasingly powerful label-free methods (5,6). Label-based quantitation methods allow a direct read out of relative peptide quantities within the same MS run and generally involve the determination of a ‘heavy to light ratio’ in the same mass spectrum. In contrast, modern, label free methods employ sophisticated bioinformatic normalization strategies to compare protein intensities between an unlimited number of MS runs. As sequencing information can be matched between runs (5), label free quantification methods can also provide a higher sensitivity compared to label based methods, which is especially helpful for the analysis of low abundant protein interactions. Following such strategies, recent large-scale studies have charted the composition of soluble complexes and their networks on a global scale (7–10).
Compared to soluble complexes, the characterization of chromatin-associated ones is much more difficult owing to the tight integrity of chromatin that needs to be disrupted, without affecting the complex of interest. Two main strategies are employed: The first one simulates the interaction between (modified) histones or DNA and protein complexes in vitro using peptide or DNA baits. The second one resembles classical chromatin immunoprecipitation (ChIP) and ChIP-derived protocols except that they are followed by MS.
In this review, we will focus on the contribution of proteomics to the epigenetics field, in particular the identification of gene regulatory complexes on chromatin. Due to limits in length, we refer the reader to recent reviews for the use of proteomics for the identification of histone PTMs and the definition of soluble forms of chromatin-associated complexes (11–13).
Chromatin Enrichments
Identification of the complete inventory of regulatory proteins and complexes associated with chromatin would be of tremendous help in understanding gene regulation and other chromatin-related processes. High resolution MS has already been applied to the DNA damage response (14,15), DNA repair (16), DNA replication (17–20) and mitosis (21). The analysis of the soluble Histone H3.1 interactome provided a comprehensive view of histone chaperones and components of the replication fork (22).
To characterize proteins responsible for gene transcription, several studies characterized interphase chromatin, starting with a pioneering study of Aebersold and colleagues in which they analysed the chromatin fraction of human B lymphocytes prior and after overexpression of the oncogene c-Myc (23). This revealed a large set of transcription factors and other chromatin-associated proteins, some of which displayed significant expression changes, including a 10-fold downregulation of the transcription factor ATF-3 and a 2-fold induction of the progesterone receptor. As part of an extensive chromatin characterization study in HeLa cells, Garcia and colleagues distinguished proteins differentially associated with euchromatin and heterochromatin by using partial MNase digestion (24). A recent study also applied MNase digestion to solubilize the chromatin fraction of embryonic stem cells (ESCs) and neural progenitor cells (NPCs), identifying a preferential chromatin association of the esBAF complex member Smarcd1 in ESCs (25).
Taking a different tack, Rappsilber and co-workers applied formaldehyde crosslinking and denaturing washing conditions to capture proteins that are tightly crosslinked to chromatin (26,27). Surprisingly, about half of these proteins did not have apparent chromatin-related functions. To define interphase chromatin-associated proteins, the authors therefore compared 28 different biological conditions, including different cell types, cell cycle phases, and drug treatments and applied a machine learning algorithm to identify proteins with common dynamic behavior that differed from contaminant proteins.
Histone Modification Readers
Histone proteins can be marked by phosphorylation, methylation, acetylation, ubiquitination and numerous other PTMs. These can directly affect chromatin structure, or be recognized by specific reader proteins, which themselves regulate chromatin structure to affect the expression of nearby genes (28). MS greatly contributed to the identification of these modifications (11), as well as of their respective reader proteins.
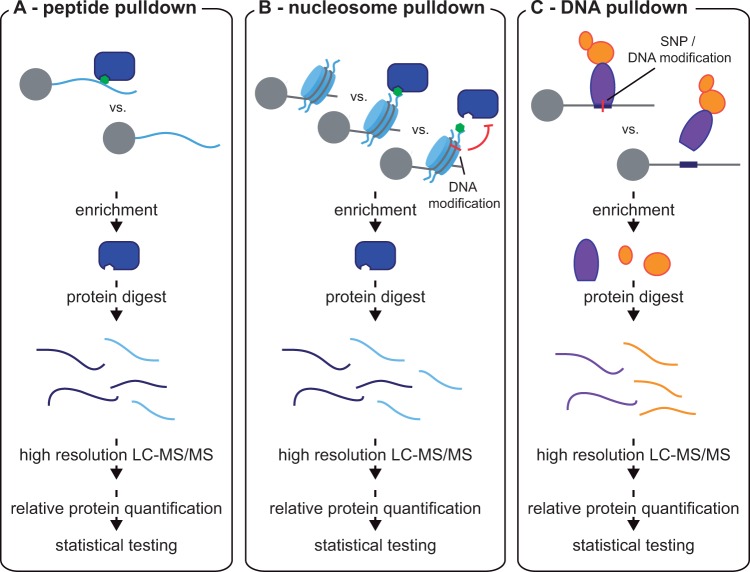
Typically, peptides representing parts of the histone tail sequence and containing one or more PTMs are linked with beads and incubated with cell extracts. Proteins specifically recognizing the PTM are enriched and then analysed by MS (Figure 1A) . Such assays identified key events such as the binding of Wdr5 and NURF to trimethylated lysine 4 on histone H3 (29,30). The introduction of quantitative proteomics in the form of SILAC labeling (4) dramatically increased sensitivity and specificity, revealing that the TFIID complex directly interacts with H3K4me3 (31) as well as interactions of other factors with trimethylated lysines (32). The same approach elegantly unraveled differential binding of different PRMT isoforms to symmetric and asymmetric dimethylation of H3R2 (33). Improvements in MS data quality and algorithms have led to a switch to label-free quantitation and this has successfully been applied to mouse tissues (34) and even birds (35). To more closely resemble binding in the context of chromatin, mono- or oligo-nucleosomes with modified histone tails have been generated by their chemical linkage to the globular domains (36–38) (Figure 1B). Although the differences in reader proteins in peptide and nucleosome pulldowns for a given histone PTM appear to be minor, an advantage of nucleosome pulldowns is the combined assessment of histone and DNA modifications. For instance, while PRC2 complex members effectively bound H3K27me3 modified nucleosomes, this interaction was lost upon introduction of DNA methylation, while the association with the ORC complex was increased (38).
Figure 1.
In vitro pulldown assays to study protein interactions with modified histone tails, DNA and assembled nucleosomes. Nuclear cell extracts are incubated with (A) peptides resembling a partial histone tail, which can carry posttranslational modifications, (B) in vitro assembled nucleosomes, harboring a DNA modification and/or modified histone tail integrated by chemical linkage, or (C) DNA baits, which harbor a specific DNA sequence, such as a disease-linked SNP or a DNA modification. Following enrichment, proteins are enzymatically digested and analysed by high resolution LC-MS/MS. Specific interactors are defined by quantitative comparison with a control by means of isotope or label-free quantitation-based methods.
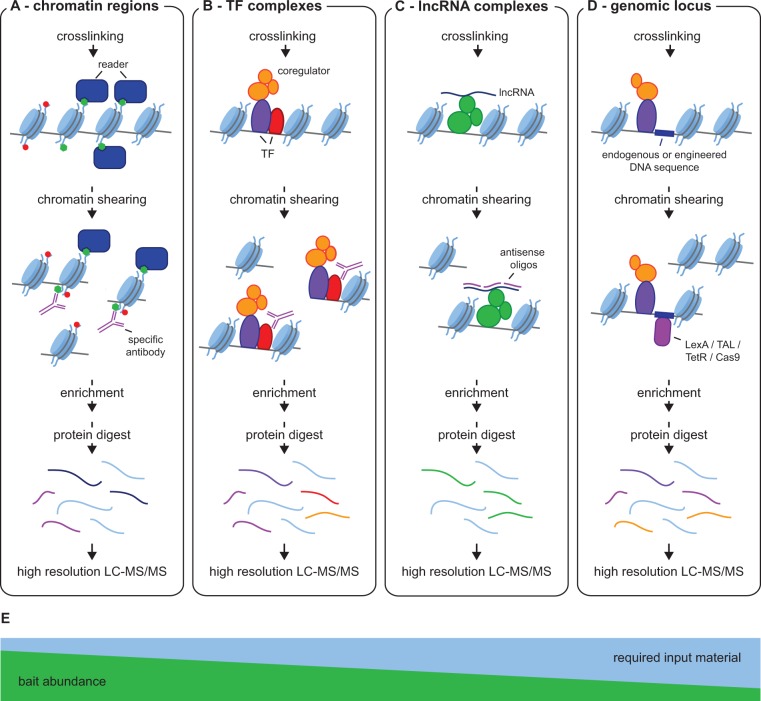
Binding of proteins to chromatin in vivo is influenced by many different factors, such as the combinatorics of different modifications on histones and DNA, the presence of DNA-binding proteins other than histone core particles and the three dimensional structure of chromatin (39). In the classical chromatin-immunoprecipitation (ChIP) method chromatin is crosslinked by formaldehyde and sheared by nuclease treatment or sonication to a length of one to three nucleosomes, followed by antibody based affinity purification and analysis of the bound DNA by PCR or next generation sequencing (ChIP-seq) (40,41). Technological improvements in MS have recently allowed the application of these ChIP protocols to elucidate chromatin associated protein complexes, a technology termed ChIP-MS (42,43), ChroP (44,45), mChIP (46,47), or RIME (48–50) (Figure 2A). Two recent papers enriched nucleosomes containing particular histone modifications to study protein interaction networks of genomic regions marked with specific histone PTMs for promoter, enhancer and heterochromatin regions in mouse ESCs (51,52). Interestingly, the same method can also identify co-occurrence of histone marks, by analysing the histone PTM levels in the precipitated material (44). The application of ChIP-MS to histone variants elucidated nucleosomal ratios and histone PTMs of mononucleosomes containing non-canonical histone variants (43).
Figure 2.
ChIP-MS based methods to identify chromatin associated protein complexes. Cells are crosslinked by formaldehyde and chromatin is prepared by sonication or nuclease digest. Following the appropriate enrichment step, proteins are quantitatively analysed. (A) ChroP (44,45), ChIP-MS (51,52): Chromatin complexes marked with histone PTMs are enriched with PTM-specific antibodies to identify proteins associated with specific chromatin regions (B) mChIP (46,47), ChIP-MS (42,43), RIME (48–50), BioTAP-XL (53,55): Transcription factors or other chromatin associated proteins are targeted via protein tags or antibodies directed towards the endogenous proteins to identify their chromatin-bound protein complexes. Note that in case of mChIP, chromatin is not crosslinked with formaldehyde, hence the method represents a native ChIP-MS protocol. (C) ChIRP-MS (60): lncRNAs associated with chromatin complexes are enriched using an array of biotinylated antisense oligonucleotides spanning the entire sequence of the lncRNA (D) Locus-specific ChIP-MS: an individual genomic locus is enriched using a complementary DNA probe [PICh (79,80)], an engineered artificial binding site together with a respective binding protein - e.g. LexA [ChAP-MS (81), iCHIP (82)] or TetR [TChP (83)] -, or proteins that directly recognize the specific target sequence - e.g. TAL [TAL-ChAP-MS (85), enChIP (86)] or Cas9-sgRNA [CRISPR-ChAP-MS (90), enCHIP (91)]. The binding protein has one or more tags that can be used for purification. (E) The relative bait abundance and amount of input material required for the different ChIP-MS based methods shown in (A-D).
ChIP-MS for Transcription Factor Complexes and Other Chromatin Components
In addition to localizing histone PTMs across the genome, ChIP-Seq can also elucidate the binding of transcription factors, coregulatory proteins and other chromatin components. While ChIP-Seq infers protein associations by correlating their peak locations, ChIP-MS directly identifies chromatin associated complexes without a priori assumptions (Figure 2B). A pioneering ChIP-MS study in yeast characterized interaction partners of histone H2A, the histone variant Htz1p and three chromatin-bound proteins, that are difficult to purify by traditional co-immunoprecipitation (Co-IP) protocols (46). In this and a following study, in which they extended the screen to 102 different chromatin associated yeast proteins including several transcription factors, the group employed TAP-tagged baits, which were precipitated using IgG-coated magnetic beads (47).
ChIP-MS also determined interactors of the MSL (male-specific lethal) complex in Drosophila, which is responsible for X-chromosome dosage compensation in male flies (42). MSL2 and 3, two subunits of the complex, were tagged with an HTB tag, which is biotinylated in vivo, transfected into S2 cells, and precipitated from cross-linked and sheared chromatin preparation with streptavidin beads. The authors identified a range of proteins involved in active gene transcription including a putative H3K36me3 binding protein, whose loss of function phenotype resulted in partial mislocalization of the MSL complex to autosomes (42). Interestingly, the H3K36me3 modification was enriched in the MSL ChIP-MS, whereas H3K4me3 was depleted, in accordance with the ChIP-seq results (42). By exchanging the HTB with a bioTAP tag (53,54), the method was recently applied to find interaction partners PRC1 and 2 complex members in human cells (55) and drosophila HP1 from cells and from different life stages of flies (56). In a variant of ChIP-MS, a TAP-based immunoprecipitation is followed by an in vitro lysine actyl-transferase (KAT) reaction in presence of isotopically labelled Acetyl-CoA (57). This allows identification of substrate-KAT relationships of chromatin associated protein complexes.
ChIP-MS for chromatin-associated factors can also be conducted without tags using antibodies directed against the endogenous protein (48–50). Carroll and co-workers focused on coregulatory proteins of oestrogen receptor (ER) and defined specific interactors by removing proteins that were identified in a control ChIP-MS experiment using total IgG (48). By further including SILAC, the authors defined specific interactions that depend on treatment with either oestrogen or the ER antagonist tamoxifen, which identified GREB1 as an oestrogen-specific ER-interacting protein (48). GREB1 ChIP-Seq revealed a strong overlap with the ER coregulators CBP and p300, while GREB1 downregulation by siRNA resulted in a displacement of those two factors from ER-binding sites. ER ChIP-MS in solid tumour samples confirmed the interaction with GREB1 in three out of six tumour samples, suggesting clinical relevance. Subsequently, the group reported an interaction of ER with progesterone receptor (PR) in response to progesterone treatment (49). Interestingly, activated PR directed ER to genomic loci such that the resulting gene expression program was associated with a good clinical outcome.
Long non-coding RNAs (lncRNAs) can directly act at the level of chromatin as functional regulators of gene expression (58,59). In analogy to ChIP-seq, the localization of those RNAs on the genome can be determined by a ChIP-like method called ChIRP (Chromatin Isolation by RNA Purification), which includes precipitation of a given lncRNA using biotinylated antisense oligos covering the whole length of the RNA sequence (60) (Figure 2C). To identify proteins associated with the Xist lncRNA, ChIRP was recently combined with MS and uncovered 81 proteins specifically interacting with Xist on chromatin. This included HnrnK and Spen (/Sharp), which were both found to participate in Xist-mediated gene silencing (61). UV crosslinking and purification under denaturing conditions has also been used on Xist (62), and strikingly, 9 of 10 interactors overlapped with the factors identified by ChIRP-MS. Hence, formaldehyde crosslinking under ChIP conditions efficiently covers both direct lncRNA interactions as well as the chromatin background. Of note, while the UV crosslinking study postulated that the gene inhibitory mechanism of Spen (/Sharp) is mediated by an interaction with Smrt and Hdac3, neither factor was identified in the UV-crosslinking RNA pulldown or in the ChIRP-MS experiment. Hence, either ChIRP-MS was not sufficiently comprehensive, or Smrt and Hdac3 are not physically associated with Xist chromatin loci.
DNA Pulldowns
For all the advantages of ChIP-MS, it does not provide locus-specificity in the genome. However, other MS methods contribute to the understanding of protein-DNA interactions, most prominently DNA pull-down experiments followed by MS (63) (Figure 1C). DNA baits containing the DNA sequence of interest or a control sequence are biotinylated and incubated with nuclear extracts to enrich for specifically interacting proteins. Such an approach successfully identified new interactors of telomeres using polymerized biotinylated double‐stranded oligonucleotides of the telomeric sequence TTAGGG as baits (64,65). Furthermore, it has shed new light on the enigma of ultra-conserved elements (UCE) in the genome (66). Analysis of a total of 193 UCE sequences between 200bp and 1000bp in length identified a set of 425 proteins that robustly associated with those elements (67). Interestingly, UCEs in non-exonic regions were most enriched in intrinsic interactors, and at the same time most refractory to the binding of PRC proteins, which was much higher on random genomic loci, proving non-exonic UCEs to be integrators of enhancer localized transcription factors.
In addition to their application in basic biology, DNA pulldowns linked to MS also have great potential in a genetic context, by identifying differential binders of single nucleotide polymorphisms (SNPs). This approach had been termed PWAS (proteome-wide analysis of SNPs). In an example from animal husbandry, PWAS determined the cause of the genetic difference responsible for the lean phenotype of European pigs, which turned out to be due to a repressor differentially binding to an imprinted locus at the IGF2 gene (68,69). In a clinical context, PWAS was first applied to identify differential interactors to SNPs that are highly associated with type 1 diabetes at the interleukin-2 receptor (CD25) locus (70). Using concatemerized DNA oligonucleotides representing the genomic sequence surrounding a particular SNP, PWAS identified four differential interactions of transcription factors at four out of 12 investigated SNPs. Note that differential binding to more than the lead SNP in a haplo-block should be investigated, as the lead SNP is not necessarily the functional one.
Another PWAS study focused on two somatic mutations in the TERT promoter region, which are frequently associated with oncogenesis (C228T and C250T) (71). Both mutations generate de novo consensus binding motifs for E-twenty-six (ETS) transcription factors (72,73), which are recognized in vivo by the transcription factor GABP (74). Performing PWAS with either of the two mutations revealed binding of the ETS transcription factors ELF1, ELF2, and ETV, while both mutations together bound GABP as a hetero-tetramer, which requires two adjacent ETS-binding motifs. Interestingly, GABP binding was even more favoured with a DNA-bait containing two native upstream ETS-binding sites in combination with the C228T mutation. This highlights the importance of the combinatorial nature of transcription factor binding in DNA pull-down assays and suggests that oligonucleotides, which only minimally cover an SNP could fail to identify biologically meaningful interactions.
To overcome limitations of SILAC labeling, which is most easily used in cell-lines, a recent study applied label-free quantification and dimethyl labelling for DNA pulldowns, enabling the measurement of DNA interactions in extracts of primary cells (75). Using nuclear extracts of peripheral blood mononuclear cells (PBMCs), this revealed allele-specific binding to a SNP related to chronic lymphocytic leukaemia (75). Such a strategy may be interesting for future PWAS studies, as it allows direct screening for differential-binding partners as long as they are expressed in available target cells.
Apart from the elucidation of sequence-specific binders, DNA pulldowns coupled to MS can identify proteins that specifically recognize DNA modifications, such as 5-methylcytosine (5-mC) (63,76). Vermeulen and co-workers characterized reader proteins of 5-mC and 5-hydroxymethylcytosine (5-hmC), as well as their oxidized derivatives 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC) in mouse ESCs, NPCs and brain tissue (77). While 5-mC is a hallmark for epigenetic silencing, its oxidized derivatives are actively involved in gene activation (78). Interestingly, this discovered not only distinct binding proteins for 5-mC and its oxidized derivatives, but also highly dynamic binding of proteins in the three different cell types, indicating specialized roles towards differentiation and pluripotency control (77).
Locus-Specific Pulldowns
Binding of protein complexes to DNA in vivo is strongly influenced by the chromatin environment. To study the protein repertoire at a particular genomic locus, one ideally would need to isolate just that region and all proteins associated with it. The main challenge is the required sensitivity, as a single copy-binding protein cannot yield more than two molecules per cell for a diploid locus (Figure 2D).
In the first locus-specific study, Kingston and colleagues used a desthiobiotinylated DNA probe complementary to telomere repeats, taking advantage of the repetitive character of telomere sequences (79). This uncovered a large set of proteins with a known or potential telomere-associated role, including the homeobox telomere-binding protein 1 (HOT1) also identified by the telomere DNA pull down approach mentioned above (64). The same group also identified proteins interacting with telomere-associated sequence (TAS) repeats in Drosophila (80).
Demonstrating proof of principle for single locus-specific ChIP MS, Tacket and colleagues genetically engineered a yeast strain containing a LexA-binding site in close vicinity to the GAL1 promoter, together with constitutive expression of the LexA-PrA fusion protein serving as affinity handle. Analysis of proteins interacting under transcriptionally active and repressive conditions identified Gal3, two subunits of the RNA polymerase complex (Rpb1 and Rpb2) and Spt16, a component of the FACT complex (81). The low number of specific gene activators and especially of known repressor proteins indicates that the detection threshold was still a limiting factor despite using 2.5*1011 cells.
Two arrays of eight LexA-binding sites flanked on each side by six copies of the chicken HS4 insulator complex were integrated into a plasmid in an attempt to study novel proteins that could interact with insulator elements (82). The genomically integrated LexA arrays were then targeted by an exogenously expressed LexA-CBP-FLAG fusion protein and anti-Flag antibodies. Subsequent analysis was done by silver staining, which is presumably the reason that only RNA helicase p68 and the nuclear matrix protein Matrin-3 were followed up on as specific interactors.
Analogously, a binding site of the tetracycline repressor protein (TetR) inserted into a human γ-globin minilocus was integrated into transgenic mice (83). Affinity purification used a triple tag (CFP, HA, Bio) TetR protein (TetR3T), transgenically inserted into mice expressing the BirA enzyme and crossed into the mice containing the γ-globin minilocus. A cell line of erythroid progenitor cells was then generated and expanded from the transgenic mice. Treating cells with doxycycline provided an elegant control as it leads to release of the TetR3T protein from the TetR binding site. Performing HA/streptavidin tandem purification from crosslinked cells, the authors identified 14 candidate proteins specifically interacting under non-doxycycline conditions, among them several known to bind to the γ-globin promoter such as GATA1 and CHD4.
Transcription activator-like (TAL) effector proteins have also been targeted to specific genomic loci (84). Tacket and colleagues used a TAL-PrA fusion construct recognizing a specific sequence of the GAL1 promoter (85). This led to the identification of the same four transcriptional activating proteins that they had identified with the LexA-PrA system. However, under repressive conditions, this system did not enrich the GAL1 locus, indicating that the TAL-PrA protein was not able to recognize its target sequence. In another study, a TAL-3xFLAG fusion protein was used to isolate telomeres from the mouse hematopoietic cell line Ba/F3 (86).
CRISPR-based methods are versatile tools for genomic engineering and have recently been extended by nuclease deficient Cas9 (dCas9) fusion proteins that are targeted to specific promoter regions to modify their gene expression (87–89). When this technology was employed to target the GAL1 locus it successfully enriched the GAL1 locus and revealed several enriched proteins related to transcription, however, not GAL3 nor the other three factors found before (90). Similar to the TAL-PrA construct, the CRISPR based method was not able to enrich the locus under transcriptional inactive conditions probably due to a lack of DNA accessibility as in the case of TAL-PrA.
The IRF1 promoter in human embryonic kidney-derived 293T cells was targeted by expressing a dCas9-3xFlag fusion protein together with a specific gRNA or control gRNA (91). Although the authors identified a range of nuclear and chromatin associated proteins, no specific transcription factors known to recognize this locus were identified.
Outlook
As we have documented here, there are now many proteomics methods to advance chromatin interaction research. The identification of reader proteins of histone PTMs and DNA sequences by pulldown approaches is most mature. PWAS enables geneticists to follow up on their SNP of interest in a more direct way than just predicting transcription factor binding by consensus sequences. When applying label free proteomics, primary cells and tissues can easily be used, while a lower limit in input material for meaningful PWAS results of around 600 µg of nuclear extract for an experiment containing technical triplicates for each of the two alleles has been reported (71).
ChIP-based techniques for targeting transcription factors even under endogenous levels are already showing great potential. Powerful label-free quantitative MS methods are making ChIP-MS increasingly applicable to clinical questions related to transcription factors in areas such as cancer or metabolic diseases. Special care should be taken when using antibodies directed against endogenous proteins as these may have off-target specificities. Fortunately, this can be excluded by elegant controls, such as knock down of the bait or a second antibody reacting with a different epitope.
Locus-specific ChIP-MS so far works well only for repeat sequences with multiple copies in the cell and existing studies targeting individual loci are primarily conceptual. This is almost entirely due to the fact that a very low number of bound proteins are extracted from each cell. Therefore, this is clearly an area where future generations of MS technology could enable major progress.
The rapidly developing CRISPR-based techniques will likely make them the methods of choice also for locus-specific ChIP-MS. Multiple guide RNAs for targeting a locus of interest either in combination or in individual pull-downs should decrease off-target binding of dCas9 with a single guide (92). This will be important to reduce false-positive hits in locus-specific ChIP-MS experiments. Studies with both TAL and dCas9 fusion proteins indicate problems accessing the target sequence under transcriptionally repressive conditions (85,90) and genomic binding of dCas9 positively correlates with DNAse hypersensitivity (92) and low nucleosome occupancy (93). Therefore, care should be taken that the guide RNA has access to the genome, which can be done by taking into account recent recommendations for effective guide RNA design.
In conclusion, chromatin-associated interaction proteomics has already yielded many new discoveries in biology. Ongoing technological advances in high sensitivity, quantitative MS, combined with ingenious genomic and biochemical strategies have even greater potential for basic research and for the clinic.
Acknowledgements
We thank our colleagues at the Department of Proteomics and Signal Transduction for help and fruitful discussions.
Conflict of Interest statement. None declared.
Funding
M.W. is supported by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (grant 01ZX1313A-2014). Funding to pay the Open Access publication charges for this article was provided by the Max Planck Society for the Advancement of Science.
References
- 1.Altelaar A.F., Munoz J., Heck A.J. (2013) Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet., 14, 35–48. [DOI] [PubMed] [Google Scholar]
- 2.Gingras A.C., Gstaiger M., Raught B., Aebersold R. (2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol., 8, 645–654. [DOI] [PubMed] [Google Scholar]
- 3.Dunham W.H., Mullin M., Gingras A.C. (2012) Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics, 12, 1576–1590. [DOI] [PubMed] [Google Scholar]
- 4.Ong S.E., Mann M. (2007) Stable isotope labeling by amino acids in cell culture for quantitative proteomics. Methods Mol. Biol., 359, 37–52. [DOI] [PubMed] [Google Scholar]
- 5.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics, 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cappadona S., Baker P.R., Cutillas P.R., Heck A.J., van Breukelen B. (2012) Current challenges in software solutions for mass spectrometry-based quantitative proteomics. Amino Acids, 43, 1087–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature, 440, 637–643. [DOI] [PubMed] [Google Scholar]
- 8.Havugimana P.C., Hart G.T., Nepusz T., Yang H., Turinsky A.L., Li Z., Wang P.I., Boutz D.R., Fong V., Phanse S., et al. (2012) A census of human soluble protein complexes. Cell, 150, 1068–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hein M.Y., Hubner N.C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I.A., Weisswange I., Mansfeld J., Buchholz F., et al. (2015) A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell, 163, 712–723. [DOI] [PubMed] [Google Scholar]
- 10.Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., et al. (2015) The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell, 162, 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang H., Lin S., Garcia B.A., Zhao Y. (2015) Quantitative proteomic analysis of histone modifications. Chem. Rev., 115, 2376–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange M., Demajo S., Jain P., Di Croce L. (2011) Combinatorial assembly and function of chromatin regulatory complexes. Epigenomics, 3, 567–580. [DOI] [PubMed] [Google Scholar]
- 13.Dasgupta S., Lonard D.M., O'Malley B.W. (2014) Nuclear receptor coactivators: master regulators of human health and disease. Ann. Rev. Med., 65, 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiacovo M.P., Elledge S.J. (2010) A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U S A., 107, 18475–18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D.R., Gidvani R.D., Ingalls B.P., Duncker B.P., McConkey B.J. (2011) Differential chromatin proteomics of the MMS-induced DNA damage response in yeast. Proteome Science, 9, 62.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raschle M., Smeenk G., Hansen R.K., Temu T., Oka Y., Hein M.Y., Nagaraj N., Long D.T., Walter J.C., Hofmann K., et al. (2015) DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science, 348, 1253671.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khoudoli G.A., Gillespie P.J., Stewart G., Andersen J.S., Swedlow J.R., Blow J.J. (2008) Temporal profiling of the chromatin proteome reveals system-wide responses to replication inhibition. Curr. Biol., 18, 838–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubota T., Hiraga S., Yamada K., Lamond A.I., Donaldson A.D. (2011) Quantitative proteomic analysis of chromatin reveals that Ctf18 acts in the DNA replication checkpoint. Mol. Cell. Proteomics, 10, M110 005561.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirbu B.M., Couch F.B., Cortez D. (2012) Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc., 7, 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alabert C., Bukowski-Wills J.C., Lee S.B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A. (2014) Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol., 16, 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohta S., Bukowski-Wills J.C., Sanchez-Pulido L., Alves Fde L., Wood L., Chen Z.A., Platani M., Fischer L., Hudson D.F., Ponting C.P., et al. (2010) The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell, 142, 810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campos E.I., Smits A.H., Kang Y.H., Landry S., Escobar T.M., Nayak S., Ueberheide B.M., Durocher D., Vermeulen M., Hurwitz J., et al. (2015) Analysis of the Histone H3.1 Interactome: A Suitable Chaperone for the Right Event. Mol. Cell, 60, 697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiio Y., Eisenman R.N., Yi E.C., Donohoe S., Goodlett D.R., Aebersold R. (2003) Quantitative proteomic analysis of chromatin-associated factors. J. Am. Soc. Mass. Spectrom., 14, 696–703. [DOI] [PubMed] [Google Scholar]
- 24.Torrente M.P., Zee B.M., Young N.L., Baliban R.C., LeRoy G., Floudas C.A., Hake S.B., Garcia B.A. (2011) Proteomic interrogation of human chromatin. PloS One, 6, e24747.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alajem A., Biran A., Harikumar A., Sailaja B.S., Aaronson Y., Livyatan I., Nissim-Rafinia M., Sommer A.G., Mostoslavsky G., Gerbasi V.R., et al. (2015) Differential association of chromatin proteins identifies BAF60a/SMARCD1 as a regulator of embryonic stem cell differentiation. Cell Rep., 10, 2019–2031. [DOI] [PubMed] [Google Scholar]
- 26.Kustatscher G., Hegarat N., Wills K.L., Furlan C., Bukowski-Wills J.C., Hochegger H., Rappsilber J. (2014) Proteomics of a fuzzy organelle: interphase chromatin. EMBO J., 33, 648–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kustatscher G., Wills K.L., Furlan C., Rappsilber J. (2014) Chromatin enrichment for proteomics. Nat. Protoc., 9, 2090–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strahl B.D., Allis C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 29.Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D. (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell, 121, 859–872. [DOI] [PubMed] [Google Scholar]
- 30.Wysocka J., Swigut T., Xiao H., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., et al. (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature, 442, 86–90. [DOI] [PubMed] [Google Scholar]
- 31.Vermeulen M., Mulder K.W., Denissov S., Pijnappel W.W., van Schaik F.M., Varier R.A., Baltissen M.P., Stunnenberg H.G., Mann M., Timmers H.T. (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell, 131, 58–69. [DOI] [PubMed] [Google Scholar]
- 32.Vermeulen M., Eberl H.C., Matarese F., Marks H., Denissov S., Butter F., Lee K.K., Olsen J.V., Hyman A.A., Stunnenberg H.G., et al. (2010) Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell, 142, 967–980. [DOI] [PubMed] [Google Scholar]
- 33.Migliori V., Muller J., Phalke S., Low D., Bezzi M., Mok W.C., Sahu S.K., Gunaratne J., Capasso P., Bassi C., et al. (2012) Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat. Struct. Mol. Biol., 19, 136–144. [DOI] [PubMed] [Google Scholar]
- 34.Eberl H.C., Spruijt C.G., Kelstrup C.D., Vermeulen M., Mann M. (2013) A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell, 49, 368–378. [DOI] [PubMed] [Google Scholar]
- 35.Bluhm A., Casas-Vila N., Scheibe M., Butter F. (2015) Reader interactome of epigenetic histone marks in birds. Proteomics, 16:427–436. [DOI] [PubMed] [Google Scholar]
- 36.Nikolov M., Stutzer A., Mosch K., Krasauskas A., Soeroes S., Stark H., Urlaub H., Fischle W. (2011) Chromatin affinity purification and quantitative mass spectrometry defining the interactome of histone modification patterns. Mol. Cell. Proteomics, 10, M110 005371.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shema-Yaacoby E., Nikolov M., Haj-Yahya M., Siman P., Allemand E., Yamaguchi Y., Muchardt C., Urlaub H., Brik A., Oren M., et al. (2013) Systematic identification of proteins binding to chromatin-embedded ubiquitylated H2B reveals recruitment of SWI/SNF to regulate transcription. Cell Rep., 4, 601–608. [DOI] [PubMed] [Google Scholar]
- 38.Bartke T., Vermeulen M., Xhemalce B., Robson S.C., Mann M., Kouzarides T. (2010) Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell, 143, 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bannister A.J., Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res., 21, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park P.J. (2009) ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet., 10, 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furey T.S. (2012) ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev., Genet., 13, 840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C.I., Alekseyenko A.A., LeRoy G., Elia A.E., Gorchakov A.A., Britton L.M., Elledge S.J., Kharchenko P.V., Garcia B.A., Kuroda M.I. (2013) Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat. Struct. Mol. Biol., 20, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Won K.J., Choi I., LeRoy G., Zee B.M., Sidoli S., Gonzales-Cope M., Garcia B.A. (2015) Proteogenomics analysis reveals specific genomic orientations of distal regulatory regions composed by non-canonical histone variants. Epigenetics Chromatin, 8, 13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soldi M., Bonaldi T. (2013) The proteomic investigation of chromatin functional domains reveals novel synergisms among distinct heterochromatin components. Mol. Cell. Proteomics, 12, 764–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soldi M., Bonaldi T. (2014) The ChroP approach combines ChIP and mass spectrometry to dissect locus-specific proteomic landscapes of chromatin. J. Vis. Exp., doi: 10.3791/51220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambert J.P., Mitchell L., Rudner A., Baetz K., Figeys D. (2009) A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol. Cell. Proteomics, 8, 870–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lambert J.P., Fillingham J., Siahbazi M., Greenblatt J., Baetz K., Figeys D. (2010) Defining the budding yeast chromatin-associated interactome. Mol. Syst. Biol., 6, 448.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohammed H., D'Santos C., Serandour A.A., Ali H.R., Brown G.D., Atkins A., Rueda O.M., Holmes K.A., Theodorou V., Robinson J.L., et al. (2013) Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep., 3, 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohammed H., Russell I.A., Stark R., Rueda O.M., Hickey T.E., Tarulli G.A., Serandour A.A., Birrell S.N., Bruna A., Saadi A., et al. (2015) Progesterone receptor modulates ERalpha action in breast cancer. Nature, 523, 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohammed H., Taylor C., Brown G.D., Papachristou E.K., Carroll J.S., D'Santos C.S. (2016) Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat. Protoc., 11, 316–326. [DOI] [PubMed] [Google Scholar]
- 51.Ji X., Dadon D.B., Abraham B.J., Lee T.I., Jaenisch R., Bradner J.E., Young R.A. (2015) Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proc. Natl. Acad. Sci. U S A, 112, 3841–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engelen E., Brandsma J.H., Moen M.J., Signorile L., Dekkers D.H., Demmers J., Kockx C.E., Ozgur Z., van I.W.F., van den Berg D.L., et al. (2015) Proteins that bind regulatory regions identified by histone modification chromatin immunoprecipitations and mass spectrometry. Nat. Commun., 6, 7155.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alekseyenko A.A., McElroy K.A., Kang H., Zee B.M., Kharchenko P.V., Kuroda M.I. (2015) BioTAP-XL: Cross-linking/Tandem Affinity Purification to Study DNA Targets, RNA, and Protein Components of Chromatin-Associated Complexes. Current Protocols in Molecular Biology/Edited by Frederick M. Ausubel … [et al.], 109, 21 30–32. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zee B.M., Alekseyenko A.A., McElroy K.A., Kuroda M.I. (2016) Streamlined discovery of cross-linked chromatin complexes and associated histone modifications by mass spectrometry. Proc. Natl. Acad. Sci. U S A, 113, 1784–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alekseyenko A.A., Gorchakov A.A., Kharchenko P.V., Kuroda M.I. (2014) Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc. Natl. Acad. Sci. U S A, 111, 2488–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alekseyenko A.A., Gorchakov A.A., Zee B.M., Fuchs S.M., Kharchenko P.V., Kuroda M.I. (2014) Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes & Dev., 28, 1445–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell L., Huard S., Cotrut M., Pourhanifeh-Lemeri R., Steunou A.L., Hamza A., Lambert J.P., Zhou H., Ning Z., Basu A., et al. (2013) mChIP-KAT-MS, a method to map protein interactions and acetylation sites for lysine acetyltransferases. Proc. Natl. Acad. Sci. U S A, 110, E1641–E1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rinn J.L., Chang H.Y. (2012) Genome regulation by long noncoding RNAs. Ann. Rev. Biochem., 81, 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinodoz S., Guttman M. (2014) Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol., 24, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chu C., Qu K., Zhong F.L., Artandi S.E., Chang H.Y. (2011) Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell, 44, 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M., Magnuson T., Heard E., Chang H.Y. (2015) Systematic discovery of Xist RNA binding proteins. Cell, 161, 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McHugh C.A., Chen C.K., Chow A., Surka C.F., Tran C., McDonel P., Pandya-Jones A., Blanco M., Burghard C., Moradian A., et al. (2015) The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature, 521, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mittler G., Butter F., Mann M. (2009) A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res., 19, 284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kappei D., Butter F., Benda C., Scheibe M., Draskovic I., Stevense M., Novo C.L., Basquin C., Araki M., Araki K., et al. (2013) HOT1 is a mammalian direct telomere repeat-binding protein contributing to telomerase recruitment. EMBO J., 32, 1681–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Casas-Vila N., Scheibe M., Freiwald A., Kappei D., Butter F. (2015) Identification of TTAGGG-binding proteins in Neurospora crassa, a fungus with vertebrate-like telomere repeats. BMC Genomics, 16, 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D. (2004) Ultraconserved elements in the human genome. Science, 304, 1321–1325. [DOI] [PubMed] [Google Scholar]
- 67.Viturawong T., Meissner F., Butter F., Mann M. (2013) A DNA-centric protein interaction map of ultraconserved elements reveals contribution of transcription factor binding hubs to conservation. Cell Rep., 5, 531–545. [DOI] [PubMed] [Google Scholar]
- 68.Markljung E., Jiang L., Jaffe J.D., Mikkelsen T.S., Wallerman O., Larhammar M., Zhang X., Wang L., Saenz-Vash V., Gnirke A., et al. (2009) ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol., 7, e1000256.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Butter F., Kappei D., Buchholz F., Vermeulen M., Mann M. (2010) A domesticated transposon mediates the effects of a single-nucleotide polymorphism responsible for enhanced muscle growth. EMBO Rep., 11, 305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Butter F., Davison L., Viturawong T., Scheibe M., Vermeulen M., Todd J.A., Mann M. (2012) Proteome-wide analysis of disease-associated SNPs that show allele-specific transcription factor binding. PLoS Genet., 8, e1002982.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Makowski M.M., Willems E., Fang J., Choi J., Zhang T., Jansen P.W., Brown K.M., Vermeulen M. (2015) An interaction proteomics survey of transcription factor binding at recurrent TERT promoter mutations. Proteomics, 16:417–426. [DOI] [PubMed] [Google Scholar]
- 72.Horn S., Figl A., Rachakonda P.S., Fischer C., Sucker A., Gast A., Kadel S., Moll I., Nagore E., Hemminki K., et al. (2013) TERT promoter mutations in familial and sporadic melanoma. Science, 339, 959–961. [DOI] [PubMed] [Google Scholar]
- 73.Huang F.W., Hodis E., Xu M.J., Kryukov G.V., Chin L., Garraway L.A. (2013) Highly recurrent TERT promoter mutations in human melanoma. Science, 339, 957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bell R.J., Rube H.T., Kreig A., Mancini A., Fouse S.D., Nagarajan R.P., Choi S., Hong C., He D., Pekmezci M., et al. (2015) Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science, 348, 1036–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hubner N.C., Nguyen L.N., Hornig N.C., Stunnenberg H.G. (2015) A quantitative proteomics tool to identify DNA-protein interactions in primary cells or blood. J. Proteome Res., 14, 1315–1329. [DOI] [PubMed] [Google Scholar]
- 76.Bartels S.J., Spruijt C.G., Brinkman A.B., Jansen P.W., Vermeulen M., Stunnenberg H.G. (2011) A SILAC-based screen for Methyl-CpG binding proteins identifies RBP-J as a DNA methylation and sequence-specific binding protein. PloS One, 6, e25884.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W., Bauer C., Munzel M., Wagner M., Muller M., Khan F., et al. (2013) Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell, 152, 1146–1159. [DOI] [PubMed] [Google Scholar]
- 78.Vasanthakumar A., Godley L.A. (2015) 5-hydroxymethylcytosine in cancer: significance in diagnosis and therapy. Cancer Genet., 208, 167–177. [DOI] [PubMed] [Google Scholar]
- 79.Dejardin J., Kingston R.E. (2009) Purification of proteins associated with specific genomic Loci. Cell, 136, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Antao J.M., Mason J.M., Dejardin J., Kingston R.E. (2012) Protein landscape at Drosophila melanogaster telomere-associated sequence repeats. Mol. Cell. Biol., 32, 2170–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Byrum S.D., Raman A., Taverna S.D., Tackett A.J. (2012) ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Rep., 2, 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fujita T., Fujii H. (2011) Direct identification of insulator components by insertional chromatin immunoprecipitation. PloS One, 6, e26109.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pourfarzad F., Aghajanirefah A., de Boer E., Ten Have S., Bryn van Dijk T., Kheradmandkia S., Stadhouders R., Thongjuea S., Soler E., Gillemans N., et al. (2013) Locus-specific proteomics by TChP: targeted chromatin purification. Cell Rep., 4, 589–600. [DOI] [PubMed] [Google Scholar]
- 84.Bogdanove A.J., Voytas D.F. (2011) TAL effectors: customizable proteins for DNA targeting. Science, 333, 1843–1846. [DOI] [PubMed] [Google Scholar]
- 85.Byrum S.D., Taverna S.D., Tackett A.J. (2013) Purification of a specific native genomic locus for proteomic analysis. Nucleic Acids Res., 41, e195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fujita T., Asano Y., Ohtsuka J., Takada Y., Saito K., Ohki R., Fujii H. (2013) Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Sci. Rep., 3, 3171.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sander J.D., Joung J.K. (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol., 32, 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C., et al. (2014) Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell, 159, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A., et al. (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Waldrip Z.J., Byrum S.D., Storey A.J., Gao J., Byrd A.K., Mackintosh S.G., Wahls W.P., Taverna S.D., Raney K.D., Tackett A.J. (2014) A CRISPR-based approach for proteomic analysis of a single genomic locus. Epigenetics, 9, 1207–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fujita T., Fujii H. (2013) Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem. Biophys. Res. Commun., 439, 132–136. [DOI] [PubMed] [Google Scholar]
- 92.Wu X., Scott D.A., Kriz A.J., Chiu A.C., Hsu P.D., Dadon D.B., Cheng A.W., Trevino A.E., Konermann S., Chen S., et al. (2014) Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol., 32, 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Horlbeck M.A., Witkowsky L.B., Guglielmi B., Replogle J.M., Gilbert L.A., Villalta J.E., Torigoe S.E., Tjian R., Weissman J.S. (2016) Nucleosomes impede Cas9 access to DNA in vivo and in vitro. eLife, 5, pii: e12677. [DOI] [PMC free article] [PubMed] [Google Scholar]