In a cohort of Ebola virus–infected patients from Monrovia, Liberia, we found that patients were 20% more likely to survive when Plasmodium species parasitemia was detected, even after controlling for Ebola viral load and age.
Keywords: coinfection, ebolavirus, plasmodium, survival
Abstract
Background. The ongoing Ebola outbreak in West Africa has resulted in 28 646 suspected, probable, and confirmed Ebola virus infections. Nevertheless, malaria remains a large public health burden in the region affected by the outbreak. A joint Centers for Disease Control and Prevention/National Institutes of Health diagnostic laboratory was established in Monrovia, Liberia, in August 2014, to provide laboratory diagnostics for Ebola virus.
Methods. All blood samples from suspected Ebola virus–infected patients admitted to the Médecins Sans Frontières ELWA3 Ebola treatment unit in Monrovia were tested by quantitative real-time polymerase chain reaction for the presence of Ebola virus and Plasmodium species RNA. Clinical outcome in laboratory-confirmed Ebola virus–infected patients was analyzed as a function of age, sex, Ebola viremia, and Plasmodium species parasitemia.
Results. The case fatality rate of 1182 patients with laboratory-confirmed Ebola virus infections was 52%. The probability of surviving decreased with increasing age and decreased with increasing Ebola viral load. Ebola virus–infected patients were 20% more likely to survive when Plasmodium species parasitemia was detected, even after controlling for Ebola viral load and age; those with the highest levels of parasitemia had a survival rate of 83%. This effect was independent of treatment with antimalarials, as this was provided to all patients. Moreover, treatment with antimalarials did not affect survival in the Ebola virus mouse model.
Conclusions. Plasmodium species parasitemia is associated with an increase in the probability of surviving Ebola virus infection. More research is needed to understand the molecular mechanism underlying this remarkable phenomenon and translate it into treatment options for Ebola virus infection.
The Ebola virus epidemic in West Africa has so far resulted in 28 646 suspected, probable, and confirmed cases, including 11 323 deaths between 26 December 2013 and 30 March 2016 [1]. Liberia, Sierra Leone, and Guinea have all been declared Ebola free, but the virus has reemerged on several occasions, with new cases most likely originating from viral persistence in survivors [1]. Malaria endemicity remains the largest public health burden in West Africa, with year-round transmission of Plasmodium species (spp) parasites to humans [2]. In Liberia, malaria is a leading cause of morbidity and mortality, affecting >1.2 million people annually [2]. Additionally, the breakdown of public health infrastructure in affected countries during the Ebola outbreak has likely resulted in an increase in malaria morbidity and mortality [3, 4]. The high rate of malaria prevalence has therefore likely resulted in coinfections with Plasmodium spp parasites and Ebola virus in a subset of patients, with unknown consequences.
In response to the expanding Ebola virus outbreak, a joint Centers for Disease Control and Prevention (CDC) and National Institutes of Health (NIH) diagnostic laboratory was established at ELWA (Eternal Love Winning Africa) in Monrovia, Liberia, in late August 2014. The laboratory provided diagnostic support to several Ebola treatment units (ETUs) and hospitals that were managing potential Ebola virus–infected patients. Here, we utilized demographic and laboratory data generated from patients admitted to the Médecins Sans Frontières (MSF) ELWA3 ETU in Monrovia with confirmed Ebola virus infection to better understand the effect of patient-level factors, including coinfection with Plasmodium spp parasites, on survival. Additionally, the effect of antimalarial treatments on Ebola survival was tested in a mouse model.
METHODS
Molecular Diagnostic Testing
Whole-blood samples were collected from patients presenting at the ETU with symptoms consistent with Ebola virus infection and transferred to the diagnostic laboratory. RNA was extracted from whole blood, and quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed to detect Ebola viremia as described previously [5]. Plasmodium spp parasitemia was detected by qRT-PCR as described previously [6, 7].
Data Collection
The human samples and accompanying metadata were exclusively collected from MSF's ELWA3 ETU in Monrovia solely as diagnostic samples for public health surveillance and not human subjects research, so institutional review board review and approval were not required. Patient data forms accompanied patient whole-blood samples submitted to the diagnostic laboratory. Patient name, age, sex, and diagnostic test results were recorded in a database. Age was further grouped into the following categories: <5 years, 5 to <12 years, 12 to <18 years, 18 to <40 years, 40 to <65 years, and ≥65 years. The clinical outcome of laboratory-confirmed Ebola virus–infected patients was determined by cross-referencing a list of deceased patients obtained from the ETU with the laboratory database. The database was transferred to an honest broker for removal of all personally identifiable information at the request of the NIH Office of Human Subjects Research. The honest broker was not involved in data collection or data analysis. For data analysis, the authors only had access to the database that was redacted by the honest broker.
Statistical Analysis
A retrospective analysis of the data collected at the CDC/NIH lab was performed. Descriptive statistics and Student t tests or χ2 tests were used to compare differences between patients with confirmed Ebola virus infection who survived and those who died. Demographic and clinical variables were modeled using univariable generalized log-binomial regression analysis to estimate risk ratios (RRs) and identify factors significantly (P < .05) associated with surviving Ebola virus infection in patients ≥5 years old (patients <5 years old were excluded from stratified analyses due to limited sample sizes preventing robust survival estimates for this group). Plasmodium parasitemia cycle threshold (Ct) values, Ebola virus Ct values, and age were categorized by the following levels for analysis: Plasmodium: Ct ≤ 20, Ct > 20 to <30, or negative (Ct ≥ 30); Ebola virus load: Ct < 25; Ct 25 to <30; or Ct 30 to <37; and age: 5 to <40 years or ≥40 years. Significant variables in univariable models were considered for inclusion in a multivariable model with survival as the outcome built using backward stepwise selection procedures; the final model was selected to minimize the Akaike information criterion. RRs and 95% confidence intervals (CI) were reported for significant variables; adjusted RRs (aRR) were reported for variables included in the multivariable model. Patients with missing data were compared to those with complete records for differences, and were excluded from respective analyses. All analyses were conducted using Statistical Analysis Software (SAS) version 9.3 (SAS Institute, Cary, North Carolina).
Animal Experiments
All animal experiments were approved by the Institutional Animal Care and Use Committee of Rocky Mountain Laboratories, NIH, and carried out by certified staff in an Association for Assessment and Accreditation of Laboratory Animal Care International–accredited facility, according to the institution's guidelines for animal use, and followed the guidelines and basic principles in the US Public Health Service Policy on Humane Care and Use of Laboratory Animals (available at http://grants.nih.gov/grants/olaw/references/PHSPolicyLabAnimals.pdf), and the Guide for the Care and Use of Laboratory Animals (available at https://grants.nih.gov/grants/olaw/Guide-for-the-Care-and-use-of-laboratory-animals.pdf).
Six-week-old BALB/c mice (Harlan, Indianapolis, Indiana) were inoculated intraperitoneally with 100 50% lethal dose of mouse-adapted Ebola virus [8]. Treatments (n = 7 animals per treatment group) were initiated 1 hour postinoculation and consisted of oral gavage with lumefantrine (350 mg/kg), artemether (100 mg/kg), lumefantrine (350 mg/kg) and artemether (100 mg/kg), artesunate (100 mg/kg), amodiaquine (120 mg/kg), or artesunate (100 mg/kg) and amodiaquine (120 mg/kg) resuspended in 100 µL of peanut oil [9, 10], or treated with vehicle alone and continued once daily for 14 days or until animals reached a predetermined humane endpoint. Treatment doses were determined through a combination of allometric scaling based on clinical dosing in humans and toxicity data in mice, when available. An untreated control group was used for comparison. Animals were euthanized when an approved, predetermined humane endpoint was reached.
RESULTS
Ebola Virus Infection
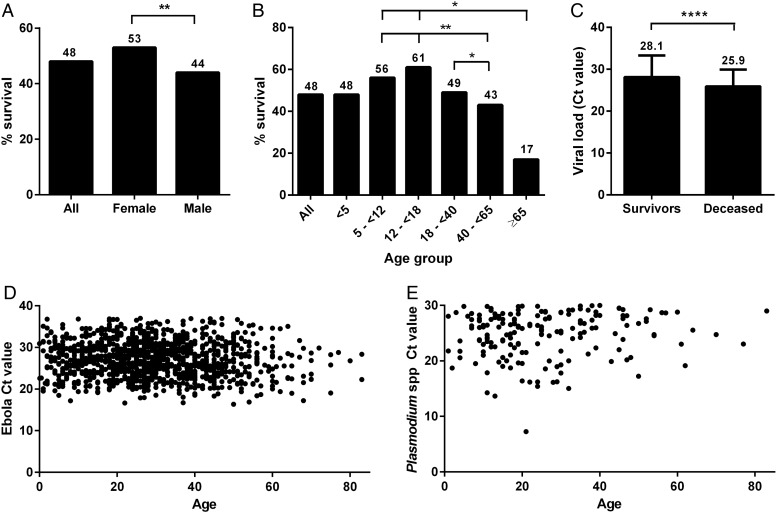
From late August 2014 until mid-February 2015, 1868 initial patient samples, collected from patients suspected of being infected with Ebola virus, were submitted to the joint CDC/NIH laboratory at ELWA by the ELWA3 ETU in Monrovia, Liberia. The laboratory confirmed Ebola virus infection in 1182 (63%) of those initial samples using qRT-PCR. Of those, 570 (48%) patients survived the Ebola virus infection. Females had greater survival than males (53% vs 44%; P = .007) (Table 1; Figure 1A), and in univariable analysis were 20% more likely to survive (RR, 1.2 [95% CI, 1.0–1.4]; Table 2), although this was no longer significant when adjusted for other variables in multivariable analysis. There were also significant differences in survival by age group (P = .007). Patients aged 5 to <40 years had higher survival (52%) than those aged ≥40 years (41%) or <5 years (48%) of age (Table 1). Patients aged 12 to <18 years experienced the highest survival rate (61%), whereas those ≥65 years had the lowest survival (17%) (Figure 1B). In univariable analysis, those aged ≥40 years were 20% less likely to survive than those aged 5 to <40 years (RR, 0.8 [95% CI, .7–.9]; Table 2).
Table 1.
Descriptive Characteristics of a Cohort of Ebola-Infected Patients by Survival Status
| Characteristic | Total Ebola-Infected Population | Survived | Deceased | P Value |
|---|---|---|---|---|
| Total population | 1182 | 570 (48) | 612 (52) | … |
| Age, y, mean ± SD (range) | 29.8 ± 15.6 (0.01–83) | 27.8 ± 14.5 (0.01–77) | 31.6 ± 16.4 (0.1–83) | .003a |
| Age group, No. (%) | .007b | |||
| <5 y | 44 (4) | 21 (48) | 23 (52) | … |
| 5 to <40 y | 773 (70) | 400 (52) | 373 (48) | … |
| ≥40 y | 293 (26) | 120 (41) | 173 (59) | … |
| Sex, female, No. (%) | 570 (52) | 300 (53) | 270 (47) | .007b |
| Ebola virus Ct value, mean ± SD | 27.0 ± 4.7 | 28.4 ± 4.3 | 25.9 ± 3.8 | <.0001a |
| Ct value <25 | 388 (33) | 136 (35) | 252 (65) | <.0001b |
| Ct value 25 to <30 | 480 (41) | 218 (45) | 262 (55) | … |
| Ct value 30 to <37 | 305 (26) | 209 (69) | 96 (31) | … |
| Plasmodium Ct value, mean ± SDc | 24.8 ± 4.0 | 24.1 ± 4.5 | 25.7 ± 3.0 | .008a |
| Plasmodium positive, No. (%) | 185 (19) | 107 (58) | 78 (42) | .007b |
| Ct value ≤20 | 29 (3) | 24 (83) | 5 (17) | … |
| Ct value >20 to <30 | 156 (16) | 83 (53) | 73 (47) | … |
Percentages were calculated out of the total number of cases with data available for age (n = 1110), sex (n = 1103), and malaria testing (n = 956); patients with missing data did not significantly differ from others with respect to survival outcomes.
Abbreviations: Ct, cycle threshold; SD, standard deviation.
a Two-sided t test.
b χ2 goodness-of-fit test.
c Only Ct values that were positive for Plasmodium (ie, <30) were included in calculations.
Figure 1.
Survival rate and patient demographics among a cohort of patients with laboratory-confirmed Ebola virus infection. Survival rates in a cohort of 1182 patients were calculated by sex (A) and age group (B). The average Ebola virus cycle threshold (Ct) value (a proxy for viral load) on admission was calculated for patients who survived vs patients who died from the infection (C). Scatterplots show the distribution of Ebola virus Ct values (D) and Plasmodium spp Ct values (E) by age in Ebola-infected patients. Of note, a high Ct value corresponds to a low viral load and vice versa. Numbers above bars indicate the percentage of patients who survived. *P < .05; **P < .01; ****P < .0001.
Table 2.
Association of Patient Demographic and Clinical Factors With Survival Among a Cohort of Ebola-Infected Patients Using Univariable and Multivariable Log-Binomial Regression Analysis With Survival as the Outcome
| Variable | Univariable Model |
Multivariable Modela |
||||
|---|---|---|---|---|---|---|
| RR | 95% CI | P Value | aRR | 95% CI | P Value | |
| Ebola Ct value | 1.3 | 1.2–1.5 | <.0001 | 1.3 | 1.2–1.4 | <.0001 |
| Plasmodium positivea | 1.2 | 1.1–1.4 | .003 | … | … | … |
| Plasmodium levela | ||||||
| Ct ≤ 20 | 1.8 | 1.5–2.2 | <.0001 | 1.4 | 1.2–1.7 | <.0001 |
| Ct > 20 to <30 | 1.1 | .97–1.4 | .1 | 1.2 | .98–1.4 | .08 |
| Negative | Ref | … | … | … | … | … |
| Age group | ||||||
| 5 to <40 y | Ref | … | … | … | … | … |
| ≥40 y | 0.8 | .7–.9 | .003 | 0.8 | .6–.9 | .005 |
| Sex (female) | 1.2 | 1.0–1.4 | .02 | … | … | … |
Abbreviations: aRR, adjusted relative risk; CI, confidence interval; Ct, cycle threshold; Ref, referent against which other groups are compared; RR, relative risk.
a Correlated variables were assessed independently from one another; only the categorical Plasmodium spp variable was included in the multivariable model to assess the effect of survival by parasitemia level.
Although the Ct value of an Ebola virus qRT-PCR is not a direct measure of infectious virus, it can be used as a proxy for viral load, with high Ct values corresponding to a low viral load and low Ct values to a high viral load; Ct values have been used as a proxy for viral load during the current as well as previous outbreaks [7, 11–17]. Ebola virus qRT-PCR Ct value upon admission was a significant predictor of survival in univariable analysis in our cohort (P < .0001; Table 2). Patients who survived had a higher average Ebola virus qRT-PCR Ct value on admission (28.4 ± 4.3) compared with those who died (25.9 ± 3.8) (P < .0001; Table 1; Figure 1C). Ebola virus qRT-PCR Ct values were evenly distributed and did not differ significantly by age (P = .2; Figure 1D). Moreover, there was an increase in survival of 30% (RR, 1.3 [95% CI, 1.3–1.5]; Table 2) with a decrease in viral load level (ie, increasing Ct value); patients with the lowest viral loads (Ct 30–37) had a survival rate of 69%, whereas those with the highest viral loads (Ct < 25) had a survival rate of only 35% (Table 1).
Effect of Plasmodium Parasitemia on Survival
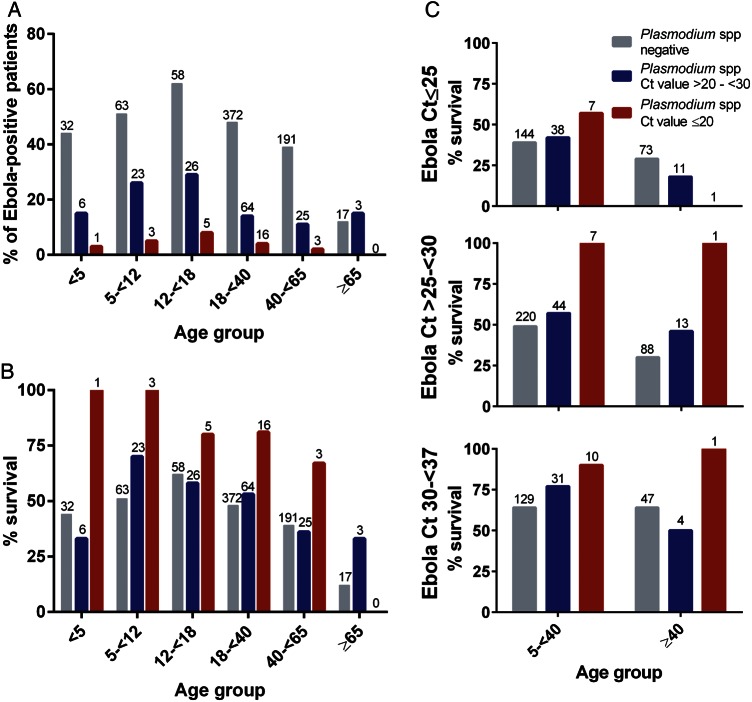
Patient samples submitted to the CDC/NIH ELWA laboratory were simultaneously tested for the presence of Plasmodium spp parasitemia by qRT-PCR. In total, 956 of 1182 laboratory-confirmed Ebola virus patients were examined for Plasmodium spp parasitemia, of whom 185 (19%) were positive. Patients with Plasmodium spp parasitemia were more likely to be younger (P = .0001) than those testing negative (Figures 1E and 2A). Overall, patients with Plasmodium spp parasitemia had a survival rate of 58% vs only 46% for those without coinfection (P = .007; Table 1). However, those with the highest level of parasitemia, as determined by a Ct value of ≤20, had 83% survival compared with 53% for those with Ct values >20 to <30 (Table 1; Figure 2B). Among this high Plasmodium spp parasitemia group, survival ranged from a low of 67% among those aged 40 to <65 years to a high of 100% among those <12 years old (Figure 2B).
Figure 2.
Increased levels of Plasmodium spp parasitemia (cycle threshold [Ct] ≤ 20) are associated with greater survival across age groups among a cohort of patients with laboratory-confirmed Ebola virus infection. A, Percentage of Ebola virus–infected patients coinfected with Plasmodium spp by age group and Plasmodium Ct value as a proxy for parasite load. B, Patients were stratified by age and Plasmodium Ct value (as a proxy for parasite load). No patients aged ≥65 years with Plasmodium Ct values ≤20 were present in this cohort. C, Patients were stratified by age, Plasmodium Ct value (as a proxy for parasite load), and Ebola virus Ct value (as a proxy for viral load), and survival rate was calculated among these groups. Patients aged <5 years were excluded from multistratified analyses due to limited sample sizes across categories preventing accurate survival estimates. Gray bars: Plasmodium spp negative; blue bars: Plasmodium spp Ct value >20–<30; red bars: Plasmodium spp Ct value ≤20. Numbers above bars indicate number of patients in this group.
After controlling for both age and Ebola Ct value, presence of Plasmodium spp parasitemia (Ct < 30) was associated with a 20% increase in survival (aRR, 1.2 [95% CI, 1.1–1.4]; P = .004) in the multivariable model relative to those who were negative for Plasmodium spp coinfection, whereas a Plasmodium spp Ct value of ≤20 was associated with a 40% increase in survival (aRR, 1.4 [95% CI, 1.2–1.7]; P < .0001; Table 2 and Figure 2C). Patients with the highest Ebola viral loads (represented by low Ct values) still experienced the lowest survival rates, with those coinfected with greater levels of Plasmodium spp (Ct value ≤20) having slightly (P > .05) higher survival (43%) than those without Plasmodium spp detected (39%). However, among patients with moderate to low Ebola Ct values (Ct ≥ 25), those with Plasmodium spp Ct values of ≤20 had nearly double the survival rate of those without Plasmodium spp detected across age groups (94% vs 54% for ages 5 to <40 years and 100% vs 41% for ages ≥40 years, respectively; P < .0001; Figure 2C). A formal interaction term between Plasmodium spp and Ebola virus Ct values was assessed in the multivariable model and was not significant (data not shown).
To exclude the possibility that Plasmodium spp coinfection resulted in patients visiting the ETU earlier and thus receiving supportive care from an earlier disease stage onward than patients without Plasmodium spp coinfection, we attempted to analyze the time between symptom onset and admission to the ETU for both groups of patients. However, this analysis proved to be problematic as admission dates were missing for a majority of patients. Instead, the time between self-reported time of symptom onset and molecular diagnosis was analyzed, assuming that any discrepancies between time of admission and time of molecular diagnosis would be similar in both patient groups, but realizing the limited value of this analysis due to inadequate reporting of symptom onset and missing or incorrect information reported in the database. The number of days between onset and molecular diagnosis did not differ significantly by survival and Plasmodium spp coinfection status (P = .2), nor was it a significant predictor in a model with survival as the outcome (P = .3) (data not shown).
Effect of Antimalarial Treatment on Ebola Survival
Although all patients at the ELWA3 ETU were treated with the antimalarial drugs artemether and lumefantrine (AL), as per World Health Organization and MSF protocol [18, 19], we wanted to evaluate if antimalarial treatments would directly affect Ebola virus replication and disease outcome. Therefore, 2 antimalarial treatment regimens, AL as well as artesunate and amodiaquine (ASAQ) were tested in the mouse model of Ebola virus infection; individual antimalarial drugs (artemether, lumefantrine, artesunate, and amodiaquine), as well as combination therapy (AL, ASAQ) were tested. Treatment was started 1 hour postinoculation with a lethal dose of mouse-adapted Ebola virus. All animals treated with antimalarial drugs, either individually or in combination, as well as vehicle-treated controls, died of the Ebola virus infection within 8 days postinoculation (Figure 3).
Figure 3.
The effect of antimalarial treatments on survival of Ebola virus infection in a lethal mouse model. Mice were inoculated with a 100 50% lethal dose of mouse-adapted Ebola virus, treatment with antimalarial compounds was started at 1 hour postinoculation, and survival was monitored. Animals were left untreated or treated with vehicle alone (left panel); treated with lumefantrine (350 mg/kg; L), artemether (100 mg/kg; A), or lumefantrine (350 mg/kg) and artemether (100 mg/kg; AL) (middle panel); or treated with artesunate (100 mg/kg; AS), amodiaquine (120 mg/kg; AQ), or artesunate (100 mg/kg) and amodiaquine (120 mg/kg; ASAQ) (right panel). Treatments were continued daily until the end of the experiment.
DISCUSSION
The Ebola virus outbreak in West Africa has had a devastating effect on the population, directly as well as indirectly through the breakdown of the public health infrastructure in the 3 main affected countries, Sierra Leone, Liberia, and Guinea. The unprecedented size of this outbreak has provided a unique opportunity to increase our understanding of the pathogenesis of Ebola virus infection in humans and to guide the development of countermeasures against Ebola virus infections. Here, we analyzed the laboratory diagnostic results and disease outcome data available from Ebola virus–infected patients admitted to the ELWA3 ETU in Monrovia, Liberia, and found an association between coinfection with Plasmodium spp parasites and surviving an Ebola virus infection. One explanation for this association could be that the antimalarial treatment provided to those patients with a positive Plasmodium spp parasitemia laboratory diagnosis had an antiviral effect on Ebola virus replication. However, all patients at the ELWA3 ETU received a course of AL combination treatment upon admission to the ETU according to MSF protocols, regardless of the outcome of the Plasmodium spp parasitemia test. Thus, the observed increased survival is unlikely to be due to a beneficial effect from the AL treatment. A field report and recent study from an ETU in Foya, Liberia, suggested an increased survival in Ebola patients treated with the antimalarial drugs ASAQ [20, 21]. Therefore, ASAQ and AL were tested in a lethal mouse model of Ebola virus infection. Neither of the antimalarial treatments affected survival in the mouse model, nor did survival increase when any of the 4 drugs were given as individual treatments. These results do not support an antiviral effect of antimalarial drugs against Ebola virus.
Increased survival among those with both Ebola virus infection and Plasmodium spp parasitemia was independent of viral load and age, suggesting that the increased survival was the result of an indirect effect on the host rather than a direct effect on Ebola virus replication. Due to the outbreak setting in which the data we analyzed here were collected, we were not able to analyze other factors that may have affected survival, such as time between symptom onset and admission to the ETU or the potential presence of comorbidities in a subset of patients. We were able to circumvent the first problem by using date of laboratory diagnosis as a proxy for admission date and we have no reason to think comorbidities would differ significantly between patients with and without Plasmodium spp coinfection. However, improvement of data collection in future outbreaks would enable a more thorough analysis of factors that may influence survival.
A correlation between coinfection with another virus and increased probability of survival of Ebola virus infection has been suggested previously. In a small cohort of Ebola virus–infected patients in Sierra Leone, a positive effect of GB virus C coinfection on survival was detected, although this effect was confounded by age [22]. GB virus C, which is not known to cause human disease, is thought to modulate the immune response to human immunodeficiency virus (HIV) and thereby attenuate HIV pathogenesis. The authors hypothesize that a similar immunomodulatory mechanism exists during Ebola virus infection.
Similarly, there are several examples of Plasmodium spp infections resulting in a suppression of the immune response to a secondary infection. Children with malaria and a respiratory infection were less likely to have pneumonia than children with a respiratory infection alone [23]. This epidemiological link was investigated experimentally by simultaneously infecting C57BL/6J mice with Plasmodium chabaudi chabaudi AS and pneumovirus of mice (PVM). In coinfected mice, weight loss due to PVM, inflammatory cytokine production, and recruitment of inflammatory cells to the lungs were reduced compared to mice infected with PVM alone. This effect may be due to an early systemic upregulation of IFN-β observed in coinfected mice that was not observed in mice infected with Plasmodium or PVM alone [24].
In children with Salmonella enterica serotype Typhimurium infection, a severe Plasmodium falciparum coinfection can turn a self-limiting infection into a life-threatening one. Based on coinfection studies in mice, it is thought that the P. falciparum infection causes a suppression of the inflammatory response through the induction of interleukin 10 with uncontrolled bacterial growth as a result [25]. Analogous to the 2 examples above, coinfection with Plasmodium spp in Ebola virus–infected patients may result in a dampening of the detrimental cytokine storm [26] and thus increase survival.
Alternatively, the induction of natural killer (NK) cells by Plasmodium infection may explain the increased survival in coinfected individuals. Recently, it was shown that cynomolgus macaques vaccinated with vesicular stomatitis virus-Ebola virus (VSV-EBOV), a replication-competent vaccine vector expressing Ebola virus glycoprotein, at 3 days prior to challenge were partially protected from lethal Ebola virus challenge. The most likely explanation for protection by VSV-EBOV in the absence of antibodies was the activation of innate immune responses and potentially NK cells induced by the VSV-EBOV infection [27]. Infection with Plasmodium spp also results in activation of NK cells (reviewed in [28]), thus providing a possible explanation for the positive effect of Plasmodium coinfection on Ebola virus survival.
The joint CDC/NIH diagnostic laboratory at ELWA was set up for diagnostic outbreak response rather than scientific research purposes. As such, the cytokine responses in whole-blood samples from patients in our cohort could not be studied due to the limited availability of equipment and reagents in the field laboratory at the time of sample collection and the lack of appropriate cold-chain conditions required for retrospective analysis.
The link between Plasmodium spp coinfection and survival of Ebola virus infection clearly warrants further investigation into the mechanism(s) underlying this phenomenon. If increased survival is indeed due to an immunomodulatory effect of Plasmodium spp coinfection, immunomodulatory drugs could be investigated as potential options for supportive therapy to alleviate Ebola virus infection once the mechanism is better understood.
Although we found a positive association between Plasmodium spp parasitemia and survival of Ebola virus infection in our cohort, this effect was not affected by treatment with antimalarials. Thus, our data currently do not warrant changing the guidelines for treatment of malaria in patients with suspected, probable, and confirmed Ebola virus infection [18, 19].
Notes
Acknowledgments. The authors thank Mulbah Jallah, Monrovia, Liberia, for his excellent support and assistance in operating the Laboratory at Eternal Love Winning Africa (ELWA), as well as Kay Menk, Dawn Clifton, and Les Shupert (all National Institute for Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH]) for logistical support. We further acknowledge the World Health Organization (WHO) Headquarters in Geneva, the WHO Regional Office for Africa (WHO/AFRO), the Ministry of Health and Social Welfare, Liberia, the Centers for Disease Control and Prevention (CDC), the NIH, and ELWA. Finally, we thank the people of Liberia for their hospitality and cooperation.
Author contributions. K. R., J. A., E. d. W., V. J. M., H. F.: conceived and designed the study, data collection and analysis, wrote the manuscript. D. F., C. O., A. M., M. O., B. J., R. J. F., J. B. P., D. S., V. O., C. O., T. H., A. G., C. M., N. v. D., G. Z., J. S., T. B., K. M., T. R., S. L. E., F. F., B. N. W., B. F.: data collection and analysis. A. G., J. E. S., G. K.: provided essential reagents. K. C. Z., S. T. N., S. B.: provided logistics support to enable outbreak response. T. G. N., F. K. B., M. M.: responsible for in-country Ebola outbreak response. R. G., J. S., M. d. S., A. S.: case management of ETU. All authors read and approved the manuscript. K. R. and E. d. W. had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Disclaimer. The funding source did not play a role in study design; collection, interpretation, and analysis of data; in the writing of this manuscript; or in the decision to submit this manuscript for publication.
Financial support. This work was partially funded by the Intramural Research Program of the NIAID/NIH.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. Ebola situation report. Available at: http://apps.who.int/iris/bitstream/10665/204714/1/ebolasitrep_30mar2016_eng.pdf?ua=1 Accessed 30 March 2016.
- 2.World Health Organization. World malaria report. Geneva: WHO, 2014:1–242.
- 3.Plucinski MM, Guilavogui T, Sidikiba S et al. . Effect of the Ebola-virus-disease epidemic on malaria case management in Guinea, 2014: a cross-sectional survey of health facilities. Lancet Infect Dis 2015; 15:1017–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker PG, White MT, Griffin JT, Reynolds A, Ferguson NM, Ghani AC. Malaria morbidity and mortality in Ebola-affected countries caused by decreased health-care capacity, and the potential effect of mitigation strategies: a modelling analysis. Lancet Infect Dis 2015; 15:825–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Wit E, Munster VJ, Rosenke K et al. . Ebola laboratory response at the Eternal Love Winning Africa campus, Monrovia, Liberia, 2014–2015. J Infect Dis 2016; doi:10.1093/infdis/jiw216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee MA, Tan CH, Aw LT et al. . Real-time fluorescence-based PCR for detection of malaria parasites. J Clin Microbiol 2002; 40:4343–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzpatrick G, Vogt F, Gbabai OB et al. . The contribution of Ebola viral load at admission and other patient characteristics to mortality in a Medecins Sans Frontieres (MSF) Ebola Case Management Centre (CMC), Kailahun, Sierra Leone, June–October, 2014. J Infect Dis 2015; 212:1752–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray M, Davis K, Geisbert T, Schmaljohn C, Huggins J. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis 1998; 178:651–61. [DOI] [PubMed] [Google Scholar]
- 9.Nontprasert A, Pukrittayakamee S, Dondorp AM, Clemens R, Looareesuwan S, White NJ. Neuropathologic toxicity of artemisinin derivatives in a mouse model. Am J Trop Med Hyg 2002; 67:423–9. [DOI] [PubMed] [Google Scholar]
- 10.Nontprasert A, Pukrittayakamee S, Nosten-Bertrand M, Vanijanonta S, White NJ. Studies of the neurotoxicity of oral artemisinin derivatives in mice. Am J Trop Med Hyg 2000; 62:409–12. [DOI] [PubMed] [Google Scholar]
- 11.Bah EI, Lamah MC, Fletcher T et al. . Clinical presentation of patients with Ebola virus disease in Conakry, Guinea. N Engl J Med 2015; 372:40–7. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez A, Lukwiya M, Bausch D et al. . Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: cellular responses, virus load, and nitric oxide levels. J Virol 2004; 78:10370–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schieffelin JS, Shaffer JG, Goba A et al. . Clinical illness and outcomes in patients with Ebola in Sierra Leone. N Engl J Med 2014; 371:2092–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Towner JS, Rollin PE, Bausch DG et al. . Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J Virol 2004; 78:4330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de La Vega MA, Caleo G, Audet J et al. . Ebola viral load at diagnosis associates with patient outcome and outbreak evolution. J Clin Invest 2015; 125:4421–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanini S, Portella G, Vairo F et al. . Blood kinetics of Ebola virus in survivors and nonsurvivors. J Clin Invest 2015; 125:4692–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Duan HJ, Chen HY et al. . Age and Ebola viral load correlate with mortality and survival time in 288 Ebola virus disease patients. Intern J Infect Dis 2015; 42:34–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention. Recommendations for managing and preventing cases of malaria in areas with Ebola. Available at: http://www.cdc.gov/vhf/ebola/outbreaks/malaria-cases.html. Accessed 14 July 2016.
- 19.Sterk E. Filovirus haemorrhagic fever guideline. Brussels: MSF, 2008.
- 20.Médecins Sans Frontières. Artesunate-amodiaquine is associated with reduced Ebola mortality, 2015. Available at: http://www.msf.org.uk/sites/uk/files/late_breaker_asaq_final.pdf Accessed 7 October 2015.
- 21.Gignoux E, Azman AS, de Smet M et al. . Effect of artesunate-amodiaquine on mortality related to Ebola virus disease. N Eng J Med 2016; 374:23–32. [DOI] [PubMed] [Google Scholar]
- 22.Lauck M, Bailey AL, Andersen KG, Goldberg TL, Sabeti PC, O'Connor DH. GB virus C coinfections in West African Ebola patients. J Virol 2015; 89:2425–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D'Acremont V, Kilowoko M, Kyungu E et al. . Beyond malaria—causes of fever in outpatient Tanzanian children. N Eng J Med 2014; 370:809–17. [DOI] [PubMed] [Google Scholar]
- 24.Edwards CL, Zhang V, Werder RB et al. . Coinfection with blood-stage Plasmodium promotes systemic type I interferon production during pneumovirus infection but impairs inflammation and viral control in the lung. Clin Vaccine Immunol 2015; 22:477–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lokken KL, Mooney JP, Butler BP et al. . Malaria parasite infection compromises control of concurrent systemic non-typhoidal Salmonella infection via IL-10-mediated alteration of myeloid cell function. PLoS Pathog 2014; 10:e1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wauquier N, Becquart P, Padilla C, Baize S, Leroy EM. Human fatal Zaire ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl Trop Dis 2010; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marzi A, Robertson SJ, Haddock E et al. . Ebola vaccine. VSV-EBOV rapidly protects macaques against infection with the 2014/15 Ebola virus outbreak strain. Science 2015; 349:739–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevenson MM, Riley EM. Innate immunity to malaria. Nat Rev Immunol 2004; 4:169–80. [DOI] [PubMed] [Google Scholar]