ABSTRACT
Master genes are known to induce the differentiation of a multipotent cell into a specific cell type. These molecules are often transcription factors that switch on the regulatory cascade that triggers cell specification. Gcm was first described as the master gene of the glial fate in Drosophila as it induces the differentiation of neuroblasts into glia in the developing nervous system. Later on, Gcm was also shown to regulate the differentiation of blood, tendon and peritracheal cells as well as that of neuronal subsets. Thus, the glial master gene is used in at least 4 additional systems to promote differentiation. To understand the numerous roles of Gcm, we recently reported a genome-wide screen of Gcm direct targets in the Drosophila embryo. This screen provided new insight into the role and mode of action of this powerful transcription factor, notably on the interactions between Gcm and major differentiation pathways such as the Hedgehog, Notch and JAK/STAT. Here, we discuss the mode of action of Gcm in the different systems, we present new tissues that require Gcm and we revise the concept of ‘master gene’.
KEYWORDS: DamID, Gcm, high-throughput screen, hedgehog, immune system, JAK/STAT, nervous system, Notch
The Gcm Cascade
Glial Cell missing/Glial cell deficient (Gcm/Glide or Gcm throughout the text) is an atypical zinc finger transcription factor essential for the differentiation of the lateral glial cells in the Drosophila embryo. gcm loss of function (LOF) embryos are devoid of glial cells, with neural precursors or neuroblasts producing essentially neurons (Fig. 1A, A′). Conversely, embryos expressing Gcm throughout the nervous system (gcm gain of function (GOF) embryos) present supernumerary glial cells with neuroblasts producing essentially glia.1-3 Due to its preponderant role in glia development, Gcm was first described and named as the master regulator of glia differentiation and indeed ectopic expression of Gcm is sufficient to induce the expression of glial markers even in the epidermis and in the mesoderm,4 much as the ectopic expression of the famous eyeless master gene is sufficient to induce the formation of ectopic eyes on wings, legs and antennae.5
Figure 1.
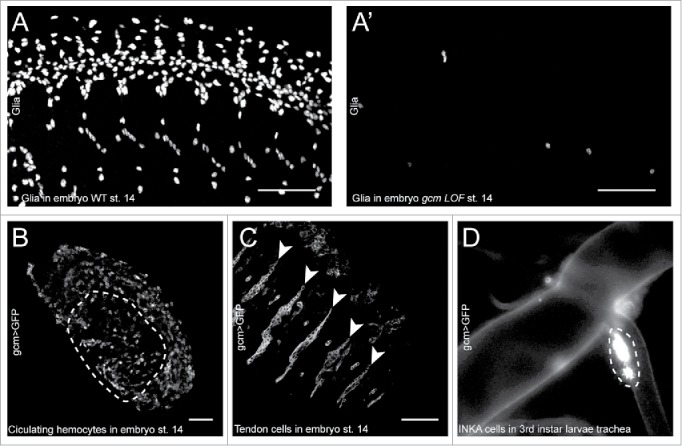
A-A′) Confocal projections of wild type (A) and gcm LOF (A′) embryos at stage 14 immunolabelled with the Repo glial marker, anterior to the left, latero-ventral views. Note the absence of glia in the gcm LOF embryo. B, C) Confocal sections of a gcm > GFP transgenic embryo at stage 14 labeled with the anti-GFP antibody, anterior to the left, dorso-lateral views. The section was acquired in the inner layers of the embryo to reveal the circulating hemocytes, outlined by a dashed line in (B) and in the outer layers to reveal the tendon cells indicated by arrowheads in (C). The scale bar in (A–C) represents 50 µm. D) Tracheal branch of a gcm > GFP 3rd instar larva analyzed by epifluorescence microscopy (200x magnification). The INKA cells are recognizable by their position and are outlined by the dashed line.
Subsequently, Gcm was shown to be expressed and required in the immune system, where it participates to the development of the embryonic professional macrophages called plasmatocytes (Fig. 1B).6-8 The role of gcm in plasmatocytes is not as essential as in the nervous system. gcm LOF animals present a deficit in plasmatocytes but not a total lack; moreover, the mutant plasmatocytes differentiate incompletely, expressing only some markers and lacking their phagocytic capacity.6,8,9 Interestingly, glia and blood represent the major phagocytic cells within and outside the nervous system respectively,10,11 a functional relation that is worth exploring in the future and that may explain the shared requirement for Gcm.
More recently, however, Gcm was also shown to be expressed in tendon cells, in peritracheal cells and in specific neuronal populations (Fig. 1C and D).12-17 Importantly, in these tissues, the gene is expressed in the differentiated cells, as opposed to the transient and early expression observed in glia and in hemocytes, already suggesting that the so-called master genes may have distinct roles depending on the cell type. In tendon cells, Gcm is necessary for their function/terminal differentiation. gcm LOF embryos display altered muscle attachment.18,19 In the trachea, gcm is expressed in the peritracheal secretory cells called INKA cells.17 The INKA cells secrete the ecdysis-triggering hormone and participate to the control of ecdysial behaviors.20 gcm LOF animals are devoid of INKA cells.17 Finally, gcm is expressed in specific embryonic and post-embryonic neuronal populations of the brain and indeed the only cell type expressing Gcm in the adult nervous system are the neuronal clusters in the brain.12-17 The neuronal expression of Gcm represents a conundrum, since the ectopic expression of Gcm throughout the neurogenic region leads to the differentiation of glia at the expense of neurons. One likely explanation is that these neurons may express Gcm only when they are already differentiated, as it is known that ectopic expression of Gcm in postmitotic neurons does not trigger their conversion into glia.21
Similar to Gcm, other “master genes” are also expressed in several cell types. Eyeless, which was first described for its role in retina development,5 is involved in the development of the mushroom body,22-27 in the development of the insulin producing neurons 25 and in the optic lobes, where it participates to the temporal cascade defining the medulla neuroblasts' fate 26,27 and regulates neuronal migration.22
…revisited by the DamID screen
To gain a better understanding of the Gcm regulatory network, we recently performed a genome-wide screen of Gcm direct targets in the whole organism using the DNA adenine methyltransferase (Dam) identification technique.28-30 This technique consists in building a fusion protein between Gcm and the Dam, expressing the fusion protein in the whole embryo and deducing the binding sites of Gcm from the adenine methylation profile of the genome. This approach allowed a comprehensive identification of the Gcm direct targets, a strong advantage over microarray assays that likely favor the identification of indirect targets. This method also allowed us to identify the potential direct targets without tissue or developmental stage biases. Typically, we identified and validated direct targets of Gcm that are found to be expressed in larvae but not in embryos. Thus, this approach allowed a comprehensive identification of the genes directly regulated by Gcm and we could validate the efficiency of the DamID by functional assays (41 genes were validated out of 47 tested).
Overall, the DamID screen revealed 1031 direct targets and gave novel insights on the biological pathways regulated by the Gcm transcription factor. The most prominent observations are that (i) Gcm targets major developmental cascades, (ii) Gcm accompanies cell specification, from the inhibition of the stem cell identity to the activation of genes necessary for the function of the differentiated cell and (iii) Gcm is present in tissues where it was never detected before.
Interaction between Gcm and major signaling pathways
One of the foremost observations of our DamID screen is the impact of Gcm on 3 major developmental pathways: JAK/STAT, Hedgehog and Notch. These pathways are activated mostly in non-cell autonomous fashion, which means that they allow a cell to sense and react to the surrounding environment. During development, these pathways are widely used to synchronize the development of specific cell types at the appropriate time and place.
The Notch pathway was extensively described during the differentiation of sensory organ precursors (SOP). Notch (N) is a transmembrane protein that is activated by adjacent cells expressing the transmembrane ligands Delta (Dl) or Serrate (Ser).31,32 In the wing disc, the SOPs are specified during the larval stage from groups of 3 to 7 cells called proneural clusters.33 One cell of the cluster expresses Dl and differentiates into the SOP. Dl activates the N pathway in the adjacent cells and leads to the inhibition of the SOP fate ensuring that only one SOP is defined per cluster.34-36 This mechanism is known as lateral inhibition. Of note, the N pathway is directly involved in gliogenesis by inducing Gcm expression in the central nervous system and repressing Gcm expression in specific lineages of the peripheral nervous system.37-39 According to the DamID screen, Gcm induces the expression of the receptor N, its ligands Ser and Dl as well as its down-stream targets the E(spl) complex.
For the JAK/STAT pathway, its non-cell autonomous activation is observed in the differentiation of plasmatocytes into lamellocytes after parasitization. The parasitoid wasps lay eggs in the Drosophila larvae and trigger an immune response.40,41 Upon parasitization, the circulating plasmatocytes secrete cytokines such as Unpaired 2 (Upd2) and Unpaired 3 (Upd3) and recruit more hemocytes.42 The cytokines activate the JAK/STAT pathway, thus inducing plasmatocytes proliferation and differentiation into lamellocytes,43-46 which then encapsulate the wasp egg.40,47 Gcm was previously described as an inhibitor of the JAK/STAT pathway,48 and the DamID screen indicates that Gcm targets the transcription factor Stat92E, the inhibitors Ptp61F and Socs36E and the cytokine Upd1.
As per the Hedgehog pathway, this non-cell autonomous signalisation is exemplified in the setting of segmentation in the Drosophila embryo. The pathway is activated by the secreted protein Hedgehog (Hh) (reviewed by Lum and Beachy 49). Hh is expressed in the posterior part of each segment of the embryo and diffuses toward the anterior part. This creates a gradient of Hh from posterior to anterior that activates the Hh target genes in a dosage dependent fashion and allows the differentiation of specific precursors according to their positions in the segment (reviewed by Sanson 50). Of note, the Hh pathway controls Gcm expression during tendon cell differentiation.19 The DamID screen revealed that Gcm induces the expression of the secreted molecule Hh, its receptor Ptc, the transmembrane protein Smo, the transcriptional regulator Ci and the down-stream targets Roadkill (Rdx) and Decapentaplegic (Dpp).
Importantly, for the 3 pathways Gcm induces the expression of the ligands (i.e. Dl/Ser, Hh, Upd1), that of the receptors and of core components (i.e., N, Ptc/Smo/Ci and Stat92E) as well as that of the down-stream targets (i.e. E(spl) complex, Rdx/Dpp), meaning that Gcm can act upstream and downstream of the pathways. In addition, the N and the Hh pathways are already known to modulate Gcm expression, which suggests the presence of feedback regulatory loops between Gcm and the different signaling pathways. Overall, these data show that Gcm interacts strongly with the 3 major signaling pathways and that it can modulate the pathways at multiple levels.
Gcm accompanies cell specification, from the inhibition of stem cell identity to the function of the differentiated cell
Gcm induces the expression of inhibitors of alternative fates and inhibitors of stem cells factors. For instance, Drosophila neuroblasts produce glia and neurons and Gcm induces the expression of the transcription factor Tramtrack (Ttk), which represses the neuronal fate,51 as well as the expression of genes repressing neuroblast proliferation (mira, lola, pros and brat).30 In the developing immune system, prohemocytes produce crystal cells and plasmatocytes and Gcm promotes the expression of the zinc finger transcription factor called U-shaped (Ush), which inhibits crystal cell formation.30,52
Gcm also induces the transcription of genes that control the final differentiation of the cells. The most described target of Gcm is Repo, a homeodomain transcription factor that remains expressed in glia until adulthood.53-56 After an initial phase of gliogenesis in which repo expression depends on Gcm, Repo maintains its own expression through autoregulation and becomes independent of Gcm.56,57 This fits with the recently emerged concept that transiently expressed master genes are relayed by their target genes that code for permanently expressed transcription factors. In line with this idea, repo mutant glia lose the expression of late glial markers (reviewed by Cattenoz and Giangrande 58).54,59 Comparable mechanisms are displayed during the specification of the muscle cells in the mesoderm: the myogenic determinant Twist (Twi) is expressed transiently and induces the expression of Myocyte enhancer factor 2 (Mef-2) that initially depends on Twi and subsequently becomes independent of it, to carry over the myogenic differentiation program.60-63
In addition to the classes of genes described above, the screen revealed that Gcm targets directly numerous genes that function in the fully differentiated cells. Typically, Gcm induces the expression of proteins working in the blood brain barrier, in axon ensheathment or in chemoattraction. These genes are necessary in glial subsets when these cells are thought to be terminally differentiated. In the immune system, Gcm is expressed at early embryonic stages and induces the expression of proteins involved in the defense against pathogen that will not be necessary before the larval stage.30 Thus, the DamID data strongly suggest that the Gcm fate determinant directly contributes to late aspects of differentiation/function by inducing the expression of effector molecules, the expression of whose could be maintained/sustained by molecules such as the Repo transcription factor in glia. A first example is provided by Frazzled (Fra), the chemoattractant receptor for the Netrin ligand. We have recently found that this receptor is expressed in wing peripheral glia where it triggers the initiation of migration. Gcm induces the expression of Fra, which is subsequently maintained by Repo (Gupta et al. Under Revision). The simplest interpretation is that migration (and Fra expression) is part of the glial specification pathway, which is hence initially triggered by Gcm. In this way, the long-lasting impact of fate determinants may ensure a safer cell specification pathway. Similar observations were made for Eyeless, which regulates the expression of the photoreceptor Rhodopsin necessary for the function but not the development of the eye.64-67
Novel tissues and molecular pathways involving Gcm
A Gene Ontology (GO) analysis was carried out on the list of genes identified by the DamID screen. In such analysis, each gene is associated to a list of terms (GO terms) summarising their functions. The comparison of the DamID list of terms with the list of terms of the whole genome allows the identification of terms that are over-represented.68 Surprisingly, the analysis suggests a role for Gcm in tissues/biological processes in which its expression or function had never been described before: the reproductive system, salivary glands, eye and heart development, further broadening the spectrum of Gcm activity (Fig. 2A). While confirmation awaits thorough functional studies, a developmental analysis of the gcm expression profile has been initiated using the G-trace approach in order to validate the DamID finding.69 This type of experiments detects the cells that express or have expressed a given gene at some point of their life. For instance, gcm G-trace analyses identify the 3rd instar larva salivary gland as a tissue expressing Gcm (Fig. 2B). Quantitative PCR assays do confirm gcm expression (normalized to Act5C and Gapdh the level is 0.00015 s.e.m. 2,93E-05, n = 3 with > 10 glands per sample) and the analysis of a gcm hypomorphic, viable allele obtained by gene conversion on a P element inserted into the gcm promoter (gcmGal4)14,70 reveals a stronger variability in the length of the salivary glands as compared to that observed in wild type animals (Fig. 2C). Of note, the size of the nuclei and the number of cells per gland do not vary significantly (Fig. 2D and E). This data suggest a potential role for Gcm in the late differentiation or function of the salivary gland. Thus, the DamID screen allowed us to discover new territories requiring Gcm and further experiments will clarify the precise role of Gcm in these tissues.
Figure 2.
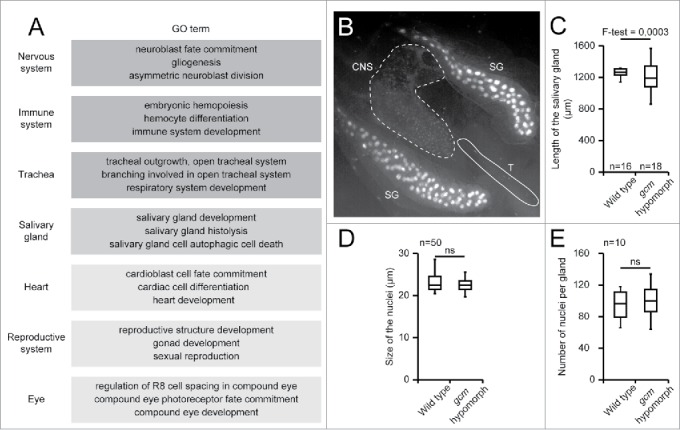
A) Tissues associated GO terms issued from the enrichment analysis carried out on the list of genes identified as Gcm direct target by the DamID screen, using DAVID.30,69,70 The list displayed was arbitrarily limited to 3 GO terms per tissue, each GO term being enriched more than 2 folds with a P-value below 0.05. Dark gray indicates tissues/biological processes already known to require Gcm, intermediate intensity gray indicates a tissue analyzed in this report, pale gray indicates tissues/biological processes still not shown to require Gcm. B) Gcm lineage analysis revealed by GFP labeling in a gcmGal4/+;gtrace/+ 3rd instar larva. Tissues were analyzed by epifluorescence microscopy (20x). Note the presence of GFP positive cells throughout the salivary glands (SG), in the central nervous system (CNS) and in the trachea (T). The signal is much stronger in salivary glands, due to chromosome polytenysation. C) Length of the salivary glands in wild type and in gcm hypomorphic 3rd instar larvae (n = number of salivary glands). F-test was used to compare the variance in the 2 genotypes. The gcm hypomorphic allele used is gcmGal4 (also called P{GawB}gcmrA87.P, Flybase ID: FBti0130081) in homozygous condition. D) Size of the nuclei (diameter) of the salivary gland cells in wild type and in gcm hypomorphic 3rd instar larvae. Size was estimated on 50 nuclei from 5 salivary glands using FIJI. 72 Student test was used to calculate the P-value. E) Number of nuclei in the salivary glands (n = 10) of wild type and gcm hypomorphic 3rd instar larvae. ns indicates that the difference is not significant.
Together with the observation that the glial master regulator is also required in tendon, neurons and peritrachea, the DamID data already indicate that the instructive role of ‘master genes' has to be reconsidered as the activity of these regulatory genes depends on cell-specific factors/epigenetic landscapes. This is exemplified by the loss of Gcm gliogenic potential in differentiated neurons and in aging neuroblasts. The neuroblasts divide several times during embryogenesis and Gcm efficiently converts early neuroblasts toward the glial fate but is unable to do so in late neuroblasts.21 Another example is provided by cell-specific manipulation of gene expression in the tendon cells, where gcm activity is tightly localized by the positional cues wingless and hh. Overexpression of gcm throughout the posterior region of the segment, where tendons cells differentiate from, leads to the expression of both tendon and glial cell markers whereas ectopic expression in the anterior region leads to the expression of glial cell markers only.19 Similar cell-specific factors may be at work in other tissues/stages (see also below). A particularly interesting question related to cell specificity concerns the factors that allow Gcm to control gliogenesis vs. hematopoiesis, which may help understanding how similar are the phagocytic cells outside and within the nervous system, an open question in vertebrates as well.
Gcm: More than the glial master gene
Altogether, the above data call for master genes having a much more refined role than expected in time and in space. They have a long lasting effect on cell differentiation, as seen for the migrating glia. They act in many more tissues than initially thought and they have a more prominent role in some tissue than in others, for example in the nervous system all lateral glia, but only few neuronal subsets require Gcm. And finally, some systems express them transiently during differentiation like glia and hemocytes whereas others express them in differentiated cells, like neurons and salivary glands. This prompts us to consider more sophisticated approaches to study Gcm function. Indeed, genes acting on a single tissue can be characterized directly by observing the phenotype of the full knock-out. Such experiments led to the definition of Gcm as the master regulator of glia differentiation.1-3 However, pleiotropic genes should be studied in a tissue/stage specific fashion to understand the molecular mechanisms at play in each system.71 Further characterization of Gcm using conditional knock down and MARCM clones will allow a complete understanding of Gcm's role in Drosophila development and will shed light on the real impact of this fate determinant.
More broadly, investigations of regulatory genes including gcm and ey clearly suggest that master genes have multiple functions in multiple tissues. These powerful transcription factors are largely “reused” in a large range of systems. This goes in line with the fact that the development of an organism requires the generation of numerous cell types with a limited number of instructive molecules, which implies the multi-usage of the most powerful of them.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank N. Di Iacovo, A. Gonzalez Garcia and the IGBMC facilities for technical assistance.
Funding
This work was supported by INSERM, CNRS, UDS, USIAS, Hôpital de Strasbourg, ARC, LIGUE and ANR grants. PB. Cattenoz was funded by the FRM and by the ANR. The IGBMC was also supported by a French state fund through the ANR labex.
References
- [1].Hosoya T, Takizawa K, Nitta K, Hotta Y. glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell 1995; 82:1025-36; PMID:7553844; http://dx.doi.org/ 10.1016/0092-8674(95)90281-3 [DOI] [PubMed] [Google Scholar]
- [2].Jones BW, Fetter RD, Tear G, Goodman CS. glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell 1995; 82:1013-23; PMID:7553843; http://dx.doi.org/ 10.1016/0092-8674(95)90280-5 [DOI] [PubMed] [Google Scholar]
- [3].Vincent S, Vonesch JL, Giangrande A. Glide directs glial fate commitment and cell fate switch between neurones and glia. Development 1996; 122:131-9; PMID:8565824 [DOI] [PubMed] [Google Scholar]
- [4].Akiyama-Oda Y, Hosoya T, Hotta Y. Alteration of cell fate by ectopic expression of Drosophila glial cells missing in non-neural cells. Dev Genes Evol 1998; 208:578-85; PMID:9811976; http://dx.doi.org/ 10.1007/s004270050217 [DOI] [PubMed] [Google Scholar]
- [5].Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 1995; 267:1788-92; PMID:7892602; http://dx.doi.org/ 10.1126/science.7892602 [DOI] [PubMed] [Google Scholar]
- [6].Bernardoni R, Vivancos V, Giangrande A. glide/gcm is expressed and required in the scavenger cell lineage. Dev Biol 1997; 191:118-30; PMID:9356176; http://dx.doi.org/ 10.1006/dbio.1997.8702 [DOI] [PubMed] [Google Scholar]
- [7].Lebestky T, Chang T, Hartenstein V, Banerjee U. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science 2000; 288:146-9; PMID:10753120; http://dx.doi.org/ 10.1126/science.288.5463.146 [DOI] [PubMed] [Google Scholar]
- [8].Alfonso TB, Jones BW. gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev Biol 2002; 248:369-83; PMID:12167411; http://dx.doi.org/ 10.1006/dbio.2002.0740 [DOI] [PubMed] [Google Scholar]
- [9].Bataille L, Auge B, Ferjoux G, Haenlin M, Waltzer L. Resolving embryonic blood cell fate choice in Drosophila: interplay of GCM and RUNX factors. Development 2005; 132:4635-44; PMID:16176949; http://dx.doi.org/ 10.1242/dev.02034 [DOI] [PubMed] [Google Scholar]
- [10].Sonnenfeld MJ, Jacobs JR. Macrophages and glia participate in the removal of apoptotic neurons from the Drosophila embryonic nervous system. J Comp Neurol 1995; 359:644-52; PMID:7499553; http://dx.doi.org/ 10.1002/cne.903590410 [DOI] [PubMed] [Google Scholar]
- [11].Shklyar B, Sellman Y, Shklover J, Mishnaevski K, Levy-Adam F, Kurant E. Developmental regulation of glial cell phagocytic function during Drosophila embryogenesis. Dev Biol 2014; 393:255-69; PMID:25046770; http://dx.doi.org/ 10.1016/j.ydbio.2014.07.005 [DOI] [PubMed] [Google Scholar]
- [12].Chotard C, Leung W, Salecker I. glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron 2005; 48:237-51; PMID:16242405; http://dx.doi.org/ 10.1016/j.neuron.2005.09.019 [DOI] [PubMed] [Google Scholar]
- [13].Colonques J, Ceron J, Tejedor FJ. Segregation of postembryonic neuronal and glial lineages inferred from a mosaic analysis of the Drosophila larval brain. Mech Dev 2007; 124:327-40; PMID:17344035; http://dx.doi.org/ 10.1016/j.mod.2007.01.004 [DOI] [PubMed] [Google Scholar]
- [14].Soustelle L, Giangrande A. Novel gcm-dependent lineages in the postembryonic nervous system of Drosophila melanogaster. Deve Dyn 2007; 236:2101-8; PMID:17654713; http://dx.doi.org/ 10.1002/dvdy.21232 [DOI] [PubMed] [Google Scholar]
- [15].Soustelle L, Trousse F, Jacques C, Ceron J, Cochard P, Soula C, Giangrande A. Neurogenic role of Gcm transcription factors is conserved in chicken spinal cord. Development 2007; 134:625-34; PMID:17215311; http://dx.doi.org/ 10.1242/dev.02750 [DOI] [PubMed] [Google Scholar]
- [16].Yoshida S, Soustelle L, Giangrande A, Umetsu D, Murakami S, Yasugi T, Awasaki T, Ito K, Sato M, Tabata T. DPP signaling controls development of the lamina glia required for retinal axon targeting in the visual system of Drosophila. Development 2005; 132:4587-98; PMID:16176948; http://dx.doi.org/ 10.1242/dev.02040 [DOI] [PubMed] [Google Scholar]
- [17].Laneve P, Delaporte C, Trebuchet G, Komonyi O, Flici H, Popkova A, D'Agostino G, Taglini F, Kerekes I, Giangrande A. The Gcm/Glide molecular and cellular pathway: new actors and new lineages. Dev Biol 2013; 375:65-78; PMID:23276603; http://dx.doi.org/ 10.1016/j.ydbio.2012.12.014 [DOI] [PubMed] [Google Scholar]
- [18].Altenhein B, Becker A, Busold C, Beckmann B, Hoheisel JD, Technau GM. Expression profiling of glial genes during Drosophila embryogenesis. Dev Biol 2006; 296:545-60; PMID:16762338; http://dx.doi.org/ 10.1016/j.ydbio.2006.04.460 [DOI] [PubMed] [Google Scholar]
- [19].Soustelle L, Jacques C, Altenhein B, Technau GM, Volk T, Giangrande A. Terminal tendon cell differentiation requires the glide/gcm complex. Development 2004; 131:4521-32; PMID:15342477; http://dx.doi.org/ 10.1242/dev.01290 [DOI] [PubMed] [Google Scholar]
- [20].O'Brien MA, Taghert PH. A peritracheal neuropeptide system in insects: Release of myomodulin-like peptides at ecdysis. J Exp Biol 1998; 201:193-209; PMID:9405303 [DOI] [PubMed] [Google Scholar]
- [21].Flici H, Erkosar B, Komonyi O, Karatas OF, Laneve P, Giangrande A. Gcm/Glide-dependent conversion into glia depends on neural stem cell age, but not on division, triggering a chromatin signature that is conserved in vertebrate glia. Development 2011; 138:4167-78; PMID:21852399; http://dx.doi.org/ 10.1242/dev.070391 [DOI] [PubMed] [Google Scholar]
- [22].Morante J, Erclik T, Desplan C. Cell migration in Drosophila optic lobe neurons is controlled by eyeless/Pax6. Development 2011; 138:687-93; PMID:21208993; http://dx.doi.org/ 10.1242/dev.056069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kurusu M, Nagao T, Walldorf U, Flister S, Gehring WJ, Furukubo-Tokunaga K. Genetic control of development of the mushroom bodies, the associative learning centers in the Drosophila brain, by the eyeless, twin of eyeless, and dachshund genes. Proc Natl Acad Sci USA 2000; 97:2140-4; PMID:10681433; http://dx.doi.org/ 10.1073/pnas.040564497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Callaerts P, Leng S, Clements J, Benassayag C, Cribbs D, Kang YY, Walldorf U, Fischbach KF, Strauss R. Drosophila Pax-6/eyeless is essential for normal adult brain structure and function. J Neurobiol 2001; 46:73-88; PMID:11153010; http://dx.doi.org/ 10.1002/1097-4695(20010205)46:2%3c73::AID-NEU10%3e3.0.CO;2-N [DOI] [PubMed] [Google Scholar]
- [25].Clements J, Hens K, Francis C, Schellens A, Callaerts P. Conserved role for the Drosophila Pax6 homolog Eyeless in differentiation and function of insulin-producing neurons. Proc Natl Acad Sci U S A 2008; 105:16183-8; PMID:18852455; http://dx.doi.org/ 10.1073/pnas.0708330105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, Morante J, Celik A, Desplan C. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 2013; 498:456-62; PMID:23783517; http://dx.doi.org/ 10.1038/nature12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Suzuki T, Takayama R, Sato M. eyeless/Pax6 controls the production of glial cells in the visual center of Drosophila melanogaster. Dev Biol 2016; 409:343-53; PMID:26670857; http://dx.doi.org/ 10.1016/j.ydbio.2015.12.004 [DOI] [PubMed] [Google Scholar]
- [28].van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet 2001; 27:304-8; PMID:11242113; http://dx.doi.org/ 10.1038/85871 [DOI] [PubMed] [Google Scholar]
- [29].van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol 2000; 18:424-8; PMID:10748524; http://dx.doi.org/ 10.1038/74487 [DOI] [PubMed] [Google Scholar]
- [30].Cattenoz PB, Popkova A, Southall TD, Aiello G, Brand AH, Giangrande A. Functional conservation of the glide/Gcm regulatory network controlling glia, hemocyte, and tendon cell differentiation in drosophila. Genetics 2016; 202:191-219; PMID:26567182; http://dx.doi.org/ 10.1534/genetics.115.182154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MAT, Artavanistsakonas S. Molecular-interactions between the protein products of the neurogenic loci notch and delta, 2 Egf-homologous genes in Drosophila. Cell 1990; 61:523-34; PMID:2185893; http://dx.doi.org/ 10.1016/0092-8674(90)90534-L [DOI] [PubMed] [Google Scholar]
- [32].Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanistsakonas S. Specific Egf repeats of notch mediate interactions with delta and serrate - implications for notch as a multifunctional receptor. Cell 1991; 67:687-99; PMID:1657403; http://dx.doi.org/ 10.1016/0092-8674(91)90064-6 [DOI] [PubMed] [Google Scholar]
- [33].Simpson P, Carteret C. Proneural clusters: equivalence groups in the epithelium of Drosophila. Development 1990; 110:927-32; PMID:2088728 [DOI] [PubMed] [Google Scholar]
- [34].Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 1998; 393:382-6; PMID:9620803; http://dx.doi.org/ 10.1038/30756 [DOI] [PubMed] [Google Scholar]
- [35].Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev 1995; 9:2598-608; PMID:7590238; http://dx.doi.org/ 10.1101/gad.9.21.2598 [DOI] [PubMed] [Google Scholar]
- [36].Oellers N, Dehio M, Knust E. bHLH proteins encoded by the Enhancer of split complex of Drosophila negatively interfere with transcriptional activation mediated by proneural genes. Mol Gen Genet 1994; 244:465-73; PMID:8078474; http://dx.doi.org/ 10.1007/BF00583897 [DOI] [PubMed] [Google Scholar]
- [37].Udolph G, Rath P, Chia W. A requirement for Notch in the genesis of a subset of glial cells in the Drosophila embryonic central nervous system which arise through asymmetric divisions. Development 2001; 128:1457-66; PMID:11262244 [DOI] [PubMed] [Google Scholar]
- [38].Van de Bor V, Giangrande A. Notch signaling represses the glial fate in fly PNS. Development 2001; 128:1381-90; PMID:11262238 [DOI] [PubMed] [Google Scholar]
- [39].Umesono Y, Hiromi Y, Hotta Y. Context-dependent utilization of Notch activity in Drosophila glial determination. Development 2002; 129:2391-9; PMID:11973271 [DOI] [PubMed] [Google Scholar]
- [40].Rizki TM, Rizki RM. Lamellocyte differentiation in Drosophila larvae parasitized by Leptopilina. Dev Comp Immunol 1992; 16:103-10; PMID:1499832; http://dx.doi.org/ 10.1016/0145-305X(92)90011-Z [DOI] [PubMed] [Google Scholar]
- [41].Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila. Dev Biol 2001; 230:243-57; PMID:11161576; http://dx.doi.org/ 10.1006/dbio.2000.0123 [DOI] [PubMed] [Google Scholar]
- [42].Markus R, Laurinyecz B, Kurucz E, Honti V, Bajusz I, Sipos B, Somogyi K, Kronhamn J, Hultmark D, Ando I. Sessile hemocytes as a hematopoietic compartment in Drosophila melanogaster. P Natl Acad Sci USA 2009; 106:4805-9; PMID:19261847; http://dx.doi.org/ 10.1073/pnas.0801766106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Stofanko M, Kwon SY, Badenhorst P. Lineage tracing of lamellocytes demonstrates Drosophila macrophage plasticity. PLoS One 2010; 5:e14051; PMID:21124962; http://dx.doi.org/ 10.1371/journal.pone.0014051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wright VM, Vogt KL, Smythe E, Zeidler MP. Differential activities of the Drosophila JAK/STAT pathway ligands Upd, Upd2 and Upd3. Cell Signal 2011; 23:920-7; PMID:21262354; http://dx.doi.org/ 10.1016/j.cellsig.2011.01.020 [DOI] [PubMed] [Google Scholar]
- [45].Hombria JC, Brown S, Hader S, Zeidler MP. Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev Biol 2005; 288:420-33; PMID:16277982; http://dx.doi.org/ 10.1016/j.ydbio.2005.09.040 [DOI] [PubMed] [Google Scholar]
- [46].Gilbert MM, Weaver BK, Gergen JP, Reich NC. A novel functional activator of the Drosophila JAK/STAT pathway, unpaired2, is revealed by an in vivo reporter of pathway activation. Mech Develop 2005; 122:939-48; PMID:15925495; http://dx.doi.org/ 10.1016/j.mod.2005.03.004 [DOI] [PubMed] [Google Scholar]
- [47].Yang HR, Kronhamn J, Ekstrom JO, Korkut GG, Hultmark D. JAK/STAT signaling in Drosophila muscles controls the cellular immune response against parasitoid infection. Embo Rep 2015; 16:1664-72; PMID:26412855; http://dx.doi.org/ 10.15252/embr.201540277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jacques C, Soustelle L, Nagy I, Diebold C, Giangrande A. A novel role of the glial fate determinant glial cells missing in hematopoiesis. Int J Dev Biol 2009; 53:1013-22; PMID:19598118; http://dx.doi.org/ 10.1387/ijdb.082726cj [DOI] [PubMed] [Google Scholar]
- [49].Lum L, Beachy PA. The Hedgehog response network: Sensors, switches, and routers. Science 2004; 304:1755-9; PMID:15205520; http://dx.doi.org/ 10.1126/science.1098020 [DOI] [PubMed] [Google Scholar]
- [50].Sanson B. Generating patterns from fields of cells. Examples from Drosophila segmentation. Embo Rep 2001; 2:1083-8; PMID:11743020; http://dx.doi.org/ 10.1093/embo-reports/kve255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Giesen K, Hummel T, Stollewerk A, Harrison S, Travers A, Klambt C. Glial development in the Drosophila CNS requires concomitant activation of glial and repression of neuronal differentiation genes. Development 1997; 124:2307-16; PMID:9199357 [DOI] [PubMed] [Google Scholar]
- [52].Fossett N, Tevosian SG, Gajewski K, Zhang Q, Orkin SH, Schulz RA. The Friend of GATA proteins U-shaped, FOG-1, and FOG-2 function as negative regulators of blood, heart, and eye development in Drosophila. Proc Natl Acad Sci U S A 2001; 98:7342-7; PMID:11404479; http://dx.doi.org/ 10.1073/pnas.131215798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Campbell G, Goring H, Lin T, Spana E, Andersson S, Doe CQ, Tomlinson A. RK2, a glial-specific homeodomain protein required for embryonic nerve cord condensation and viability in Drosophila. Development 1994; 120:2957-66; PMID:7607085 [DOI] [PubMed] [Google Scholar]
- [54].Halter DA, Urban J, Rickert C, Ner SS, Ito K, Travers AA, Technau GM. The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development 1995; 121:317-32; PMID:7768175 [DOI] [PubMed] [Google Scholar]
- [55].Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev 1994; 8:981-94; PMID:7926782; http://dx.doi.org/ 10.1101/gad.8.8.981 [DOI] [PubMed] [Google Scholar]
- [56].Lee BP, Jones BW. Transcriptional regulation of the Drosophila glial gene repo. Mech Dev 2005; 122:849-62; PMID:15939231; http://dx.doi.org/ 10.1016/j.mod.2005.01.002 [DOI] [PubMed] [Google Scholar]
- [57].Flici H, Cattenoz PB, Komonyi O, Laneve P, Erkosar B, Karatas OF, Reichert H, Berzsenyi S, Giangrande A. Interlocked loops trigger lineage specification and stable fates in the Drosophila nervous system. Nat Commun 2014; 5:4484; PMID:25066644; http://dx.doi.org/ 10.1038/ncomms5484 [DOI] [PubMed] [Google Scholar]
- [58].Cattenoz PB, Giangrande A. New insights in the clockwork mechanism regulating lineage specification: Lessons from the drosophila nervous system. Dev Dynam 2015; 244:332-41; PMID:25399853; http://dx.doi.org/ 10.1002/dvdy.24228 [DOI] [PubMed] [Google Scholar]
- [59].Kerr KS, Fuentes-Medel Y, Brewer C, Barria R, Ashley J, Abruzzi KC, Sheehan A, Tasdemir-Yilmaz OE, Freeman MR, Budnik V. Glial wingless/Wnt regulates glutamate receptor clustering and synaptic physiology at the Drosophila neuromuscular junction. J Neurosci 2014; 34:2910-20; PMID:24553932; http://dx.doi.org/ 10.1523/JNEUROSCI.3714-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Taylor MV, Beatty KE, Hunter HK, Baylies MK. Drosophila Mef2 is regulated by twist and is expressed in both the primordia and differentiated cells of the embryonic somatic, visceral and heart musculature. Mech Develop 1995; 50:29-41; PMID:7605749; http://dx.doi.org/ 10.1016/0925-4773(94)00323-F [DOI] [PubMed] [Google Scholar]
- [61].Furlong EEM, Andersen EC, Null B, White KP, Scott MP. Patterns of gene expression during Drosophila mesoderm Development. Science 2001; 293:1629-33; PMID:11486054; http://dx.doi.org/ 10.1126/science.1062660 [DOI] [PubMed] [Google Scholar]
- [62].Baylies MK, Bate M. twist: A myogenic switch in Drosophila. Science 1996; 272:1481-4; PMID:8633240; http://dx.doi.org/ 10.1126/science.272.5267.1481 [DOI] [PubMed] [Google Scholar]
- [63].Cripps RM, Lovato TL, Olson EN. Positive autoregulation of the Myocyte enhancer factor-2 myogenic control gene during somatic muscle development in Drosophila. Dev Biol 2004; 267:536-47; PMID:15013812; http://dx.doi.org/ 10.1016/j.ydbio.2003.12.004 [DOI] [PubMed] [Google Scholar]
- [64].Punzo C, Plaza S, Seimiya M, Schnupf P, Kurata S, Jaeger J, Gehring WJ. Functional divergence between eyeless and twin of eyeless in Drosophila melanogaster. Development 2004; 131:3943-53; PMID:15253940; http://dx.doi.org/ 10.1242/dev.01278 [DOI] [PubMed] [Google Scholar]
- [65].Sheng G, Thouvenot E, Schmucker D, Wilson DS, Desplan C. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: evidence for a conserved function in photoreceptors. Genes Dev 1997; 11:1122-31; PMID:9159393; http://dx.doi.org/ 10.1101/gad.11.9.1122 [DOI] [PubMed] [Google Scholar]
- [66].Punzo C, Kurata S, Gehring WJ. The eyeless homeodomain is dispensable for eye development in Drosophila. Genes Dev 2001; 15:1716-23; PMID:11445545; http://dx.doi.org/ 10.1101/gad.196401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Nfonsam LE, Cano C, Mudge J, Schilkey FD, Curtiss J. Analysis of the transcriptomes downstream of Eyeless and the Hedgehog, Decapentaplegic and Notch signaling pathways in Drosophila melanogaster. PLoS One 2012; 7:e44583; PMID:22952997; http://dx.doi.org/ 10.1371/journal.pone.0044583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al.. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics 2000; 25:25-9; PMID:10802651; http://dx.doi.org/ 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Evans CJ, Olson JM, Ngo KT, Kim E, Lee NE, Kuoy E, Patananan AN, Sitz D, Tran P, Do MT, et al.. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods 2009; 6:603-5; PMID:19633663; http://dx.doi.org/ 10.1038/nmeth.1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Paladi M, Tepass U. Function of Rho GTPases in embryonic blood cell migration in Drosophila. J Cell Sci 2004; 117:6313-26; PMID:15561773; http://dx.doi.org/ 10.1242/jcs.01552 [DOI] [PubMed] [Google Scholar]
- [71].Chatterjee S, Sivakamasundari V, Yap SP, Kraus P, Kumar V, Xing X, Lim SL, Sng J, Prabhakar S, Lufkin T. In vivo genome-wide analysis of multiple tissues identifies gene regulatory networks, novel functions and downstream regulatory genes for Bapx1 and its co-regulation with Sox9 in the mammalian vertebral column. BMC Genomics 2014; 15:1072; PMID:25480362; http://dx.doi.org/ 10.1186/1471-2164-15-1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al.. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9:676-82; PMID:22743772; http://dx.doi.org/ 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]


