Summary
Background
Recent analyses of de novo DNA mutations in modern humans have suggested a nuclear substitution rate that is approximately half that of previous estimates based on fossil calibration. This result has led to suggestions that major events in human evolution occurred far earlier than previously thought.
Result
Here we use mitochondrial genome sequences from 10 securely dated ancient modern humans spanning 40,000 years as calibration points for the mitochondrial clock, thus yielding a direct estimate of the mitochondrial substitution rate. Our clock yields mitochondrial divergence times that are in agreement with earlier estimates based on calibration points derived from either fossils or archaeological material. In particular, our results imply a separation of non-Africans from the most closely related sub-Saharan African mitochondrial DNAs (haplogroup L3) of less than 62,000-95,000 years ago.
Conclusion
Though single loci like mitochondrial DNA (mtDNA) can only provide biased estimates of population split times, they can provide valid upper bounds; our results exclude most of the older dates for African and non-African split times recently suggested by de novo mutation rate estimates in the nuclear genome.
Introduction
Differences in DNA sequences correspond to nucleotide substitutions that have accumulated since their split from a most recent common ancestor (MRCA). When the average number of substitutions occurring per unit time can be determined, the “molecular clock” rate can be estimated. Under the assumption of constant rates of change amongst lineages, molecular clocks have been used to estimate divergence times between closely related species, or between populations. Fossil evidence has been frequently used to estimate a date for the MRCA of two related groups, thus providing a calibration point for the molecular clock. The sparseness of the fossil record, however, poses limitations on the reliability of such estimates. For example, in human evolution, no fossil has yet been identified to represent the uncontested MRCA for humans and chimpanzees or other closely related primate species. As a consequence, the nuclear and mitochondrial mutation rates for the human lineage have been heavily debated[1].
Recent analyses of de novo substitutions from genome sequencing of parent and offspring trios allow the direct calculation of nuclear substitution rates per generation. This alternative to the fossil calibration of the human molecular clock is arguably more accurate. Surprisingly, publications using this approach have recently pointed to de novo rates that are about half the value of those previously determined from fossil calibrations[1-5]. A slower substitution rate has important implications for inferring the timing of key events in human evolution such as our divergence from our common ancestor with chimpanzees, our divergence from Neandertals and Denisovans, and the migration of modern humans from one region or habitat to another. Taking these new rates into consideration, most date estimates would be pushed back by a factor of two, for example yielding a West African / non-African split date of 90-130 kya, which is up to 60 kya older than some previous estimates[1].
Attempts at calculating the human mitochondrial DNA (mtDNA) substitution rate have relied on either estimates derived from fossil calibration or archaeological evidence of founding migrations; however, reliance on a single calibration date can easily lead to a biased rate estimate[6]. Estimates of substitution rates for the coding region of the mtDNA have ranged from 1.26×10−8 substitutions per site per year[7], calibrated with a chimpanzee-human divergence time of 6.5 million years before present (BP), to 1.69×10−8, inferred from an accepted date of 45 kya for the peopling of Oceania[8]. This 45kya date is based on radiocarbon dated archaeological material, thus providing a calibrated age for the haplogroup (hg) Q lineage that is unique to contemporary populations from this region. Soares et al.[9] considered both the coding and non-coding regions in their estimate to accommodate the effects of natural selection, and arrived at a rate of 1.67×10−8 substitutions per site per year using a calibration point of 7 million years BP for the chimpanzee-human split.
An alternative approach for obtaining greater precision in measuring substitution rates is through the analysis of genetic data from ancient samples for which reliable radiocarbon dates are available. Ancient humans are well-suited to provide calibration points for the human mitochondrial molecular clock: reliable radiocarbon dates are available for many specimens, hence the number of substitutions that have accumulated amongst lineages can be directly translated into the number of substitutions per site per year. Branch shortening, the effect of fewer substitutions on extinct branches when visually depicted on a phylogenetic tree, is commonly observed in phylogenetic studies of ancient humans[10], and directly corresponds to the number of substitutions that are absent in the ancient human but present in the derived state in the descendant groups.
Here we use the complete or nearly complete mitochondrial genomes from 10 ancient modern humans for which reliable radiocarbon dates are available to calculate the human mtDNA substitution rate directly. This strategy circumvents the limitations imposed by the use of indirect measures of substitution rates such as those obtained via fossil calibration. The samples used in this analysis span 40,000 years of human history and originate from Europe and Eastern Asia. We use our substitution rate to estimate the dates of major human evolutionary events in the last 200,000 years. Of particular note, our rate suggests an upper bound on the split between non-Africans and sub-Saharan Africans of less than 95,000 years ago. Even though this estimate is a conservative upper bound because a single locus can only establish a biased estimate of a split time, it is on the extreme lower end population split times estimated from de novo substitution rates for nuclear DNA[1].
Results
DNA Preservation and Contamination
High throughput sequencing of the enriched libraries yielded between 4,898 and 27,022,382 sequences for each sample. These sequences were used as input for the iterative mapping assembler MIA[11]. For each sample, between 0 and 74,435 unique human mtDNA fragments mapped against the revised Cambridge Reference Sequence (rCRS). Complete or nearly complete mitochondrial genomes with at least 3-fold coverage could be reconstructed for 11 individuals (Table 1) including the sample from China. The average length of the obtained mtDNA fragments ranged from 54 to 77bp (Table S6). Using a previously published contamination estimation method[11], four samples showed a low percentage of inconsistent fragments suggesting that the DNA originated from a single biological source, while the other sequences did not contain enough unique diagnostic substitutions to assess contamination using this method. Our more powerful Bayesian contamination estimate (SI text 5) suggests that all samples with the exception of Oberkassel 999 are uncontaminated to within the limits of our resolution (Table 1). On this basis, we excluded Oberkassel 999 from subsequent analyses. To further evaluate the authenticity of the ancient DNA we calculated the proportion of nucleotide misincorporations arising from DNA damage, a quantity that is known to increase over time after the death of an individual[12] and has been used as an indication of authenticity in previous work[10]. It was suggested that bone samples 100 years and older have a minimum of 20% C to T misincorporations concentrated at the 5’ end of the molecule[13]. Using this criterion, we excluded Paglicci Str. 4b from further analysis as the rate of C to T misincorporation at the 5’-end was only 8.8%, thus making an ancient origin for the DNA in this sample uncertain[14].
Table 1.
Samples analyzed in this study
| Sample |
14C age (calBP) |
Lab Number | nt covered at 3- fold coverage (% of mtDNA) |
Contamination based on [34] |
Contamination (likelihood estimate) |
C to T misincorporation at 5’ end |
Hg |
|---|---|---|---|---|---|---|---|
| Tianyuan 1301[29]a | 39,475 ± 645 | BA-03222 | 16559 (99,9%) | 0-4.7% | 0.9 – 4.7% | 31.5 | B |
| Kostenki 14[10]a | 37,985 ± 665 | OxA-X-2395-15 | 16568 (100%) | 0 – 7% | 1.7 – 8.5% | 38 | U2 |
| Dolni Vestonice 13a,b | 31,155 ± 85 | GrN-14,831 | 16570 (100%) | 0 – 2.3% | 0.9 – 2.4% | 30.1 | U8 |
| Dolni Vestonice 14a,b | 31,155 ± 85 | GrN-14,831 | 16530 (99,8%) | 0 – 100% | 1.9 – 9.2% | 24.4 | U |
| Dolni Vestonice 15b | 31,155 ± 85 | GrN-14,831 | 16051 (96,9%) | 0 – 100% | 0 – 3.9% | 20.1 | U |
| Oberkassel 998a,b | 14,020 ± 150 | OxA-4790 | 16570 (100%) | 0 – 100% | 0.5 – 2.4% | 36.0 | U5b1 |
| Oberkassel 999b | 13,430 ± 140 | OxA-4792 | 16560 (99,9%) | 0 – 100% | 3.8 – 8.3% | 31.5 | U5b1 |
| Continenza 7b | N/A | 16478 (99,5%) | 0.3 – 2.8% | 0.9 – 7.2% | 29.1 | U5b2b1 | |
| Paglicci Accesso sala 2 Rimb | N/A | 16548 (99,9%) | 0 – 5.3% | 0.3 – 2.8% | 57.1 | U2′3′4′7′8′9 | |
| Paglicci Str. 4bb | N/A | 16509 (99,6%) | 0 – 4.8% | 1.5 – 5.1% | 8.8 | H1 | |
| Boshan 11a,b | 8,180 ± 140 | MAMS-13530 | 16559 (99,9%) | 0 – 1.8% | 47.9 | B4c1a | |
| Loschboura,b | 8054 ± 127 | OxA-7338 | 16569 (100%) | 0 – 0.5% | 1.3 – 1.9% | 27.6 | U5b1a |
| Iceman[30]a | 4,550 | OxA-3371–6 OxA-3419-21 |
16576 (100%) | na | na | na | K1 |
| Saqqaq[31]a | 3,600-4,170 | OxA-20656 | 16568 (100%) | na | na | na | D2a1 |
| Cro-Magnon 1a,b | 690 ± 39 | OxA-V-2321-38 | 16499 (99,6%) | 0 – 7.7% | 0.3 – 3.3% | na | T2b1 |
Sequence used for estimating substitution rate via linear regression and Bayesian framework.
Sequence presented in this study.
Evolutionary analysis
All but one of the ancient modern human sequences from Europe belonged to mtDNA hg U, thus confirming previous findings that hg U was the dominant type of mtDNA before the spread of agriculture into Europe[15]. The exception was the Cro-Magnon 1 sample which belonged to the derived hg T2b1, an unexpected hg given the putative 30,000 year age of the sample[16]. Since the radiocarbon date for this specimen was obtained from an associated shell[16], we AMS dated the sample itself. Surprisingly, the sample had a much younger age of about 700 years, suggesting a medieval origin. Consequently, this bone fragment has now been removed from the Cro-Magnon collection at the Musée de l'Homme in Paris. Attempts to directly date other remains from the Cro-Magnon type collection unfortunately failed. The good molecular preservation of our sample for both DNA and AMS dating, in contrast, suggests that this particular bone has a different origin than the other remains in the collection.
For the remaining eight ancient Europeans, we built a phylogenetic tree for hg U, which included 63 contemporary European mtDNAs falling into this haplogroup. The tree clearly shows that all four Paleolithic pre-Last Glacial Maximum (LGM) samples display a short branch compared to the four ancient post-LGM samples (Figure S2). Predictably, the older samples Dolni Vestonice 14 and 15 fall in a basal position relative to the contemporary mtDNA hg U5. The Tianyuan sequence from Eastern China falls basal to the contemporary hg B, common in most parts of Eastern Asia, Oceania, and the Americas. The mtDNA genetic diversity that we measure in early modern Europeans is about two-fold less than the mtDNA diversity in today's Europeans, but about 1.5 times higher than that measured in Neandertals contemporary with these early modern humans[17] (excluding the older Mezmaiskaya individual) (Table S3). While these measurements provisionally suggest that a higher population size might have contributed to early modern humans out-competing Neandertals after their arrival in Europe, there are caveats to this analysis. First, we have a limited sample size of ancient specimens. Second, we have sampled from several different time periods, a practice which overestimates actual genetic diversity[17], though not in the Neanderthal population owing to its restricted mitochondrial diversity over time. Third, our sampling is non-random; for example, we included several individuals from within the same burial site (Dolni Vestonice), where maternal relatedness would give an underestimate of true diversity. More data are necessary to provide a definitive assessment of the genetic diversity of these prehistoric populations.
Substitution rate estimates
For the linear regression approach we estimate a substitution rate of 1.92×10−8 per site and year (1.16-2.68×10−8 95% CI) for the whole mtDNA and 1.25±0.68 10−8 per site and year (0.57-1.93 ×10−8 95% CI) for the coding region.
For the Bayesian approach, the final model was chosen based on a Log10 of the Bayes factor (BF) being >1.3. The best fit comparison of the results of the Bayesian MCMC analysis, calibrated with the fossil ages, favors the constant population size model over the exponential growth model (Log10 BF = 1087.2>>1.3). Although values obtained with the relaxed clock model fit the data better (Log10 BF = 3.6>1.3), the ML likelihood test does not reject the null hypothesis of a constant substitution rate across the tree topology. Using the constant size model and a relaxed clock we thus estimate a substitution rate of 2.67×10−8 substitutions per site per year (2.16-3.16 ×10−8 95% HPD) for the whole mtDNA genome and 1.57×10−8 substitutions per site per year for the coding region (1.17-1.98 ×10−8 95% HPD) (Table 2). The substitution rates for the mtDNA coding region and whole mtDNA largely overlap with the above results when the four radiocarbon-dated Neandertals are included alongside the 10 ancient modern humans (SI text 6, Table S5). In theory, mitochondrial substitution rates could have changed between Neandertals and modern humans, though we do not detect evidence of this because inclusion of Neandertal data does not lead to a rejection of the molecular clock. Regardlesss, we use only the substitution rates calculated with radiocarbon dated ancient modern humans to calculate modern human mtDNA divergence times, as this presumably affords greater accuracy.
Table 2.
Inferred TMRCA for all modern humans and mean substitution obtained for various subsets of the mtDNA genome assuming a constant population size and relaxed molecular clock
| mtDNA partition | TMRCA | μ/ site / year (units of 10−8) | ||||
|---|---|---|---|---|---|---|
| best estimate | lower 2.5% | upper 2.5% | best estimate | lower 2.5% | upper 2.5% | |
| whole mtDNA | 157,000 | 120,000 | 197,000 | 2.67 | 2.16 | 3.16 |
| Coding region | 178,000 | 126,000 | 236,000 | 1.57 | 1.17 | 1.98 |
| 1st-2nd Codon | 207,000 | 78,800 | 382,000 | 0.82 | 0.30 | 1.37 |
| 3rd Codon | 233,000 | 134,000 | 356,000 | 3.27 | 1.94 | 4.62 |
Haplogroup divergence time estimates
Using the substitution rate for the whole mtDNA genome obtained by Bayesian estimation, the time of the MRCA for all modern humans was estimated at 157 kya (120 – 197 kya 95% HPD). Our rate also implies a split of all non-African hgs from the closest widespread sub-Saharan African hg (L3) of 78.3 kya (62.4 - 94.9 kya). The MRCA of hg Q, often referred to as a maximum age for the settlement of Australia, was calculated at 42 kya (30 – 54.9 kya). The TMRCA of hg U5, often argued to have evolved within the first early modern humans in Europe[18] was calculated at 29.6 kya (22.7 – 37.2 kya).
To test if the inferred mutation rates are dependent on a single directly dated mtDNA sequence (which in principle could have an inaccurate carbon date), we carried out a Bayesian MCMC analysis for the coding regions with a constant size model and relaxed clock using each mtDNA sequence older than 4,000 years independently as separate tip calibrations. The results range from 1.14×10−8 substitutions per site per year for the Kostenki specimen to 4.5×10−8 substitutions per site per year for the Boshan specimen (Table S1). The confidence intervals for all samples overlap the value obtained for the whole dataset, suggesting that no single sample is driving our overall mutation rate estimate.
Discussion
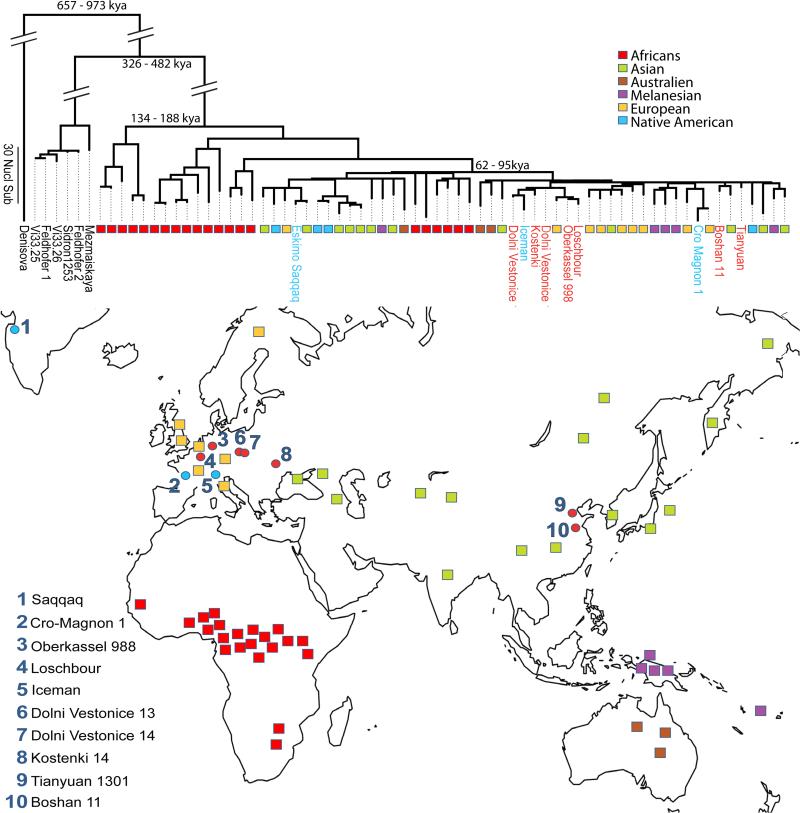
We were able to reconstruct three complete and six nearly complete mitochondrial genomes from ancient human remains found in Europe and Eastern Asia that span 40,000 years of human history. All Paleolithic and Mesolithic European samples belong to mtDNA hg U, as was previously suggested for pre-Neolithic Europeans[15]. Two of the three individuals from the Dolni Vestonice triple burial associated with the pre-ice age Gravettian culture, namely 14 and 15, show identical mtDNAs, suggesting a maternal relationship. Furthermore, both individuals display a mitochondrial sequence that falls basal in a phylogenetic tree compared to the post-ice age hunter gatherer samples from Italy and central Europe, as well as the contemporary mtDNA hg U5 (Figure 1). It has been argued that hg U5 is the most ancient sub-haplogroup of the U lineage, originating among the first early modern humans in Europe[18]. Our results support this hypothesis since we find that the two Dolni Vestonice individuals radiocarbon dated to 31.5 kya carry a type of mtDNA that is as yet uncharacterized, sits close to the root of hg U, and carries two mutations that are specific to hg U5. With our recalibrated molecular clock, we date the age of the U5 branch to approximately 30 kya, thus predating the LGM. Since the majority of late Paleolithic and Mesolithic mtDNAs analyzed to date fall on one of the branches of U5 (see also ref. [15]), our data provide strong support for maternal genetic continuity between the pre- and post-ice age European hunter-gatherers from the time of first settlement to the onset of the Neolithic. U4, another hg commonly found in Mesolithic hunter gatherers[15], has so far not been sequenced in a Paleolithic individual, and we find hgs U8 and U2 in pre-LGM individuals but not in later hunter-gatherers. At present, the genetic data on Upper Paleolithic and especially pre-ice age populations are too sparse to comment on whether or not this is representative of a change in the genetic structure of the population, perhaps caused by a bottleneck during the LGM and a subsequent repopulation from glacial refugia.
Figure 1. Tree for 54 present-day humans, 10 ancient modern humans, and 7 archaic humans.
The phylogeny in the top panel was constructed using Maximum Parsimony and rooted using midpoint rooting. The branches for present-day humans do not all end at the same point giving a sense of the inherent uncertainty in time measurements based on mtDNA due to its limited sequence span. However, the consistent shortening of the branches of ancient humans relative to their closest present-day human relatives is apparent in the figure. This is the basis for our clock calibration. Pre- and post-Neolithic ancient samples are indicated as red and blue circles, respectively, and colored squares indicate the geographical origin of 54 present-day humans that we co-analyzed with them. Date estimates for major divergence events are shown at the nodes. In the bottom panel we show a map giving geographical origin of the samples.
Using ancient mtDNA sequences from securely dated archaeological samples as calibration points has allowed us to obtain an estimate of the mtDNA substitution rate that is more reliable than the existing estimates based on calibration from the fossil and archaeological records. We arrive at a rate of 1.57×10−8 substitutions per site per year for the coding region and 2.67×10−8 substitutions per site per year for the whole molecule, which is approximately 1.6-fold higher than the fossil calibrated rate[7]. Our inferred substitution rate from the whole mtDNA implies a coalescence date for all modern human mtDNAs of 120-197 kya and of 62-95 kya for hg L3, the lineage from which all non-African mtDNA hgs descend. This places a conservative upper bound of 95 kya for the time of the last major gene exchange between non-African and sub-Saharan African populations. It is important to recognize that this divergence time may merely represent the most recent gene exchanges between the ancestors of non-Africans and the most closely related sub-Saharan Africans, and thus may reflect only the most recent population split in a long, drawn out process of population separation[1]. Nevertheless, the fact that hg L3 is currently so widespread within Africa suggests that the split dated by L3 is likely to be one of the most important ones in that history of separation, giving rise to lineages that contributed substantial fractions to the ancestry of both present-day sub-Saharan African and present-day non-African populations.
While our estimate for the population divergence of non-African and sub-Saharan Africans has a small overlap with those calculated from the de novo genomic rates, which range from 90 – 130 kya1, the ca. 30,000 year difference in the mean divergence time obtained via the different methods is worthy of discussion. We believe this discrepancy is unlikely to be explained by differences in the inheritance patterns between the two parts of the genome (exclusively maternal vs. biparental), or by differences in generation times of males and females[19] as branch shortening is calculated in years and thus not affected by changes in generation times. We note that our calculated dates are more consistent with some interpretations of the fossil record: for example, the low nuclear mutation rates from the de novo investigations imply a date of human-orangutan speciation that is at least ten million years older than what is supported by the fossil record[20], whereas our dates are in accord with a more recent ape speciation[21, 22]. One possible reason for the discrepancies between our inferences and those that have been made based on de novo mutation rates in the nuclear genome, is the possibility of a substantial rate of false-negative mutations in the de novo data sets (due to the intense filtering that these studies need to apply to discriminate false-positive mutations from true positives). It is also possible that the filtering applied in the de novo mutation rate estimation studies has excluded subsets of the genome that are more mutable and that have been included in sequence divergence calculations; it is important to estimate mutation rate and sequence divergence in the same subsets of the genome to properly calibrate time estimates and this has not been done as far as we are aware. An important direction for future research will be to compare the new de novo rates with those estimated based on patterns of substitutions observed over time in the nuclear genome as determined from ancient sequences[23], using methods similar to those employed here. This would also help to identify any demographic signals that were imperceptible in the current analysis due to our exclusive use of mitochondrial genomes.
Experimental Procedures
Samples, DNA extraction, and molecular processing
DNA extraction was performed on skeletal remains from 53 humans from Europe. Descriptions for these can be found in the SI text 1), as well as Tables 1 and Table S6. Details of DNA extraction, enrichment, and Illumina sequencing are available in the SI text 2, 3). The GenBank accession number of the mtDNA consensus sequences determined in this study are KC521454 (Boshan 11), KC521455 (Loschbour), KC521456 (Cro-Magnon 1), KC521457 (Oberkassel 998), KC521458 (DolniVestonice 14) and KC521459 (DolniVestonice 13).
Phylogenetic analysis
The consensus sequences were assigned to haplogroups (Table S2) according to Phylotree.org[24] with a custom PERL script. They were aligned using the software MUSCLE[25]. MEGA 5[26] was used to calculate mean pairwise differences and to generate a Maximum Parsimony tree, which included the sequences obtained here along with previously published early modern human mtDNAs with radiocarbon dates, 54 contemporary modern human mtDNAs from a worldwide distribution[27], six Neandertal, and one Denisovan mtDNA[17, 28] (Fig. 1).
Estimation of substitution rates
For the direct calculation of the human mtDNA substitution rate we used ten samples. These included six samples (out of a total of 54 ancient modern human remains mentioned above) that were reliably dated and for which complete or near complete mtDNA sequences had been generated at a minimum of 3-fold coverage. The remaining four samples came from previously published early modern human mtDNA sequences. The specific sequences we analyzed were Dolni Vestonice 13 and 14, Tianyuan[29], Boshan, Cro-Magnon 1, Oberkassel 998, Kostenki[10], Iceman[30], Saqqaq[31] and Loschbour (Table 1). Radiocarbon dates were calibrated with OxCal 4.1 with the dataset INTCAL09.
The complete mtDNAs were aligned using the software MUSCLE with a worldwide dataset of 311 contemporary mtDNAs[25]. Both ancient Italian samples had no reliable 14C date and were therefore not used to estimate the rate of mtDNA substitutions in humans.
To calculate the substitution rate we used a linear regression model as well as a Bayesian analysis. For the linear regression, nucleotide distances (Figure S1 A,B) to the common ancestor of all mtDNAs that fall into haplogroup R were calculated for all applicable ancient and modern-day sequences using the software MEGA5. Suitable ancient samples were identified as those with a minimum of 99.5% of base positions covered 3-fold, and which were descendents of the R lineage, thus excluding the Saqqaq individual. The number of substitutions per year was then obtained as the slope of the regression of radiocarbon age and nucleotide distance (Figure S1 C,D). The obtained rate was divided by the number of positions to calculate the rate per site per year. While the regression approach is straightforward, it does not make the most efficient use of the available information as it weights all samples equally despite their shared evolutionary history. Our second approach avoids this problem by calculating substitution rates in a Bayesian framework. Using the software package BEAST[32], we explicitly accounted for the shared phylogeny of the 311 mtDNAs and 10 ancient human mtDNAs. The general time reversible sequence substitution model with a fixed fraction of invariable sites and gamma distributed rates (GTR+I+G) was used as this model is the best-fit to our data according to Modeltest and PAUP*[33]. In addition, two different models of rate variation among branches were investigated: a strict clock and an uncorrelated lognormal-distributed relaxed clock. For these two models, both a constant population size coalescent and an exponential growth coalescent were used as tree priors. Thus, we analyzed 4=2×2 models in total. For each model, two MCMC runs were carried out with 30,000,000 iterations each, sampling every 1000 steps. The first 6,000,000 iterations were discarded as burn-in. For each model both independent runs were combined resulting in 48,000,000 iterations.
To calculate substitution rates for various partitions of the mtDNA, MCMC runs for all four models were carried out on four subsets of the mtDNA alignments. The first corresponded to the whole mtDNA sequence, the second to the coding region (position 577-16023), the third to the first and second codon positions of the protein-coding genes, and the fourth to the third codon position.
Archaic humans
As we do not know the radiocarbon age for some of the complete Neandertal mtDNAs and the Denisovan individual, the TMRCA of modern and archaic human mtDNAs was estimated from our inferred substitution rate. For this approach we aligned 54 modern human mtDNAs from a worldwide dataset with six Neandertal mtDNAs, one Denisovan, a chimpanzee and a bonobo using MUSCLE. A molecular clock test was performed using the MEGA5 by comparing the maximum likelihood value for the given topology with and without the molecular clock constraints under the General Time Reversible model (+G+I). Differences in evolutionary rates among sites were modeled using a discrete Gamma (G) distribution that allowed for invariant (I) sites. The null hypothesis of equal evolutionary rate throughout the tree was not rejected (P = 0.181, SI text 4, Table S4). Hence, we used the calculated substitution rate to extrapolate the TMRCA of modern and archaic human mtDNAs. All positions containing gaps and missing data were eliminated. There were a total of 16,518 positions in the final dataset. To calculate mtDNA divergence times the determined substitution rate computed for the whole mtDNA genome (from the previous section, based on 10 ancient mtDNAs) was used as a prior in the Bayesian software package BEAST.
Supplementary Material
Table 3.
TMRCA of different haplogroups
| mtDNA | Whole molecule | 95% HPD | Coding region | 95% HPD | ||
|---|---|---|---|---|---|---|
| Hg Group | TMRCA (KBP) | lower | upper | TMRCA (KBP) | lower | upper |
| A | 33.7 | 22.4 | 45.1 | 28.6 | 17.0 | 41.2 |
| C | 24.2 | 16.6 | 32.5 | 24.2 | 14.7 | 34.9 |
| D | 37.8 | 27.6 | 49.1 | 45.3 | 28.7 | 62.7 |
| H | 23.9 | 16.6 | 32.3 | 20.7 | 12.5 | 29.6 |
| J | 26.0 | 16.4 | 36.4 | 27.1 | 13.9 | 42.2 |
| L3 | 78.3 | 62.4 | 94.9 | 89.7 | 66.8 | 116.4 |
| M+N | 77.0 | 61.4 | 93.2 | 88.2 | 64.5 | 114.5 |
| P | 53.6 | 43.1 | 65.5 | 60.9 | 41.6 | 83.2 |
| Q | 42.0 | 30.0 | 54.9 | 39.8 | 25.6 | 55.7 |
| T | 21.1 | 13.0 | 29.8 | 17.4 | 9.0 | 26.8 |
| U5 | 29.6 | 22.7 | 37.2 | 34.4 | 22.8 | 47.7 |
| U6 | 35.7 | 24.7 | 47.6 | 31.8 | 20.4 | 44.6 |
| U | 52.1 | 44.3 | 60.9 | 56.1 | 44.0 | 70.5 |
| V | 13.5 | 7.1 | 20.3 | 15.5 | 7.5 | 24.5 |
Highlights.
Direct calculation of human mtDNA mutation rates, using complete mtDNAs reconstructed from 10 radiocarbon dated ancient modern humans. This circumvents the need for relying on traditional fossil or archaeological calibration points.
Improved molecular estimates for human evolutionary events, such as our divergence from Neandertal and Densiovan, as well as more reliable dates for major human migrations such as the Out of Africa movement and the settlement of Australia.
Acknowledgments
We are grateful to the following people for providing samples, support and advice during the course of the project: Janet Kelso, Matthias Meyer, Verena Schünemann, Tomislav Maricic, David Serre, the late Andre Langaney, Dominique Delsate, Ivor Jankovic, Pavao Rudan and Xing Gao. We furthermore thank Chris Stringer and the anonymous reviewer for comments on improvements of the manuscript. This study was funded by the DFG (KR 4015/1-1), the Chinese Academy of Sciences Strategic Priority Research Program (Grant XDA05130202), the Basic Research Data Projects (Grant 2007FY110200) of the Ministry of Science and Technology of China, NIH AI049334 , NIH grant GM100233, NSF HOMINID grant 1032255, SSHRC grant 756-2011-5010, and the Max Planck Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Scally A, Durbin R. Revising the human mutation rate: implications for understanding human evolution. Nat Rev Genet. 2012;13:745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 2.Awadalla P, Gauthier J, Myers RA, Casals F, Hamdan FF, Griffing AR, Cote M, Henrion E, Spiegelman D, Tarabeux J, et al. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. American journal of human genetics. 2010;87:316–324. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consortium GP. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Wong WS, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endicott P, Ho SY, Metspalu M, Stringer C. Evaluating the mitochondrial timescale of human evolution. Trends in ecology & evolution. 2009;24:515–521. doi: 10.1016/j.tree.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, et al. Natural selection shaped regional mtDNA variation in humans. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedlaender J, Schurr T, Gentz F, Koki G, Friedlaender F, Horvat G, Babb P, Cerchio S, Kaestle F, Schanfield M, et al. Expanding Southwest Pacific mitochondrial haplogroups P and Q. Molecular biology and evolution. 2005;22:1506–1517. doi: 10.1093/molbev/msi142. [DOI] [PubMed] [Google Scholar]
- 9.Soares P, Ermini L, Thomson N, Mormina M, Rito T, Rohl A, Salas A, Oppenheimer S, Macaulay V, Richards MB. Correcting for purifying selection: an improved human mitochondrial molecular clock. American journal of human genetics. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krause J, Briggs AW, Kircher M, Maricic T, Zwyns N, Derevianko A, Paabo S. A complete mtDNA genome of an early modern human from Kostenki, Russia. Curr Biol. 2010;20:231–236. doi: 10.1016/j.cub.2009.11.068. [DOI] [PubMed] [Google Scholar]
- 11.Green RE, Malaspinas AS, Krause J, Briggs AW, Johnson PL, Uhler C, Meyer M, Good JM, Maricic T, Stenzel U, et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briggs AW, Stenzel U, Johnson PL, Green RE, Kelso J, Prufer K, Meyer M, Krause J, Ronan MT, Lachmann M, et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14616–14621. doi: 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawyer S, Krause J, Guschanski K, Savolainen V, Paabo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PloS one. 2012;7:e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoneking M, Krause J. Learning about human population history from ancient and modern genomes. Nature Reviews Genetics. 2011;12:603–614. doi: 10.1038/nrg3029. [DOI] [PubMed] [Google Scholar]
- 15.Bramanti B, Thomas MG, Haak W, Unterlaender M, Jores P, Tambets K, Antanaitis-Jacobs I, Haidle MN, Jankauskas R, Kind CJ, et al. Genetic Discontinuity Between Local Hunter-Gatherers and Central Europe's First Farmers. Science. 2009 doi: 10.1126/science.1176869. [DOI] [PubMed] [Google Scholar]
- 16.Henry-Gambier D. Les fossiles de Cro-Magnon (Les Eyzies-de-Tayac, Dordogne): Nouvelles donnees sur leur Position chronologique et leur attribution culturelle. Bull. et Mém. de la Société d’Anthropologie de Paris. 2002;14:89–112. [Google Scholar]
- 17.Briggs AW, Good JM, Green RE, Krause J, Maricic T, Stenzel U, Lalueza-Fox C, Rudan P, Brajkovic D, Kucan Z, et al. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science. 2009;325:318–321. doi: 10.1126/science.1174462. [DOI] [PubMed] [Google Scholar]
- 18.Richards MB, Macaulay VA, Bandelt HJ, Sykes BC. Phylogeography of mitochondrial DNA in western Europe. Ann Hum Genet. 1998;62:241–260. doi: 10.1046/j.1469-1809.1998.6230241.x. [DOI] [PubMed] [Google Scholar]
- 19.Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 20.Sun JX, Helgason A, Masson G, Ebenesersdottir SS, Li H, Mallick S, Gnerre S, Patterson N, Kong A, Reich D, et al. A direct characterization of human mutation based on microsatellites. Nature genetics. 2012;44:1161–1165. doi: 10.1038/ng.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobolth A, Dutheil JY, Hawks J, Schierup MH, Mailund T. Incomplete lineage sorting patterns among human, chimpanzee, and orangutan suggest recent orangutan speciation and widespread selection. Genome research. 2011;21:349–356. doi: 10.1101/gr.114751.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews P, Cronin JE. The relationships of sivapithecus and ramapithecus and the evolution of the orangutan. Nature. 1982;297:541–546. doi: 10.1038/297541a0. [DOI] [PubMed] [Google Scholar]
- 23.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 25.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingman M, Kaessmann H, Paabo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 28.Krause J, Fu Q, Good JM, Viola B, Shunkov MV, Derevianko AP, Paabo S. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 2010;464:894–897. doi: 10.1038/nature08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Q, Meyer M, Gao X, Stenzel U, Burbano HA, Kelso J, Paabo S. DNA analysis of an early modern human from Tianyuan Cave, China. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1221359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ermini L, Olivieri C, Rizzi E, Corti G, Bonnal R, Soares P, Luciani S, Marota I, De Bellis G, Richards MB, et al. Complete mitochondrial genome sequence of the Tyrolean Iceman. Curr Biol. 2008;18:1687–1693. doi: 10.1016/j.cub.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 31.Gilbert MT, Kivisild T, Gronnow B, Andersen PK, Metspalu E, Reidla M, Tamm E, Axelsson E, Gotherstrom A, Campos PF, et al. Paleo-Eskimo mtDNA genome reveals matrilineal discontinuity in Greenland. Science. 2008;320:1787–1789. doi: 10.1126/science.1159750. [DOI] [PubMed] [Google Scholar]
- 32.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Posada D. Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr Protoc Bioinformatics. 2003 doi: 10.1002/0471250953.bi0605s00. Chapter 6, Unit 6 5. [DOI] [PubMed] [Google Scholar]
- 34.Green RE, Briggs AW, Krause J, Prufer K, Burbano HA, Siebauer M, Lachmann M, Paabo S. The Neandertal genome and ancient DNA authenticity. EMBO J. 2009;28:2494–2502. doi: 10.1038/emboj.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.