Abstract
Mitochondria are bioenergetic, biosynthetic and signaling organelles that are integral in stress sensing to allow for cellular adaptation to the environment. Therefore, it is not surprising that mitochondria are important mediators of tumorigenesis, as this process requires flexibility to adapt to cellular and environmental alterations in addition to cancer treatments. Multiple aspects of mitochondrial biology beyond bioenergetics support transformation including mitochondrial biogenesis and turnover, fission and fusion dynamics, cell death susceptibility, oxidative stress regulation, metabolism, and signaling. Thus, understanding mechanisms of mitochondrial misregulation during tumorigenesis will be critical for the next generation of cancer therapeutics.
Introduction
Historical Perspective
Louis Pasteur identified the importance of oxygen consumption in 1861, finding that yeast divided more in the presence of oxygen and that oxygen inhibited fermentation, an observation known as the ‘Pasteur effect’. The discovery of mitochondria in the 1890s, described cytologically by both Richard Altmann and Carl Benda, began to shed light on this observation, and in 1913 the biochemist Otto Warburg linked cellular respiration to grana derived from guinea pig liver extracts (Ernster and Schatz, 1981). Warburg stated that the granules functioned to enhance the activity of iron-containing enzymes and involved a transfer to oxygen (Ernster and Schatz, 1981). In the following decades, many scientists elucidated the machinery that drives mitochondrial respiration, including tricarboxylic acid (TCA) cycle and fatty acid β-oxidation enzymes in the mitochondrial matrix that generate electron donors to fuel respiration, and electron transport chain (ETC) complexes and ATP synthase in the inner mitochondrial membrane (IMM) that carry out oxidative phosphorylation (Ernster and Schatz, 1981). This biochemical understanding of mitochondrial oxidative phosphorylation gave mechanistic insight into the Pasteur effect, which could be reconstituted by adding purified, respiring liver mitochondria to glycolytic tumor supernatants and observing inhibited fermentation (Aisenberg et al., 1957). The ability of mitochondria to inhibit a glycolytic system suggested an active and direct role for mitochondria in regulating oxidative versus glycolytic metabolism (Aisenberg et al., 1957).
Warburg’s seminal discovery that cancer cells undergo aerobic glycolysis, which refers to the fermentation of glucose to lactate in the presence of oxygen as opposed to the complete oxidation of glucose to fuel mitochondrial respiration, brought attention to the role of mitochondria in tumorigenesis (Warburg, 1956). While the ‘Warburg effect’ is an undisputed feature of many (but not all) cancer cells, Warburg’s reasoning that it stemmed from damaged mitochondrial respiration caused immediate controversy (Weinhouse, 1956). We now understand that while damaged mitochondria drive the Warburg effect in some cases, many cancer cells that display Warburg metabolism possess intact mitochondrial respiration with some cancer subtypes actually depending on mitochondrial respiration. Decades of studies on mitochondrial respiration in cancer have set the framework for a new frontier focused on additional functions of mitochondria in cancer, which have identified pleiotropic roles of mitochondria in tumorigenesis.
A major function of mitochondria is ATP production, hence its nickname ‘powerhouse of the cell’. However, mitochondria perform many roles beyond energy production, including the generation of reactive oxygen species (ROS), redox molecules and metabolites, regulation of cell signaling and cell death and biosynthetic metabolism. These multifaceted functions of mitochondria in normal physiology make them important cellular stress sensors, and allow for cellular adaptation to the environment. Mitochondria similarly impart considerable flexibility for tumor cell growth and survival in otherwise harsh environments such as during nutrient depletion, hypoxia and cancer treatments, and are therefore key players in tumorigenesis.
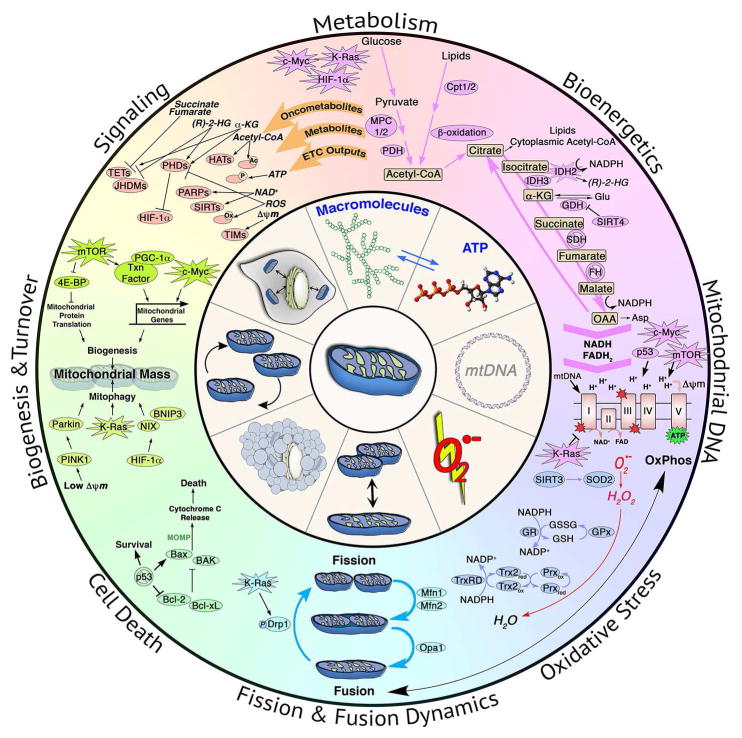
There is no simple canon for the role of mitochondria in cancer development. Instead the mitochondrial function in cancers varies depending upon genetic, environmental and tissue-of-origin differences between tumors. It is clear that the biology of mitochondria in cancer is central to our understanding of cancer biology, as many classical cancer hallmarks result in altered mitochondrial function. This review will summarize functions of mitochondria biology that contribute to tumorigenesis, which include mitochondrial biogenesis and turnover, fission and fusion dynamics, cell death, oxidative stress, metabolism and bioenergetics, signaling and mitochondrial DNA (Figures 1 and 2).
Figure 1. Mitochondria and Cancer.
The role of mitochondrial metabolism, bioenergetics, mtDNA, oxidative stress regulation, fission and fusion dynamics, cell death regulation, biogenesis and turnover and signaling in tumorigenesis.
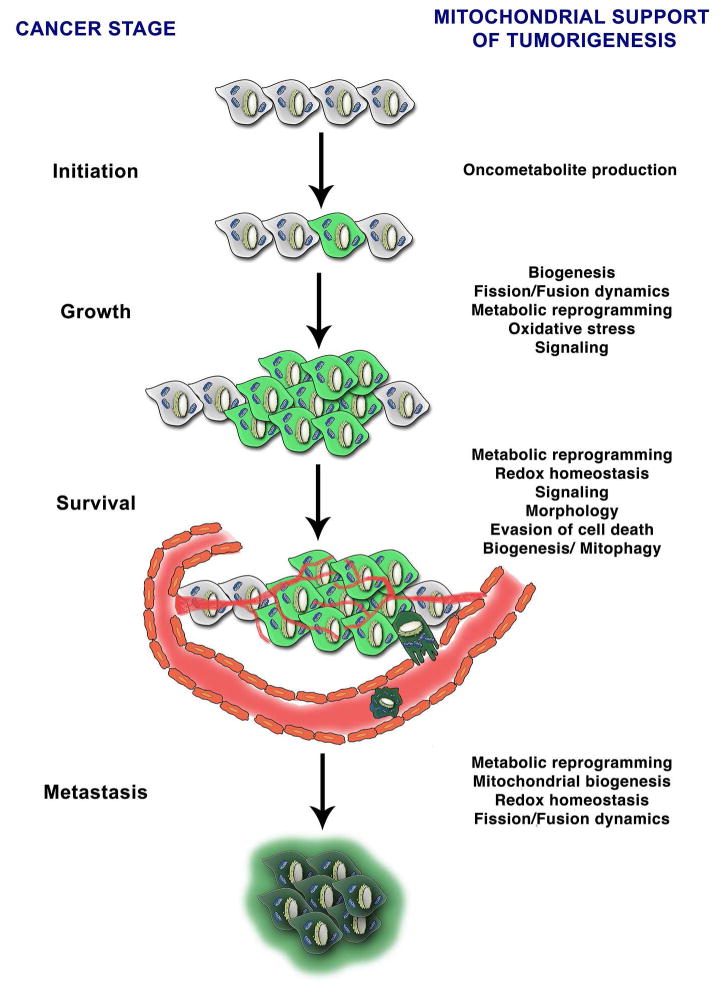
Figure 2. Mitochondria and stages of tumorigenesis.
Mitochondrial biology supports tumorigenesis at multiple stages. Mutations in mitochondrial enzymes generate oncometabolites that result in tumor initiation. Mitochondrial metabolic reprogramming, oxidative signaling and signaling can promote tumor growth and survival. Mitochondria additionally regulate redox homeostasis, susceptibility to cell death via alterations in morphology to promote cell survival. Alterations in mitochondrial mass via regulation of biogenesis and mitophagy also contribute to survival depending on cancer type. Mitochondrial metabolic reprogramming, biogenesis, redox homeostasis and dynamics also contribute to metastatic potential of cancer cells.
Mitochondrial Biogenesis and Turnover
Mitochondrial mass is dictated by two opposing pathways, biogenesis and turnover, and has emerged as both a positive and negative regulator of tumorigenesis. The role of mitochondrial biogenesis in cancer is dictated by many factors, including metabolic state, tumor heterogeneity, tissue type, microenvironment and tumor stage. Additionally, mitophagy, the selective autophagic pathway for mitochondrial turnover, maintains a healthy mitochondrial population. Importantly, misregulation of both mitochondrial biogenesis and mitophagy are central to key oncogenic signaling pathways.
Transcriptional and signaling networks regulating biogenesis
Mitochondrial biogenesis is regulated by transcriptional programs that coordinate induction of both mitochondrial and nuclear localized genes that encode mitochondrial proteins. The transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α) is a central regulator of mitochondrial biogenesis through interactions with multiple transcription factors (Tan et al., 2016). PGC-1α levels often reveal tumor reliance on mitochondrial mass, with high PGC-1α expression resulting in a dependence on mitochondrial respiration (Tan et al., 2016). In contrast, PGC-1α acts as a tumor suppressor in some cancer types, with overexpression resulting in induction of apoptosis (Tan et al., 2016). Additionally, PGC-1α is downregulated in hypoxia inducible factor-1 alpha (HIF-1α) activated renal cell carcinomas, reinforcing a switch to glycolytic metabolism in low oxygen conditions (LaGory et al., 2015; Zhang et al., 2007). Therefore, it is important to identify factors that contribute to the dichotomous effect of PGC-1α on tumor viability, as this has the potential to identify specific susceptibilities for cancer subtypes.
PGC-1α dependent mitochondrial biogenesis may also support anchorage-independent cancer cell growth, a key step in metastasis. Proteomic analysis identified upregulation of mitochondrial proteins involved in metabolism and biogenesis upon low attachment culture conditions (Lamb et al., 2014). Additionally, increased mitochondrial mass co-enriched with tumor initiating activity in patient derived breast cancer lines, which could be blocked by PGC-1α inhibition (De Luca et al., 2015). These findings remain relevant in vivo, as circulating tumor cells (CTCs) developed from primary orthotopic breast tumors show increased mitochondrial biogenesis and respiration, with PGC-1α silencing resulting in decreased CTCs and metastasis (LeBleu et al., 2014). Thus, PGC-1α dependent mitochondrial biogenesis may contribute to tumor metastatic potential.
A key activator of mitochondrial biogenesis in cancer is c-Myc, a transcription factor that globally regulates cell cycle, growth, metabolism and apoptosis. Over 400 mitochondrial genes are identified as c-Myc targets, and initial studies demonstrated that gain/loss of Myc increases/reduces mitochondrial mass respectively. (Li et al., 2005). In normal physiology, c-Myc couples mitochondrial biogenesis with cell cycle progression. However, elevated mitochondrial biogenesis due to oncogenic c-Myc increases cellular biosynthetic and respiratory capacity by upregulating mitochondrial metabolism to support rapid proliferation, complementing c-Myc’s effects on stimulating cell cycle progression and glycolytic metabolism to coordinate rapid cell growth (Figure 3).
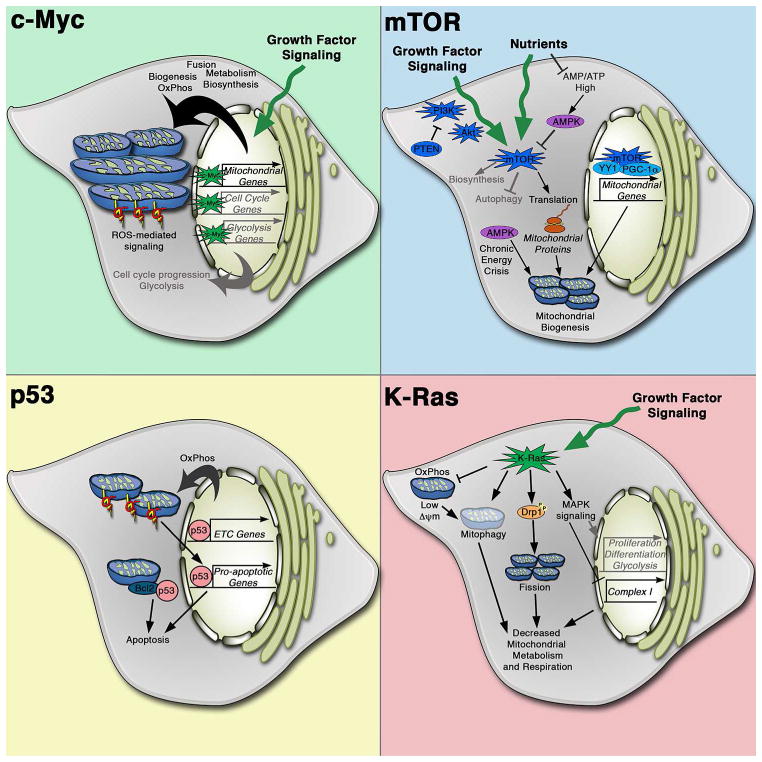
Figure 3. Effects of classical oncogenic and tumor suppressive pathways on mitochondrial biology.
Key mechanisms of mitochondrial regulation by c-MYC, K-RAS, PI3K and p53 signaling pathways. Through transcriptional regulation, c-Myc induces mitochondrial biogenesis and metabolism in addition to its stimulation of cell cycle progression and glycolysis. c-Myc promotes mitochondrial fusion and respiration, which can result in increased ROS production and oxidative signaling. Hyperactive PI3K signaling through either PI3K mutation or loss/mutatoin of the PTEN tumor suppressor results in mTOR activation, which is additionally regulated by nutrient availability, to regulate cell growth. mTOR promotes mitochondrial biogenesis both transcriptionally and translationally. Low nutrient conditions that result in a high AMP/ATP ratio result in AMPK activation, which opposes the mTOR pathway. During chronic nutrient deprivation, AMPK can also promote mitochondrial biogenesis to allow for metabolic flexibility. Loss of p53 promotes survival not only via transcriptional regulation of cell death programs but also through direct interactions with Bcl-2 proteins at the mitochondria. p53 can also induce mitochondrial respiration to promote tumorigenesis by allowing for metabolic flexibility. Oncogenic K-Ras mutations result in a coordinated program of mitochondrial regulation, inhibiting respiration through multiple mechanisms as well as promoting mitochondrial fission and mitophagy.
Another effector of mitochondrial biogenesis is the mammalian target of rapamycin (mTOR) signaling pathway, which is critical for cellular growth and energy homeostasis and is misregulated in many diseases including cancer. mTOR regulates mitochondrial biogenesis both transcriptionally via PGC-1α/Yin Yang 1 (YY1) activation, resulting in mitochondrial gene expression, and translationally via repression of inhibitory 4E-binding proteins (4E-BPs) that downregulate translation of nuclear-encoded mitochondrial proteins (Morita et al., 2015) (Figure 3).
The transcriptional networks regulating biogenesis impact therapeutic outcomes by providing cancer cells with metabolic flexibility to adapt to targeted treatments and tumor microenvironments. In B-Raf or N-Ras mutant melanomas, resistance to MEK inhibitors was partially due to a switch to oxidative metabolism mediated by PGC-1α upregulation and was overcome by mTORC1/2 inhibition, which repressed PGC-1α expression (Gopal et al., 2014; Haq et al., 2013). Likewise, in a mouse model of K-Ras mutant pancreatic ductal adenocarcinoma, cells that survive oncogene ablation have increased PGC-1α expression and mitochondrial function and the reliance on mitochondrial respiration resulted in sensitivity to oxidative phosphorylation inhibitors (Viale et al., 2014). Cancer cells can adapt their mitochondrial function according to the specific stress. For example, c-Myc upregulation and glycolytic gene expression enables resistance to metformin, a complex I inhibitor, in pancreatic cancer cells, which actively utilize mitochondrial respiration due to PGC-1α expression (Sancho et al., 2015). Similarly, c-Myc dependent mitochondrial biogenesis is normally opposed by the HIF-1α signaling pathway, but this balance is altered during oncogenic c-Myc driven transformation (Dang et al., 2008). Therefore, an important consideration in cancer therapeutics will be addressing routes of bioenergetic plasticity provided by mitochondria.
Mitophagy
Clearance of damaged mitochondria via mitophagy is critical for cellular fitness since dysfunctional mitochondria can impair ETC function and increase oxidative stress. A major trigger for mitophagy is via the PTEN-induced putative kinase 1 (PINK1)/Parkin pathway. This pathway is activated upon mitochondrial membrane depolarization, a signal of mitochondrial dysfunction that results from multiple causes including lack of reducing equivalents, hypoxia and impaired electron transport. An alternate pathway for mitophagy induction is through the HIF-1α target genes BCL2 and adenovirus E1B 19 kDa-interacting protein 3 (BNIP3) and BNIP3-like (BNIP3L/NIX), which inhibit mitochondrial respiration during hypoxic conditions that could result in excessive ROS.
Is mitophagy beneficial or harmful to cancers? Similar to autophagy, which is shown to be both pro- and anti-tumorgenic based on context, the function of mitophagy in transformation likely depends on tumor stage (Mancias and Kimmelman, 2016). Mitophagy-deficient Parkin null mice develop spontaneous hepatic tumors, and Parkin loss increases tumorigenesis in multiple cancer models (Matsuda et al., 2015). Additionally, BNIP3 and NIX are identified as tumor suppressors in multiple cancer models (Chourasia et al., 2015). Thus, in certain stages of tumorigenesis, decreased mitophagy may allow for a permissive threshold of dysfunctional mitochondria to persist, generating increased tumor-promoting ROS or other tumorigenic mitochondrial signals. In contrast, established tumors may require mitophagy for stress adaptation and survival. Supporting this concept, BNIP3 is induced in patient glioblastoma samples in response to hypoxia caused by anti-angiogenic therapy and combinatorial angiogenesis and autophagy inhibition had a potent anti-tumor effect in xenograft glioma models (Hu et al., 2012). Additionally, oncogenic K-Ras driven transformation upregulates mitophagy for the clearance of dysfunctional mitochondria, and the accumulation of dysfunctional mitochondria switches adenoma tumor fate to benign oncocytomas instead of carcinomas (Guo et al., 2013).
Fission and Fusion Dynamics
Mitochondria are extremely dynamic and the balance of fission and fusion dictates their morphology. A critical step in mitochondrial membrane fission is dynamin-related protein-1 (Drp1) recruitment to mitochondria and interaction with its outer mitochondria membrane (OMM) receptors, where it causes membrane constriction fueled by GTPase activity. Drp1 mitochondrial translocation and activity is regulated by phosphorylation mediated by multiple kinases that respond to distinct cell cycle and stress conditions (Mishra and Chan, 2016). The mitofusins, Mfn1 and Mfn2, along with optic atrophy-1 (Opa1) mediate mitochondrial fusion. Mitochondria exist as either fused, tubular networks or as fragmented granules depending on cellular state, with mitochondrial metabolism, respiration and oxidative stress regulating fission/fusion machinery (Mishra and Chan, 2016). Mitochondrial morphology also affects susceptibility to mitophagy and apoptosis (Kasahara and Scorrano, 2014).
Multiple studies have demonstrated an imbalance of fission and fusion activities in cancer, with elevated fission activity and/or decreased fusion resulting in a fragmented mitochondrial network (Senft and Ronai, 2016). Importantly, restoration of fused mitochondrial networks in these studies, through either Drp1 knockdown/ inhibition or Mfn2 overexpression, impaired cancer cell growth, suggesting that mitochondrial network remodeling is important in tumorigenesis. Increased Drp1 expression is associated with a migratory phenotype in multiple cancer types, further highlighting the role of mitochondrial dynamics in metastasis (Senft and Ronai, 2016).
Altered mitochondrial dynamics are a key feature of K-Ras dependent cellular transformation, with oncogenic K-Ras stimulating mitochondrial fragmentation via ERK1/2-mediated phosphorylation of Drp1 (Kashatus et al., 2015; Serasinghe et al., 2015). Knockdown or inhibition of Drp1 renders cells resistant to oncogenic K-Ras mediated transformation and impairs tumor growth (Kashatus et al., 2015). Additionally, remodeling of the mitochondrial network upon oncogenic K-Ras expression affected mitochondrial function, decreasing membrane potential and increasing ROS generation (Serasinghe et al., 2015). Thus, K-Ras mediated mitochondrial network remodeling creates a state of upregulated tumorigenic stimuli to support cellular transformation. c-Myc also affects mitochondrial dynamics by altering the expression levels of multiple fission and fusion proteins (Graves et al., 2012). However, the net effect causes mitochondrial fusion (von Eyss et al., 2015), and further studies are needed to understand the differential effects of oncogenic signaling pathways on mitochondrial dynamics.
Cell Death
A hallmark of cancers is their ability to evade cell death, a phenomenon tightly linked to mitochondria. The pro-apoptotic Bcl-2 family members BAX and BAK are recruited to the OMM and oligomerize to mediate mitochondrial outer membrane permeabilization (MOMP), resulting in pore formation and cytochrome c release from mitochondria into the cytosol to activate caspases, the executors of programmed cell death. During normal physiology, anti-apoptotic family members such as Bcl-2 and Bcl-XL bind and inhibit BAX/BAK. Tumor cells escape apoptosis by downregulating pro-apoptotic Bcl-2 genes and/or upregulating anti-apoptotic Bcl-2 genes, achieved through multiple mechanisms reviewed elsewhere (Lopez and Tait, 2015). The balance of pro- and anti-apoptotic proteins affects a cancer cell’s susceptibility to apoptotic stimuli and may predict how a tumor will respond to chemotherapy (Sarosiek et al., 2013).
Mitochondrial shape also dictates apoptotic susceptibility, as Drp1 loss delays cytochrome c release and apoptotic induction, although follow-up work indicated that fission was not required for Bax/Bak-mediated apoptosis (Martinou and Youle, 2011). Instead, a GTPase-independent function of Drp1 in membrane remodeling and hemifusion results in Bax oligomerization and subsequent MOMP, indicating that Drp1 can promote apoptosis independent of fission (Martinou and Youle, 2011). The importance of mitochondrial shape in apoptosis is further demonstrated by Mfn-1 loss-induced mitochondrial hyperfragmentation causing resistance to apoptotic stimuli due to loss of Bax interaction with the mitochondrial membranes. In this study, Drp1 inhibition rescued sensitivity to apoptotic stimuli by restoring a balanced mitochondrial network (Renault et al., 2015). Additionally, Mfn1 is a target of the MEK/ERK signaling pathway - phosphorylated Mfn1 inhibits mitochondria fusion and interacts with BAK to stimulate its oligomerization and subsequent MOMP (Pyakurel et al., 2015). Therefore, while fission and fusion do not necessarily regulate apoptosis per se, a balance of these activities appears to generate a mitochondrial shape that supports interactions with pro-apoptotic Bcl2 proteins.
Oxidative Stress
ROS, in the form of superoxide and hydroxyl free radicals and hydrogen peroxide, are produced from physiological metabolic reactions. Mitochondria are major contributors to cellular ROS and have multiple antioxidant pathways to neutralize ROS including superoxide dismutase (SOD2), glutathione, thioredoxin and peroxiredoxins. The early observation that cancer cells have high ROS levels led to an overly simple hypothesis that inhibiting ROS could be a successful therapeutic strategy. However, a more complex picture is emerging, where ROS stimulates signaling and proliferation, and the concomitant upregulation of antioxidant pathways prevents ROS-mediated cytotoxicity and may even enhance tumor survival (Shadel and Horvath, 2015; Sullivan and Chandel, 2014).
Multiple physiological reactions, including electron transport by the ETC and NAD(P)H oxidases result in ROS production, and these are often exacerbated during tumorigenesis by oncogenic signaling, ETC mutations, and hypoxic microenvironments. High levels of ROS contribute to oxidation of macromolecules, such as lipids, proteins and DNA, and can contribute to genomic instability to promote transformation. However, modest elevations of ROS observed in many tumors can regulate cell signaling via cysteine oxidation. Indeed, H2O2 inactivates the tumor suppressor PTEN by oxidizing active site cysteine residues, causing the formation of a disulfide bond, which prevents PTEN from inactivating the PI3K pathway (Sullivan and Chandel, 2014). Since ROS can inactivate protein tyrosine phosphatases through oxidation of cysteine residues, ROS may have many yet to be discovered effects on diverse, mitogen-activated pathways that are normally inhibited by phosphatases (Sullivan and Chandel, 2014). ROS-mediated regulation of oncogenic signaling also effects metastasis – oxidation of cysteines in Src increased its oncogenic ability, promoting tumor cell migration and metastasis in multiple tumor types, and these phenotypes were blocked by a ROS scavenger (Porporato et al., 2014).
In response to elevated ROS, many tumors upregulate protective antioxidant pathways. For example, oncogenic K-Ras, B-raf and c-Myc actively inhibit ROS through regulation of nuclear factor (erythroid-derived 2)-like 2 (NRF2), a transcriptional regulator of the antioxidant response, to promote tumorigenesis (DeNicola et al., 2011). Similarly, a study in melanoma found that circulating tumor cells had higher levels of NADPH than primary tumor sites, presumably to combat the increased ROS caused by stress of metastasis (Piskounova et al., 2015). In this system, antioxidants promoted distant metastasis, while folate pathway inhibition prevented metastasis due to decreased NADPH production, but had no effect on the primary tumor. Similarly, antioxidant treatment increased the number of metastasis in a mouse model of malignanat melanoma, causing increased invasiveness dependent on glutathione synthesis cells (Le Gal et al., 2015). Thus, successful tumors maintain ROS levels within a window that stimulates proliferation without causing cytotoxicity. The balance of ROS production and antioxidant expression is critical for maintaining this tumor-promoting ROS level.
The requirement for upregulated antioxidant pathways may be an Achilles heel for tumor cells; combination therapy using glutathione and thioredoxin pathway inhibitors has promising results in vitro and in vivo in breast cancer models (Harris et al., 2015). Targeting other aspects of mitochondrial metabolism that contribute to redox regulation has also been proven to be a successful anti-cancer strategy. For example, inhibition of glutamate dehydrogenase 1 (GDH1) increased ROS by reducing levels of fumarate, an activator of antioxidant glutathione peroxidase 1 (GPx) to slow cancer growth (Jin et al., 2015).
Metabolism
One hallmark of tumors is metabolic reprogramming, which supports macromolecule synthesis, bioenergetic demand and cellular survival (Pavlova and Thompson, 2016). Mitochondria are hubs for metabolic reactions and drive this reprogramming through multiple mechanisms.
Alterations in glucose utilization
Many tumors divert glycolytic intermediates into the pentose phosphate pathway, serine biosynthesis and lipid biosynthesis, as opposed to complete oxidation by mitochondrial respiration. In some tumors, this is achieved by limiting pyruvate utilization by mitochondria. The availability of pyruvate for mitochondrial oxidation is regulated by pyruvate kinase (PKM), which catalyzes the final step of glycolysis to generate pyruvate. Cancers specifically upregulate the PKM2 isoform, which has low activity, allowing upstream glycolytic intermediates to accumulate and be used for anabolic processes (Christofk et al., 2008). Additionally, the mitochondrial pyruvate carrier (MPC1 and MPC2) is either lost or downregulated in a number of cancers (Schell et al., 2014). MPC re-expression had a profound effect on reducing tumor growth in xenograft models, suggesting that the expression of this fuel gatekeeper is an important determinant of growth (Yang et al., 2014). Furthermore, MPC loss stimulates compensatory pathways that maintain fuel oxidation by the TCA cycle, including upregulation of glutaminolysis and the use of fatty acids and branched chain amino acids, demonstrating mitochondrial metabolic flexibility (Schell et al., 2014; Vacanti et al., 2014; Yang et al., 2014). Thus, mitochondria remain functional during aerobic glycolysis, and mitochondrial-dependent metabolic reprogramming can support bioenergetic homeostasis during Warburg metabolism.
Reprogramming of amino acid metabolism
Glutamine can be a substrate for TCA cycle oxidation and a starting material for macromolecule synthesis (DeBerardinis et al., 2007). The amide nitrogen on glutamine is used in purine synthesis and glutamine-derived carbons are used in pyrimidine synthesis, amino acid synthesis and lipid synthesis. Catabolism of glutamine, termed glutaminolysis, is elevated in many glutamine-addicted tumors and is often driven by c-Myc upregulation of glutaminase (GLS), which converts glutamine to glutamate and ammonia (Stine et al., 2015). Glutamate is oxidized to α-ketoglutarate (α-KG) by GDH, providing an entry point into the TCA cycle. This process is inhibited by the mitochondrial-localized sirtuin, SIRT4, a tumor suppressor in multiple cancer models. SIRT4 expression in B cell lymphoma cells downregulates glutamine uptake and inhibits growth whereas SIRT4 loss in an Eμ-myc B cell lymphoma model increases glutamine consumption and accelerates tumorigenesis (Jeong et al., 2014). In addition, transaminases utilize glutamate nitrogen to couple α-KG production to synthesis of non-essential amino acids. Transaminases, but not GDH, are upregulated in 3D cultures of proliferating mammary epithelial cells compared to quiescent cells, suggesting that this pathway is important during cancer cell proliferation to support biosynthesis (Coloff et al., 2016).
In tumor cells with dysfunctional mitochondria due to ETC or TCA cycle enzyme mutations, a fraction of glutamine-derived α-KG undergoes reductive carboxylation to support biosynthesis and redox homeostasis (Mullen et al., 2012). This pathway is dependent on the NADP+/NADPH-utilizing isocitrate dehydrogenase isoforms IDH1 (cytosolic) and IDH2 (mitochondrial), catalyzing the reverse reaction of isocitrate production from α-KG. Reductive carboxylation can also support anchorage independent tumor growth in spheroid cultures by mitigating mitochondrial ROS in a coordinated cycle where cytosolic reductive carboxylation by IDH1 supports mitochondrial oxidative metabolism and NADPH production by IDH2 (Jiang et al., 2016).
As nutrients are oxidized to produce biosynthetic precursors, electrons are removed from carbon. Therefore, electron acceptors can quickly become limiting in highly proliferating cells. This observation was highlighted in a series of studies demonstrating that beyond ATP production, mitochondrial respiration is required to replenish electron-accepting cofactors NAD+ and FAD (Birsoy et al., 2015; Sullivan et al., 2015). Interestingly, when mitochondrial respiration is impaired, rather than ATP, the electron acceptors are most limiting for de novo synthesis of aspartate, a key amino acid required for protein and nucleotide synthesis.
Aside from coordinating fuel oxidation, mitochondria contribute to tumor progression through nucleotide synthesis via one-carbon metabolism. The mitochondrial folate synthesis pathway consists of serine hydroxylmethyltransferase (SHMT2) and MTHFD2. A meta-analysis of gene expression profiles identified MTHFD2 as overexpressed in many human tumors and further studies revealed its importance in survival of cancer cells (Nilsson et al., 2014). Unlike the cytosolic arm of folate metabolism that primarily uses serine, the mitochondrial arm also uses glycine as a carbon source, a potential vulnerability for cancers that upregulate this pathway. Metabolic profiling of NCI-60 lines revealed high correlation of proliferation with glycine consumption along with the increase in SHMT2, MTHFD2, and MTHFD1L (Jain et al., 2012). While the cytosolic pathway can compensate for loss of mitochondrial 1C metabolism, cells become dependent on extracellular serine and glycine for growth and are thus susceptible to inhibition of serine catabolism, highlighting the important of mitochondrial 1C metabolism in supporting tumorigenesis during nutrient deprivation (Ducker et al., 2016). For example, SHMT2 is expressed in ischaemic tumor zones, providing proliferative advantage under hypoxic conditions (Kim et al., 2015). Additionally SHMT2 regulation of serine metabolism also contributes to NADPH production and detoxification of ROS under hypoxia, a function important for survival of MYC-driven cancers (Ye et al., 2014).
Lipid metabolism
Unlike other fuels, lipid utilization in cancer is less defined at the molecular level. Cancer-specific alterations of lipid metabolism seem to be unique to tumor type, allowing for some cancers to upregulate fatty acid oxidation (FAO) while others are more dependent on lipid synthesis. Upregulation of lipogenesis is postulated to be a common feature across most tumors, in part to produce membranes for proliferation (Currie et al., 2013). Inhibition of ATP-citrate lyase (ACLY), which converts mitochondrial-derived citrate to acetyl-CoA in the cytoplasm to support lipogenesis, impairs tumorigenesis in multiple models (Currie et al., 2013). In contrast certain cancer types including lymphomas and leukemias rely primarily on FAO for ATP production (Carracedo et al., 2013). Additionally, FAO may be a preferred fuel choice for cancers undergoing stress as it is a crucial survival mechanism for breast cancer cells undergoing loss of attachment to the extracellular matrix (Carracedo et al., 2013). However, mechanisms that upregulate FAO in cancers remain poorly understood. In one example, tumor cell upregulation of the brain-specific isoform of carnitine palmitoyltransferase (Cpt-1c), required for mitochondrial FA import, resulted in increased FAO and ATP production and resistance to metabolic stress (Carracedo et al., 2013). Moreover, increased FAO may confer benefits beyond ATP generation such as maintaining redox homeostasis (Carracedo et al., 2013). Finally, production of acetyl-CoA from oxidized fatty acids could be used for epigenetic remodeling of chromatin, subsequently causing lasting changes in metabolism.
Studying cancer metabolism in vivo
Recent work has highlighted the importance of studying cancer metabolism in models comparable to the in vivo disease. For example, while glutamine fuels TCA cycle anaplerosis in vitro, this is not necessarily true of all tumors in vivo. Studies comparing the fate of labeled glucose and glutamine in mouse models of K-Ras- driven non-small cell lung cancer showed minimal contribution of glutamine to TCA cycle intermediates (Davidson et al., 2016). Additionally, studies in glioblastoma cells showed that glutamine dependent anaplerosis was not required for growth, with cells secreting glutamate even under glutamine starvation conditions (Tardito et al., 2015). In this study, glutamine synthase (GS) expression sustained growth and purine nucleotide biosynthesis during glutamine starvation. Furthermore, primary patient derived glioma stem-like cells grew independently of glutamine supplementation. These studies highlight the importance of understanding in vivo metabolic requirements of tumor cells when designing therapeutic strategies.
Mitochondrial Signaling
Mitochondrial biology and tumorigenic signaling intersect at multiple levels. First, classical oncogenic signaling pathways alter mitochondrial functions to support tumorigenesis. Second, direct signals from mitochondria affect cellular physiology and tumorigenesis. Finally, mutations in mitochondrial enzymes can result in oncometabolite production, a novel set of mitochondrial signaling molecules that function in tumor initiation.
Classical oncogenic and tumor suppressive pathways regulate mitochondrial biology
The resurgence of mitochondrial research has led to the discovery that established tumor suppressors and oncogenes directly regulate mitochondrial biology. Several hallmark cancer signaling pathways that alter mitochondrial biology to promote transformation are described herein (Figure 3).
In addition to promoting mitochondrial biogenesis, numerous studies have linked c-Myc with mitochondrial metabolism in cancer. The importance of mitochondrial metabolism in c-Myc driven growth was demonstrated in a functional screen of Myc responsive cDNAs to rescue cell growth of c-Myc null cells. The screen identified SHMT2, the first reaction in mitochondrial one-carbon metabolism, as the only target that could partially rescue growth (Nikiforov et al., 2002). While the tumorigenic contribution of increased mitochondrial biogenesis and metabolism in oncogenic c-Myc driven cancers is difficult to separate from its global upregulation of transcription, suppression of glutaminolysis can inhibit proliferation of c-Myc driven lymphoma cells (Jeong et al., 2014; Le et al., 2012).
An important signaling pathway in hypoxic tumor microenvironments is mediated by HIF-1α, which upregulates glycolytic metabolism in low oxygen conditions and inhibits mitochondrial respiration (Mucaj et al., 2012). Mitochondrial derived ROS also regulates the HIF-1α pathway via inhibition of prolyl hydroxylases (PHDs), negative regulators of HIF signaling. SIRT3, a mitochondrial deacetylase, is an important regulator of this pathway by maintaining ROS and redox homeostasis via deacetylation and activation of mitochondrial SOD2 and IDH2, and indirectly through transcriptional upregulation of antioxidant pathways (Bause and Haigis, 2013). SIRT3-dependent reduction in mitochondrial ROS results in HIF-1α degradation, limiting glycolysis and the Warburg effect in tumors (Bell et al., 2011; Finley et al., 2011).
In addition to the pleotropic effects of oncogenic K-Ras signaling on proliferation, apoptosis and metabolism, oncogenic K-Ras results in a coordinated program of mitochondrial regulation that supports transformation (Pylayeva-Gupta et al., 2011). Multiple K-Ras-dependent mechanisms can downregulate mitochondrial respiration including upregulation of mitochondrial fission (Kashatus et al., 2015; Serasinghe et al., 2015), transcriptional downregulation of complex I (Wang et al., 2015) and ERK-phosphorylation dependent mitochondrial translocation of phosphoglycerate kinase I (PGK1) (Li et al., 2016). Oncogenic K-Ras also promotes upregulation of mitophagy to preserve mitochondrial function under starvation conditions. Autophagy inhibition in cancers with active K-Ras results in a decline in mitochondrial respiration, TCA metabolite and energy levels during starvation, which may be important for tumor cell survival in nutrient-depleted microenvironments (Guo et al., 2011).
The PI3K/Akt signaling pathway stimulates cell growth and is often activated in cancer either through oncogenic mutations in signaling kinases or loss/ mutation of the PTEN tumor suppressor, a key phosphatase that shuts off this pathway (Papa et al., 2014). Although PI3K signaling is induces cell growth and upregulates glycolysis, metabolic adaptation via a switch to mitochondrial oxidative phosphorylation can mediate resistance to PI3K inhibitors, undermining the effectiveness of PI3K-specific targeted therapy (Ghosh et al., 2015). A major downstream effector of active PI3K/Akt signaling is mTOR, which participates in mTORC1 and mTORC2 signaling complexes to couple nutrient and growth factor sensing to cellular growth through regulation of translation, anabolic metabolism and autophagy (Dibble and Cantley, 2015). In addition to regulating mitochondrial biogenesis, mTORC1 stimulates multiple mitochondrial metabolic pathways. The transcriptional repression of SIRT4 downstream of mTORC1 activity results in GDH activation to upregulate glutaminolysis (Csibi et al., 2013). mTORC1 also induces the mitochondrial folate pathway to promote de novo purine synthesis via activation of the transcription factor ATF4 to result in upregulation of MTHFD2 expression (Ben-Sahra et al., 2016).
The AMP-regulated kinase (AMPK) signaling network is activated by during low energy conditions, directly inhibiting multiple targets including mTORC1 to restore energy homeostasis. AMPK is a critical downstream target of the liver kinase B1 (LKB1) tumor suppressor, which is mutated in the inherited cancer disorder Peutz-Jeghers syndrome, and mediates many LKB1 tumor suppressive functions (Faubert et al., 2015). However, AMPK loss does not fully recapitulate LKB1 loss and AMPK has both pro and anti-tumorigenic effects, which appear dependent on the presence of other oncogenic drivers as well as tumor stage (Faubert et al., 2015). While uncoupling proliferation from energy sensing allows for unhindered proliferation in the presence of oncogenic growth signaling, AMPK functions in metabolic adaptation and mitochondrial homeostasis can be beneficial in established tumors. For example, AMPK promotes mitophagy through phosphorylation of ULK kinases and is required for cell survival during starvation (Faubert et al., 2015). Additionally, AMPK activation in response to ETC dysfunction results in mitochondrial fragmentation through direct phosphorylation of mitochondrial fission factor, an OMM receptor for Drp1 (Toyama et al., 2016). Additionally, sustained energy deprivation can result in AMPK mediated upregulation of mitochondrial biogenesis via PGC-1α – allowing the cell further metabolic plasticity (Faubert et al., 2015).
p53 is a commonly mutated tumor suppressor and has been extensively studied due to its transcriptional regulation of cell cycle and apoptotic genes. It is now appreciated that p53 also has functions in the regulation of cellular metabolism via transcriptional activation of metabolic genes (Berkers et al., 2013). p53 limits glycolysis and drives transcription of genes required for ETC assembly and maintenance (Berkers et al., 2013). However, more recent work has suggested an alternate side to p53’s role in tumorigenesis, with its ability to allow for adaptation to metabolic stress resulting in pro-survival effects in tumor cells. These pro-survival effects are partially accomplished through upregulation of mitochondrial FAO and respiration, allowing cancer cells to adapt to starvation conditions (Jiang et al., 2015). In addition to transcriptional regulation of mitochondrial activity, p53 also directly functions at the mitochondria to induce apoptosis in response to stress via interactions with Bcl-2 family members (Vaseva and Moll, 2009). Tumor-derived p53 mutations no longer interact with Bcl2 and do not trigger mitochondrial outer membrane permeabilization (Vaseva and Moll, 2009). Thus, in addition to effect its transcriptional activity, p53 mutations can also promote cancer survival through direct mitochondrial functions.
Mitochondrial retrograde signals
Mitochondria are important stress sensors, and retrograde signaling from the mitochondria allows the cell to adapt to its environment. Metabolites generated by mitochondrial metabolic pathways including the TCA cycle, β-oxidation, and the ETC, affect both nuclear gene transcription via chromatin modification as well as cytosolic signaling pathways. For example, the TCA cycle intermediate α-KG is a cosubstrate for many enzymes in the cytoplasm and nucleus including the PHD family, which inhibits HIF-1α signaling by promoting its degradation, and the 10–11-translocation methylcytosine dioxygenase (TET) and Jumunji-C histone demethylase (JHDM) families of chromatin modifying enzymes. In the case of chromatin regulation, glutamine derived α-KG contributes to TET-dependent demethylation reactions (Carey et al., 2015). Additional mitochondrial regulation of chromatin occurs through histone acetylation. ACLY-dependent production of acetyl-CoA from mitochondrial-derived citrate is used by histone acetyl transferases (HATs) and oncogenic signaling pathways modify histone acetylation patterns in a ACLY dependent manner (Lee et al., 2014; Wellen et al., 2009). In addition to chromatin modification, acetyl-CoA generated from mitochondrial-derived citrate is used for the acetylation of many cytosolic and mitochondrial proteins to modulate protein activity. Thus, mitochondrial derived metabolites can effect signaling pathways, nuclear transcription, and chromatin modification.
In addition to signaling molecules, readouts of mitochondrial integrity including Δψm and MOMP also function as important signals, enabling the cell to respond to unhealthy/ dysfunctional mitochondria. Since the membrane potential generated by healthy mitochondria is required for protein import into the mitochondrial matrix and intermembrane space via the TIM22 and TIM23 translocator complexes, loss of membrane potential impairs import. If the defect in protein import is severe, the cell can initiate mitophagy to clear these unhealthy mitochondria as discussed above. Additionally, ATP generation by the ETC is an important signaling output with diminished ETC activity increasing the AMP/ATP ratio to activate AMPK signaling. ETC dysfunction can also result in decreased NAD+ levels, a co-substrate for both the sirtuin and poly(ADP-ribose) protein families, which have many functions in tumorigenesis (German and Haigis, 2015; Vyas and Chang, 2014). Finally, ROS regulates cytosolic signaling networks to promote tumorigenesis (as discussed above).
Mitochondrial oncometabolites
Dominant mutations in mitochondrial enzymes led to the exciting identification of mitochondrial-derived signaling molecules, termed oncometabolites. Mutant versions of cytoplasmic and mitochondrial IDH isoforms, found in a striking 10% of acute myeloid leukemias and 70% of glioblastomas, reduce α-KG to generate the oncometabolite (R)-2-hydroxyglutarate ((R)-2-HG) (Dang et al., 2009; Ward et al., 2010). In addition, loss of function of TCA cycle enzymes succinate dehydrogenase (SDH) and fumarate hydratase (FH), underlying the inherited cancer predispositions Hereditary Paraganglioma Syndrome and Hereditary Leiomyomatosis and Renal-Cell Cancer syndrome respectively, result in the accumulation of metabolic intermediates succinate and fumarate, which function as oncometabolites when in excess.
A major mode of action of these oncometabolites is owed to their structural similarity to α-KG, allowing them to act as competitive inhibitors of α-KG-dependent enzymes including the TET and JHDM families of chromatin modifying enzymes and the PHD family (Nowicki and Gottlieb, 2015). Inhibition of TET activity leads to hypermethylation of CpG islands, found near gene promoters, which results in gene silencing (Nowicki and Gottlieb, 2015). Additionally, repressive histone methylation marks on H3K9 and H3K27 are observed in IDH1 and IDH2 mutant gliomas due to JHDM inhibition (Lu et al., 2012). Therefore, through the production of oncometabolites, mitochondria exert strong influence on chromatin structure, and can result in tumor initiation. Both succinate and fumarate accumulation stabilize HIF-1α via PHD inhibition, reinforcing the Warburg effect (MacKenzie et al., 2007; Nowicki and Gottlieb, 2015). In contrast, (R)-2-HG activates PHD enzymes, diminishing HIF-1α levels, which resulted in the enhancement of proliferation of astrocytes (Koivunen et al., 2012). (R)-2-HG alone reversibly recapitulates the effects of IDH mutation on leukemogenesis while its enantiomer (S)-2-HG had no effect even though it more potently inhibited TET2 and PHDs, suggesting that differential requirements for HIF-1α depending on cell type can influence neoplasia (Losman et al., 2013).
FH deficiency also supports tumorigenesis independently of α-KG/HIF-1α. The high level of fumarate accumulation in FH-deficient tumors/cells results in increased protein succinylation through the covalent modification of fumarate on cysteines. Cysteine succinylation inhibits Kelch-like ECH-associated protein 1 (KEAP1), a negative regulator of Nrf2, to result in upregulation of antioxidant pathways (Adam et al., 2011). Additionally, accumulated fumarate can bind to glutathione to generate succinylated glutathione, an alternate glutathione reductase substrate that decreases NADPH and increases ROS levels (Sullivan et al., 2013). Thus, FH deficiency can alter redox homeostasis to promote tumorigenesis.
Mitochondrial DNA mutations
The presence of a separate mitochondrial genome adds to the unique and complex biology of this organelle, as mutations in mtDNA impact tumorigenesis. Mitochondria contain multiple copies of a single circular DNA molecule that encodes for 13 ETC subunits, mitochondrial ribosomes and tRNAs. In addition to distinct mtDNA haplotypes that exist among different human populations, many germline and somatic mtDNA mutations associated with cancer risk have been identified (van Gisbergen et al., 2015). Differences in mtDNA copy number are implicated in tumorigenesis, although both low and high copy numbers have been associated with various cancers, similar to the varying associations between mitochondrial biogenesis and tumorigenesis. Although the functional consequence of many of these polymorphisms/ mutations is not well understood, some mutations occur in ETC genes and can result in increased oxidative stress due to ETC dysfunction to promote tumorigenesis. Since mitochondria contain multiple copies of the mtDNA genome, cells are either homoplasmic or heteroplasmic regarding their mtDNA composition, with mutant copies of the genome spreading through the mitochondrial network through fission and fusion cycles. In this way, dominant mtDNA mutations become established in a clonal cell population. mtDNA mutations and haplotypes associated with various cancer types are reviewed elsewhere (van Gisbergen et al., 2015).
Concluding Remarks
Mitochondria are complex organelles that influence cancer initiation, growth, survival and metastasis, and many facets of mitochondrial biology beyond energy production actively contribute to tumorigenesis. These include mitochondrial mass, dynamics, cell death regulation, redox homeostasis, metabolic regulation and signaling. The interplay between these aspects of mitochondrial biology results in coordinated programs of mitochondrial regulation of cellular physiology and highlights the pleiotropic functions of mitochondria in cancer. Additionally, similar to the transforming discoveries of oncogenic mutations in growth factor signaling pathways, mutations in mitochondrial metabolic enzymes are a exciting new frontier in cancer biology.
The flexibility that mitochondria bestow tumor cells, including alterations in fuel utilization, bioenergetics, cell death susceptibility, and oxidative stress, allows for survival in the face of adverse environmental conditions such as starvation and during chemotherapeutic and targeted cancer treatments. Therefore, in order to effectively treat cancer, the escape routes to therapeutic interventions provided by mitochondria must also be considered – future studies into combination therapies that remove this flexibility will be important to advance cancer treatments.
Acknowledgments
We apologize for all the primary literature and work we could not cite due to space limitations; this review is not meant to be a comprehensive summary of all the work done in the field of mitochondrial functions in cancer. We thank Jonathan Coloff, Lydia Finley, Karina Gonzalez, Jessica Spinelli and Alison Ringel for discussion on the manuscript. S.V. is supported by a postdoctoral fellowship from the American Cancer Society (127097-PF-14-255-01-TBE). E.Z. is supported by a postdoctoral fellowship from the American Heart Association (15POST25560077). M.C.H. is supported by the Ludwig Center at Harvard, the Paul F. Glenn Foundation, and the National Institute of Diabetes and Digestive and Kidney Diseases (1R01DK103295-01A1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam J, Hatipoglu E, O’Flaherty L, Ternette N, Sahgal N, Lockstone H, Baban D, Nye E, Stamp GW, Wolhuter K, Stevens M, Fischer R, Carmeliet P, Maxwell PH, Pugh CW, Frizzell N, Soga T, Kessler BM, El-Bahrawy M, Ratcliffe PJ, Pollard PJ. Renal Cyst Formation in Fh1-Deficient Mice Is Independent of the Hif/Phd Pathway: Roles for Fumarate in Keap1 Succination and Nrf2 Signaling. Cancer Cell. 2011;20:524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aisenberg AC, Reinafarje B, Potter VR. Studies on the Pasteur Effect. I. General Observations. J Biol Chem. 1957;224:1099–1113. [PubMed] [Google Scholar]
- Bause AS, Haigis MC. Sirt3 Regulation of Mitochondrial Oxidative Stress. Exp Gerontol. 2013;48:634–639. doi: 10.1016/j.exger.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Bell EL, Emerling BM, Ricoult SJ, Guarente L. Sirt3 Suppresses Hypoxia Inducible Factor 1alpha and Tumor Growth by Inhibiting Mitochondrial Ros Production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Hoxhaj G, Ricoult SJ, Asara JM, Manning BD. Mtorc1 Induces Purine Synthesis through Control of the Mitochondrial Tetrahydrofolate Cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. Metabolic Regulation by P53 Family Members. Cell Metab. 2013;18:617–633. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular Alpha-Ketoglutarate Maintains the Pluripotency of Embryonic Stem Cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Cantley LC, Pandolfi PP. Cancer Metabolism: Fatty Acid Oxidation in the Limelight. Nat Rev Cancer. 2013;13:227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourasia AH, Boland ML, Macleod KF. Mitophagy and Cancer. Cancer Metab. 2015;3:4. doi: 10.1186/s40170-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 Splice Isoform of Pyruvate Kinase Is Important for Cancer Metabolism and Tumour Growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Coloff JL, Murphy JP, Braun CR, Harris IS, Shelton LM, Kami K, Gygi SP, Selfors LM, Brugge JS. Differential Glutamate Metabolism in Proliferating and Quiescent Mammary Epithelial Cells. Cell Metab. 2016;23:867–880. doi: 10.1016/j.cmet.2016.03.016. [DOI] [PubMed] [Google Scholar]
- Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, Henske EP, Haigis MC, Cantley LC, Stephanopoulos G, Yu J, Blenis J. The Mtorc1 Pathway Stimulates Glutamine Metabolism and Cell Proliferation by Repressing Sirt4. Cell. 2013;153:840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie E, Schulze A, Zechner R, Walther TC, Farese RV., Jr Cellular Fatty Acid Metabolism and Cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Kim JW, Gao P, Yustein J. The Interplay between Myc and Hif in Cancer. Nat Rev Cancer. 2008;8:51–56. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-Associated Idh1 Mutations Produce 2-Hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, Bauer MR, Jha AK, O’Brien JP, Pierce KA, Gui DY, Sullivan LB, Wasylenko TM, Subbaraj L, Chin CR, Stephanopolous G, Mott BT, Jacks T, Clish CB, Vander Heiden MG. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca A, Fiorillo M, Peiris-Pages M, Ozsvari B, Smith DL, Sanchez-Alvarez R, Martinez-Outschoorn UE, Cappello AR, Pezzi V, Lisanti MP, Sotgia F. Mitochondrial Biogenesis Is Required for the Anchorage-Independent Survival and Propagation of Stem-Like Cancer Cells. Oncotarget. 2015;6:14777–14795. doi: 10.18632/oncotarget.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond Aerobic Glycolysis: Transformed Cells Can Engage in Glutamine Metabolism That Exceeds the Requirement for Protein and Nucleotide Synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, Scrimieri F, Winter JM, Hruban RH, Iacobuzio-Donahue C, Kern SE, Blair IA, Tuveson DA. Oncogene-Induced Nrf2 Transcription Promotes Ros Detoxification and Tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Cantley LC. Regulation of Mtorc1 by Pi3k Signaling. Trends Cell Biol. 2015;25:545–555. doi: 10.1016/j.tcb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD. Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab. 2016 doi: 10.1016/j.cmet.2016.09.011. [DOI] [PubMed] [Google Scholar]
- Ernster L, Schatz G. Mitochondria: A Historical Review. J Cell Biol. 1981;91:227s–255s. doi: 10.1083/jcb.91.3.227s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Vincent EE, Poffenberger MC, Jones RG. The Amp-Activated Protein Kinase (Ampk) and Cancer: Many Faces of a Metabolic Regulator. Cancer Lett. 2015;356:165–170. doi: 10.1016/j.canlet.2014.01.018. [DOI] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, Haigis MC. Sirt3 Opposes Reprogramming of Cancer Cell Metabolism through Hif1alpha Destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German NJ, Haigis MC. Sirtuins and the Metabolic Hurdles in Cancer. Curr Biol. 2015;25:R569–583. doi: 10.1016/j.cub.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh JC, Siegelin MD, Vaira V, Faversani A, Tavecchio M, Chae YC, Lisanti S, Rampini P, Giroda M, Caino MC, Seo JH, Kossenkov AV, Michalek RD, Schultz DC, Bosari S, Languino LR, Altieri DC. Adaptive Mitochondrial Reprogramming and Resistance to Pi3k Therapy. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/dju502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal YN, Rizos H, Chen G, Deng W, Frederick DT, Cooper ZA, Scolyer RA, Pupo G, Komurov K, Sehgal V, Zhang J, Patel L, Pereira CG, Broom BM, Mills GB, Ram P, Smith PD, Wargo JA, Long GV, Davies MA. Inhibition of Mtorc1/2 Overcomes Resistance to Mapk Pathway Inhibitors Mediated by Pgc1alpha and Oxidative Phosphorylation in Melanoma. Cancer Res. 2014;74:7037–7047. doi: 10.1158/0008-5472.CAN-14-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JA, Wang Y, Sims-Lucas S, Cherok E, Rothermund K, Branca MF, Elster J, Beer-Stolz D, Van Houten B, Vockley J, Prochownik EV. Mitochondrial Structure, Function and Dynamics Are Temporally Controlled by C-Myc. PLoS One. 2012;7:e37699. doi: 10.1371/journal.pone.0037699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E. Activated Ras Requires Autophagy to Maintain Oxidative Metabolism and Tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, Snyder E, Santanam U, Dipaola RS, Jacks T, Rabinowitz JD, White E. Autophagy Suppresses Progression of K-Ras-Induced Lung Tumors to Oncocytomas and Maintains Lipid Homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, Wargo JA, Song JS, Fisher DE, Arany Z, Widlund HR. Oncogenic Braf Regulates Oxidative Metabolism Via Pgc1alpha and Mitf. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, Yung KY, Brenner D, Knobbe-Thomsen CB, Cox MA, Elia A, Berger T, Cescon DW, Adeoye A, Brustle A, Molyneux SD, Mason JM, Li WY, Yamamoto K, Wakeham A, Berman HK, Khokha R, Done SJ, Kavanagh TJ, Lam CW, Mak TW. Glutathione and Thioredoxin Antioxidant Pathways Synergize to Drive Cancer Initiation and Progression. Cancer Cell. 2015;27:211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- Hu YL, DeLay M, Jahangiri A, Molinaro AM, Rose SD, Carbonell WS, Aghi MK. Hypoxia-Induced Autophagy Promotes Tumor Cell Survival and Adaptation to Antiangiogenic Treatment in Glioblastoma. Cancer Res. 2012;72:1773–1783. doi: 10.1158/0008-5472.CAN-11-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite Profiling Identifies a Key Role for Glycine in Rapid Cancer Cell Proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SM, Lee A, Lee J, Haigis MC. Sirt4 Protein Suppresses Tumor Formation in Genetic Models of Myc-Induced B Cell Lymphoma. J Biol Chem. 2014;289:4135–4144. doi: 10.1074/jbc.M113.525949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, LaGory EL, Kenzelmann Broz D, Bieging KT, Brady CA, Link N, Abrams JM, Giaccia AJ, Attardi LD. Analysis of P53 Transactivation Domain Mutants Reveals Acad11 as a Metabolic Target Important for P53 Pro-Survival Function. Cell Rep. 2015;10:1096–1109. doi: 10.1016/j.celrep.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, Terada LS, Adams ND, McCabe MT, Pietrak B, Schmidt S, Metallo CM, Dranka BP, Schwartz B, DeBerardinis RJ. Reductive Carboxylation Supports Redox Homeostasis During Anchorage-Independent Growth. Nature. 2016;532:255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Li D, Alesi GN, Fan J, Kang HB, Lu Z, Boggon TJ, Jin P, Yi H, Wright ER, Duong D, Seyfried NT, Egnatchik R, DeBerardinis RJ, Magliocca KR, He C, Arellano ML, Khoury HJ, Shin DM, Khuri FR, Kang S. Glutamate Dehydrogenase 1 Signals through Antioxidant Glutathione Peroxidase 1 to Regulate Redox Homeostasis and Tumor Growth. Cancer Cell. 2015;27:257–270. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara A, Scorrano L. Mitochondria: From Cell Death Executioners to Regulators of Cell Differentiation. Trends Cell Biol. 2014;24:761–770. doi: 10.1016/j.tcb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, Counter CM, Kashatus DF. Erk2 Phosphorylation of Drp1 Promotes Mitochondrial Fission and Mapk-Driven Tumor Growth. Mol Cell. 2015;57:537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato RL, Chudnovsky Y, Pacold ME, Chen WW, Cantor JR, Shelton LM, Gui DY, Kwon M, Ramkissoon SH, Ligon KL, Kang SW, Snuderl M, Vander Heiden MG, Sabatini DM. Shmt2 Drives Glioma Cell Survival in Ischaemia but Imposes a Dependence on Glycine Clearance. Nature. 2015;520:363–367. doi: 10.1038/nature14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, Travins J, Weiss S, Looper R, Ligon KL, Verhaak RG, Yan H, Kaelin WG., Jr Transformation by the (R)-Enantiomer of 2-Hydroxyglutarate Linked to Egln Activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGory EL, Wu C, Taniguchi CM, Ding CK, Chi JT, von Eyben R, Scott DA, Richardson AD, Giaccia AJ. Suppression of Pgc-1alpha Is Critical for Reprogramming Oxidative Metabolism in Renal Cell Carcinoma. Cell Rep. 2015;12:116–127. doi: 10.1016/j.celrep.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R, Harrison H, Hulit J, Smith DL, Lisanti MP, Sotgia F. Mitochondria as New Therapeutic Targets for Eradicating Cancer Stem Cells: Quantitative Proteomics and Functional Validation Via Mct1/2 Inhibition. Oncotarget. 2014;5:11029–11037. doi: 10.18632/oncotarget.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, Zimmerman LJ, Liebler DC, Slebos RJ, Lorkiewicz PK, Higashi RM, Fan TW, Dang CV. Glucose-Independent Glutamine Metabolism Via Tca Cycling for Proliferation and Survival in B Cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gal K, Ibrahim MX, Wiel C, Sayin VI, Akula MK, Karlsson C, Dalin MG, Akyurek LM, Lindahl P, Nilsson J, Bergo MO. Antioxidants Can Increase Melanoma Metastasis in Mice. Sci Transl Med. 2015;7:308re308. doi: 10.1126/scitranslmed.aad3740. [DOI] [PubMed] [Google Scholar]
- LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, Asara JM, Kalluri R. Pgc-1alpha Mediates Mitochondrial Biogenesis and Oxidative Phosphorylation in Cancer Cells to Promote Metastasis. Nat Cell Biol. 2014;16:992–1003. 1001–1015. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, Worth AJ, Yuan ZF, Lim HW, Liu S, Jackson E, Aiello NM, Haas NB, Rebbeck TR, Judkins A, Won KJ, Chodosh LA, Garcia BA, Stanger BZ, Feldman MD, Blair IA, Wellen KE. Akt-Dependent Metabolic Reprogramming Regulates Tumor Cell Histone Acetylation. Cell Metab. 2014;20:306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc Stimulates Nuclearly Encoded Mitochondrial Genes and Mitochondrial Biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, He J, Hunter T, Wang L, Lu Z. Mitochondria-Translocated Pgk1 Functions as a Protein Kinase to Coordinate Glycolysis and the Tca Cycle in Tumorigenesis. Mol Cell. 2016;61:705–719. doi: 10.1016/j.molcel.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J, Tait SW. Mitochondrial Apoptosis: Killing Cancer Using the Enemy Within. Br J Cancer. 2015;112:957–962. doi: 10.1038/bjc.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley GS, Root DE, Ebert BL, Kaelin WG., Jr (R)-2-Hydroxyglutarate Is Sufficient to Promote Leukemogenesis and Its Effects Are Reversible. Science. 2013;339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O’Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. Idh Mutation Impairs Histone Demethylation and Results in a Block to Cell Differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie ED, Selak MA, Tennant DA, Payne LJ, Crosby S, Frederiksen CM, Watson DG, Gottlieb E. Cell-Permeating Alpha-Ketoglutarate Derivatives Alleviate Pseudohypoxia in Succinate Dehydrogenase-Deficient Cells. Mol Cell Biol. 2007;27:3282–3289. doi: 10.1128/MCB.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias JD, Kimmelman AC. Mechanisms of Selective Autophagy in Normal Physiology and Cancer. J Mol Biol. 2016;428:1659–1680. doi: 10.1016/j.jmb.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinou JC, Youle RJ. Mitochondria in Apoptosis: Bcl-2 Family Members and Mitochondrial Dynamics. Dev Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Nakanishi A, Minami A, Wada Y, Kitagishi Y. Functions and Characteristics of Pink1 and Parkin in Cancer. Front Biosci (Landmark Ed) 2015;20:491–501. doi: 10.2741/4321. [DOI] [PubMed] [Google Scholar]
- Mishra P, Chan DC. Metabolic Regulation of Mitochondrial Dynamics. J Cell Biol. 2016;212:379–387. doi: 10.1083/jcb.201511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Hulea L, Larsson O, Pollak M, St-Pierre J, Topisirovic I. Mtor Coordinates Protein Synthesis, Mitochondrial Activity and Proliferation. Cell Cycle. 2015;14:473–480. doi: 10.4161/15384101.2014.991572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucaj V, Shay JE, Simon MC. Effects of Hypoxia and Hifs on Cancer Metabolism. Int J Hematol. 2012;95:464–470. doi: 10.1007/s12185-012-1070-5. [DOI] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive Carboxylation Supports Growth in Tumour Cells with Defective Mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, O’Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD. A Functional Screen for Myc-Responsive Genes Reveals Serine Hydroxymethyltransferase, a Major Source of the One-Carbon Unit for Cell Metabolism. Mol Cell Biol. 2002;22:5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic Enzyme Expression Highlights a Key Role for Mthfd2 and the Mitochondrial Folate Pathway in Cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki S, Gottlieb E. Oncometabolites: Tailoring Our Genes. FEBS J. 2015;282:2796–2805. doi: 10.1111/febs.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa A, Wan L, Bonora M, Salmena L, Song MS, Hobbs RM, Lunardi A, Webster K, Ng C, Newton RH, Knoblauch N, Guarnerio J, Ito K, Turka LA, Beck AH, Pinton P, Bronson RT, Wei W, Pandolfi PP. Cancer-Associated Pten Mutants Act in a Dominant-Negative Manner to Suppress Pten Protein Function. Cell. 2014;157:595–610. doi: 10.1016/j.cell.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, Morrison SJ. Oxidative Stress Inhibits Distant Metastasis by Human Melanoma Cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porporato PE, Payen VL, Perez-Escuredo J, De Saedeleer CJ, Danhier P, Copetti T, Dhup S, Tardy M, Vazeille T, Bouzin C, Feron O, Michiels C, Gallez B, Sonveaux P. A Mitochondrial Switch Promotes Tumor Metastasis. Cell Rep. 2014;8:754–766. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- Pyakurel A, Savoia C, Hess D, Scorrano L. Extracellular Regulated Kinase Phosphorylates Mitofusin 1 to Control Mitochondrial Morphology and Apoptosis. Mol Cell. 2015;58:244–254. doi: 10.1016/j.molcel.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. Ras Oncogenes: Weaving a Tumorigenic Web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault TT, Floros KV, Elkholi R, Corrigan KA, Kushnareva Y, Wieder SY, Lindtner C, Serasinghe MN, Asciolla JJ, Buettner C, Newmeyer DD, Chipuk JE. Mitochondrial Shape Governs Bax-Induced Membrane Permeabilization and Apoptosis. Mol Cell. 2015;57:69–82. doi: 10.1016/j.molcel.2014.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, Barneda D, Sellers K, Campos-Olivas R, Grana O, Viera CR, Yuneva M, Sainz B, Jr, Heeschen C. Myc/Pgc-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab. 2015;22:590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- Sarosiek KA, Chi X, Bachman JA, Sims JJ, Montero J, Patel L, Flanagan A, Andrews DW, Sorger P, Letai A. Bid Preferentially Activates Bak While Bim Preferentially Activates Bax, Affecting Chemotherapy Response. Mol Cell. 2013;51:751–765. doi: 10.1016/j.molcel.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell JC, Olson KA, Jiang L, Hawkins AJ, Van Vranken JG, Xie J, Egnatchik RA, Earl EG, DeBerardinis RJ, Rutter J. A Role for the Mitochondrial Pyruvate Carrier as a Repressor of the Warburg Effect and Colon Cancer Cell Growth. Mol Cell. 2014;56:400–413. doi: 10.1016/j.molcel.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senft D, Ronai ZA. Regulators of Mitochondrial Dynamics in Cancer. Curr Opin Cell Biol. 2016;39:43–52. doi: 10.1016/j.ceb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serasinghe MN, Wieder SY, Renault TT, Elkholi R, Asciolla JJ, Yao JL, Jabado O, Hoehn K, Kageyama Y, Sesaki H, Chipuk JE. Mitochondrial Division Is Requisite to Ras-Induced Transformation and Targeted by Oncogenic Mapk Pathway Inhibitors. Mol Cell. 2015;57:521–536. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Horvath TL. Mitochondrial Ros Signaling in Organismal Homeostasis. Cell. 2015;163:560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. Myc, Metabolism, and Cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Chandel NS. Mitochondrial Reactive Oxygen Species and Cancer. Cancer Metab. 2014;2:17. doi: 10.1186/2049-3002-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Martinez-Garcia E, Nguyen H, Mullen AR, Dufour E, Sudarshan S, Licht JD, Deberardinis RJ, Chandel NS. The Proto-Oncometabolite Fumarate Binds Glutathione to Amplify Ros-Dependent Signaling. Mol Cell. 2013;51:236–248. doi: 10.1016/j.molcel.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Luo X, Xiao L, Tang M, Bode AM, Dong Z, Cao Y. The Role of Pgc1alpha in Cancer Metabolism and Its Therapeutic Implications. Mol Cancer Ther. 2016;15:774–782. doi: 10.1158/1535-7163.MCT-15-0621. [DOI] [PubMed] [Google Scholar]
- Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, Miletic H, Sakariassen PO, Weinstock A, Wagner A, Lindsay SL, Hock AK, Barnett SC, Ruppin E, Morkve SH, Lund-Johansen M, Chalmers AJ, Bjerkvig R, Niclou SP, Gottlieb E. Glutamine Synthetase Activity Fuels Nucleotide Biosynthesis and Supports Growth of Glutamine-Restricted Glioblastoma. Nat Cell Biol. 2015;17:1556–1568. doi: 10.1038/ncb3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama EQ, Herzig S, Courchet J, Lewis TL, Jr, Loson OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC, Shaw RJ. Metabolism. Amp-Activated Protein Kinase Mediates Mitochondrial Fission in Response to Energy Stress. Science. 2016;351:275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacanti NM, Divakaruni AS, Green CR, Parker SJ, Henry RR, Ciaraldi TP, Murphy AN, Metallo CM. Regulation of Substrate Utilization by the Mitochondrial Pyruvate Carrier. Mol Cell. 2014;56:425–435. doi: 10.1016/j.molcel.2014.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gisbergen MW, Voets AM, Starmans MH, de Coo IF, Yadak R, Hoffmann RF, Boutros PC, Smeets HJ, Dubois L, Lambin P. How Do Changes in the Mtdna and Mitochondrial Dysfunction Influence Cancer and Cancer Therapy? Challenges, Opportunities and Models. Mutat Res Rev Mutat Res. 2015;764:16–30. doi: 10.1016/j.mrrev.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Vaseva AV, Moll UM. The Mitochondrial P53 Pathway. Biochim Biophys Acta. 2009;1787:414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, Kost-Alimova M, Muller F, Colla S, Nezi L, Genovese G, Deem AK, Kapoor A, Yao W, Brunetto E, Kang Y, Yuan M, Asara JM, Wang YA, Heffernan TP, Kimmelman AC, Wang H, Fleming JB, Cantley LC, DePinho RA, Draetta GF. Oncogene Ablation-Resistant Pancreatic Cancer Cells Depend on Mitochondrial Function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eyss B, Jaenicke LA, Kortlever RM, Royla N, Wiese KE, Letschert S, McDuffus LA, Sauer M, Rosenwald A, Evan GI, Kempa S, Eilers M. A Myc-Driven Change in Mitochondrial Dynamics Limits Yap/Taz Function in Mammary Epithelial Cells and Breast Cancer. Cancer Cell. 2015;28:743–757. doi: 10.1016/j.ccell.2015.10.013. [DOI] [PubMed] [Google Scholar]
- Vyas S, Chang P. New Parp Targets for Cancer Therapy. Nat Rev Cancer. 2014;14:502–509. doi: 10.1038/nrc3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Song M, Zeng ZL, Zhu CF, Lu WH, Yang J, Ma MZ, Huang AM, Hu Y, Huang P. Identification of Ndufaf1 in Mediating K-Ras Induced Mitochondrial Dysfunction by a Proteomic Screening Approach. Oncotarget. 2015;6:3947–3962. doi: 10.18632/oncotarget.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the Origin of Cancer Cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB. The Common Feature of Leukemia-Associated Idh1 and Idh2 Mutations Is a Neomorphic Enzyme Activity Converting Alpha-Ketoglutarate to 2-Hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhouse S. On Respiratory Impairment in Cancer Cells. Science. 1956;124:267–269. doi: 10.1126/science.124.3215.267. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. Atp-Citrate Lyase Links Cellular Metabolism to Histone Acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Ko B, Hensley CT, Jiang L, Wasti AT, Kim J, Sudderth J, Calvaruso MA, Lumata L, Mitsche M, Rutter J, Merritt ME, DeBerardinis RJ. Glutamine Oxidation Maintains the Tca Cycle and Cell Survival During Impaired Mitochondrial Pyruvate Transport. Mol Cell. 2014;56:414–424. doi: 10.1016/j.molcel.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, Qing G, Liu Z, Simon MC, Rabinowitz JD, Thompson CB. Serine Catabolism Regulates Mitochondrial Redox Control During Hypoxia. Cancer Discov. 2014;4:1406–1417. doi: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. Hif-1 Inhibits Mitochondrial Biogenesis and Cellular Respiration in Vhl-Deficient Renal Cell Carcinoma by Repression of C-Myc Activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]