Abstract
Background
Early childhood malnutrition affects 113 million children worldwide, impacting health and increasing vulnerability for cognitive and behavioral disorders later in life. Molecular signatures after childhood malnutrition, including the potential for intergenerational transmission, remain unexplored.
Methods
We surveyed blood DNA methylomes (~483,000 individual CpG sites) in 168 subjects across two generations (G1,G2), including 50 G1 individuals hospitalized during the first year of life for moderate to severe protein energy malnutrition, then followed up to 48 years in the Barbados Nutrition Study. Attention deficits and cognitive performance were evaluated with Connors Adult Attention Rating Scale (CAARS) and Wechsler Abbreviated Scale of Intelligence (WASI). Expression of nutrition-sensitive genes was explored by qRT-PCR in rat prefrontal cortex.
Results
We identified altogether 134 nutrition-sensitive differentially methylated genomic regions (DMR), with the overwhelming majority (87%) specific for G1. Multiple neuropsychiatric risk genes, including COMT, IFNG, MIR200B, SYNGAP1 and VIP2R showed associations of specific methyl-CpGs with attention and IQ. Interferon Gamma (IFNG) expression was decreased in prefrontal cortex of rats showing attention deficits after developmental malnutrition.
Conclusions
Early childhood malnutrition entails long lasting epigenetic signatures associated with liability for attention and cognition, and limited potential for intergenerational transmission.
Keywords: Epigenetics, DNA Methylation, Childhood Malnutrition, Attention deficits, Cognition, Prefrontal Cortex
INTRODUCTION
Malnutrition in infancy and early childhood is a major public health challenge, affecting an estimated 113 million children worldwide and causing significant morbidity and mortality(1). This includes a poorly understood long-lasting vulnerability to psychiatric disease, with increased risk for attention deficits, personality disorders and impaired cognition, even decades after exposure(2–4). The underlying mechanisms remain poorly understood. Of note, (mal)nutrition during early prenatal periods could affect DNA methylation, a key mechanism for epigenetic regulation affecting genome organization and function without altering underlying DNA sequence(5). However, to date, it remains unclear whether or not there are long-lasting epigenetic changes in humans exposed to malnutrition in infancy, and whether such nutrition-sensitive DNA methylation signatures are associated with changes in brain function and behavior. Furthermore, nutrition-related effects on DNA methylation, particularly those operating around the time of conception, could potentially be passed on through the germline and affect offspring metabolic function and health(6, 7). However, it remains unclear whether epigenetic alterations exist in the offspring of a parent exposed to malnutrition in infancy, similar to the multi-generational transmission of DNA methylation signatures after maternal separation stress in postnatal mice(8, 9).
To gain first insights into these questions, we interrogated, on a genome-wide scale, blood DNA methylation in 168 subjects across two generations (referred to hereafter as G1 and G2). These included 50 G1 generation individuals who suffered from moderate-severe protein energy malnutrition during the first year after birth and their G2 offspring (Figure 1). Our population is unique, because it has been monitored and examined for 48 years, in the context of the Barbados Nutrition Study (BNS). The study, launched in 1967, provided critical insight into the longitudinal course of malnutrition-related neuropsychological functioning and vulnerability to psychiatric and medical disease2–4. Here, we provide multiple evidence that infant malnutrition triggers long-lasting DNA methylation changes associated with liability for defective attention and cognition in adult life.
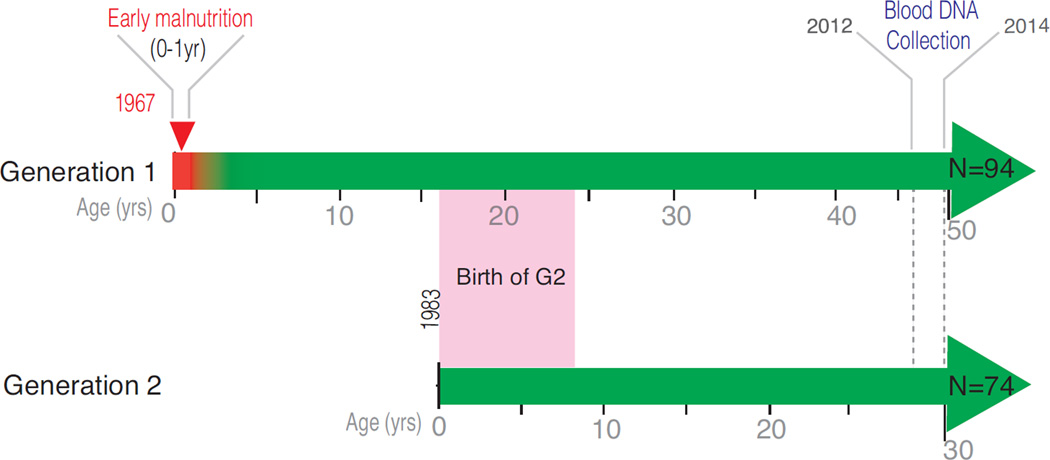
Figure 1. Overview and time line of the BNS DNA methylation study.
Generation G1 (N=94) included 44 controls and 50 subjects who were malnourished during the first year of life. Subjects with a history of early life malnutrition then enrolled in a Government–sponsored intervention program that included nutrition education, food subsidies, routine health care, home visits and a pre-school program that extended to 12 years of age. Approximately 44 years after exposure to malnutrition, blood was drawn by venipuncture for the DNA methylation scan. Generation G2 (N=74), all offspring of the parental G1 (44 G2 MAL, 30 G2 CON subjects), consistently had adequate nutritional status across the lifespan, and ranged from age 16–30 years when blood was drawn for DNA profiling.
METHODS
The research design and sample recruitment for the Barbados Nutrition Study have been described elsewhere(2, 4, 10). Epigenetic data were collected in the original cohort and their offspring between 2012 and 2014, when the G1 participants were in their fifth decade of life and their offspring were young adults (Figure 1). The current study included 94 adult G1 participants (N=50 MAL (malnourished in the first year of life); N= 44 CON (controls) with epigenetic and behavioral data. Also included are epigenetic data from 74 G2 children of G1 participants (N=44 offspring of the MAL group; N=30 offspring of the CON group). The total sample included 168 individuals (Table 1). Bisulfite-treated DNA from blood (venipuncture) was assessed with the Infinium HumanMethylation450 BeadChip (Illumina). Attention and cognition were assessed with Connors Adult Attention Rating Scale Self-Report Screening Version(11) and Wechsler Abbreviated Scale of Intelligence (WASI(12)). Childhood Standard of Living (SES) was assessed using an Ecology Questionnaire whose 50 items queried conditions in the household and the educational and employment history of the parents(13). Supplemental Methods provide details on 1. Study cohort, 2. blood sample collection, 3. Methylation profiling, 4. Data processing/statistical analysis, 5. Comparison of previously malnourished (MAL) and control (CON) groups, 6. Cognitive/behavioral measures, 7. Methylation/phenotype correlations, 8. ZFP57 genotyping, 9. Bisulfite pyrosequencing, 10. Rat studies.
Table 1.
Demographic Characteristics of the Study Population
| Generation 1 (G1) | Generation 2 (G2) | |||
|---|---|---|---|---|
| MAL (N=50) | CON (N=44) | MAL (44) | CON (N=30) | |
| Sex | ||||
| Males, n (%) | 23 (46.0) | 24 (54.6) | 18 (40.9) | 17 (56.7) |
| Females, n (%) | 27 (54.0) | 20 (45.4) | 26 (59.1) | 13 (43.3) |
| Mean age (SD) | 44.6 (1.8) | 43.9 (2.0) | 20.8 (3.5) | 21.6 (4.1) |
RESULTS
More than 100 genomic loci show DNA methylation changes after early childhood malnutrition
We profiled genome-wide DNA methylation in blood samples of 168 participants enrolled in the BNS (Figure 1), including 50 subjects with moderate-severe protein energy malnutrition during the first year of life (MAL) and 44 matched controls (CON) from the G1 generation as well as 74 of their G2 offspring (44 MAL and 30 CON) (Table 1). Differences in cohort demographics, including age, sex ratios, blood count/differential were minimal and without significance (Table 1, Supplemental Table S1). Genome-wide DNA methylation profiles, interrogated via Illumina Infinium HumanMethylation450, queried ~483,000 individual CpG sites genome-wide (covering ~2% of all CpG sites in the genome)(14). We retained 461,272 autosomal and 11,021 chr. X probes after removing probes with low signal intensity and those that were within 5 bp vicinity to common SNP, as such variants can introduce biases in probe performance. However, larger windows (>5bp from the CpG tested) tend to have minimal impact(15). For each sample, methylation values for individual CpG sites were measured as β-values ranging from 0 to 1, corresponding to completely unmethylated (0) and fully methylated (1) sites, respectively. Because cis-regulatory mechanisms generally encompass multiple CpGs at a given locus, we used a 1 kb sliding window approach(16) to identify methylation differences between cohorts, rather than focusing on isolated changes in single CpGs. Differentially methylated regions (DMR) at 1 kb sliding windows were determined for the G1 (MAL vs. CON) and G2 cohort (offspring of MAL vs. offspring of CON) separately. The 1kb sliding window was choosen because of the highly correlative structure of methylation values maintained for probes with separation up to 1kb(16), and because CpG methylation LD (linkage disequilibrium correlation) typically extends for ~500bp(17), corresponding to the 1kb window centered on the CpG in question. White blood cell composition, including neutrophil, lymphocyte, monocyte, eosinophil and basophil proportions, was indistinguishable between groups (Supplemental Table S1). Because each of our samples underwent a full blood count count prior to performing DNA extraction, we were able to include the potential effect of blood cell heterogeneity using these direct measurements for the five main blood cell types in our statistical analysis and to perform a Likelihood Ratio test to analyze cell types in our statistical analses (see Supplemental Methods). These findings, taken together effectively rules out cell type differences as confound of the DNA methylome analyses. Because direct measurement of blood cell composition was incorporated into our statistical analyses as the most robust approach for accounting for this co-variate, indirect modeling (18) was not required.
For G1, we identified 102 autosomal DMRs, comprising altogether 1000 CpGs, of which 609 were hyperand 391 hypomethylated in MAL subjects. 100/102 DMRs, or 98%,maintained significance when adjusted for childhood socioeconomic status (Supplemental Table S2). For G2, we identified 16 DMR, containing 202 CpGs, of which 104 were hyper- and 98 hypomethylated in offspring of MALs (Supplemental Table S3). The aforementioned analyses were limited to autosomal probes, to avoid confounds due to X inactivation-related DNA methylation differences between males and females(19). In addition, we also analyzed childhood malnutrition-sensitive loci on the X, identifying 15 X-linked DMRs in G1 males (Supplemental Table S4), and three X-DMRs in G2 males (Supplemental Table S5). Notably, X chromosome methylation at baseline is inherently more variable in females than males(19). Unsurprisingly, there was only a single X-linked DMR in G1 females (Supplemental Table S6), and none in G2. In addition, we report the top 100 single probe levels (sorted by p-value) that are differentially methylated in G1 MAL vs. CON (Supplemental Table S7) and G2 MAL vs. CON (Supplemental Table S8).
Enrichment patterns of nutrition-sensitive DNA methylation sites
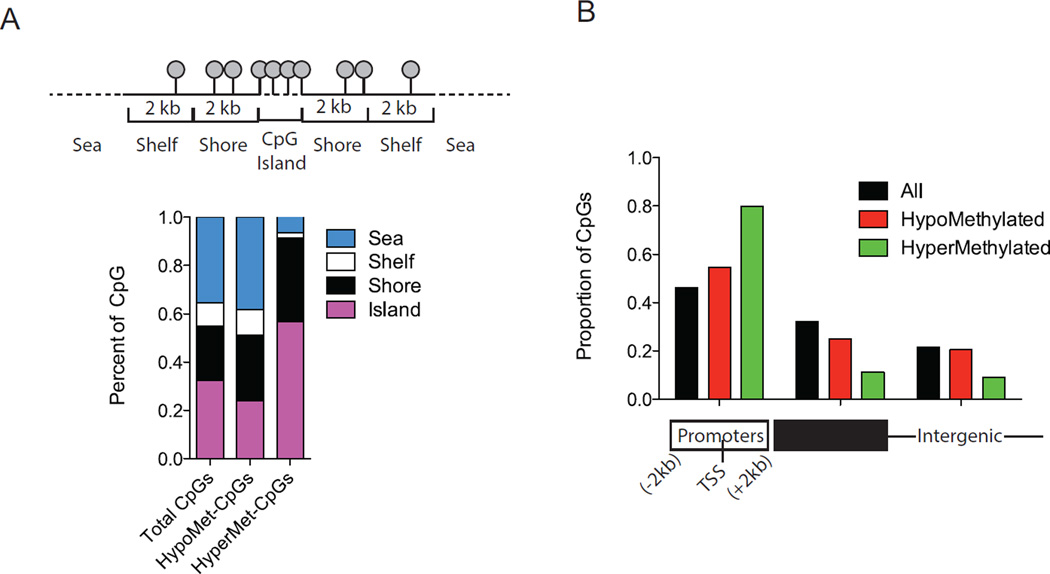
We compared, across all G1 and G2 DMRs, the distribution of hypo- and hypermethylated DMRs to the background CpG distribution in the Illumina 450K array, which has preferential probe placement in gene promoters, CpG islands, and putative regulatory elements. Strikingly, CpG islands showed an approximately 2-fold, significant (P<10−39) overrepresentation among hypermethylated DMRs, together with mild underrepresentation among hypomethylated DMRs (Figure 2A). Consistent with the majority of CpG islands being found in close proximity to 5’ regulatory sequences involved in transcriptional control(20), we also observed a 2-fold enrichment for gene promoter sequence specifically among hypermethylated DMRs (P<10−71), together with a significant underrepresentation of gene bodies (P<10−38) and intergenic sequences (P<10−18) (Figure 2B). In addition, CpG shores showed preferential enrichment both among hyper- (P<10−12) and hypomethylated (P<0.05) DMRs, which is interesting given that shores often harbor cis-regulatory sequences associated with dynamic DNA methylation and modulation of gene expression(21, 22). These changes were highly specific, because CpG shelves and sea lacked enrichment (Figure 2A). We constructed a genome-wide map of all autosomal DMRs (from Supplemental Tables S2,S3) positioned within 2kb of an annotated gene transcription start site (Figure 3). In G1 MAL, the majority of promoter-associated DMRs were hypermethylated (Supplemental Table S2, compare columns G and H), including genes with a critical role in neurodevelopment, and multiple regulators of cortical and striatal monoamine signaling (COMT, DCTN1-AS1) and connectivity (MIR200B, FOXP2, RAB3B) (Figure 3, Supplemental Table S9).
Figure 2. Genome-wide distribution of DNA methylation changes after malnutrition.
Distribution of CpG sites (A) in CpG islands, shores, shelves and sea and (B) relative to RefSeq gene promoters, gene bodies and intergenic regions. CpGs in hypomethylated and hypermethylated DMRs are compared to all autosomal CpGs (total CpGs) on the Illumina array using Pearson’s chi-square test. Notice pronounced (approximately 2-fold) overrepresentation of hypermethylated DMRs at CpG islands and gene promoters.
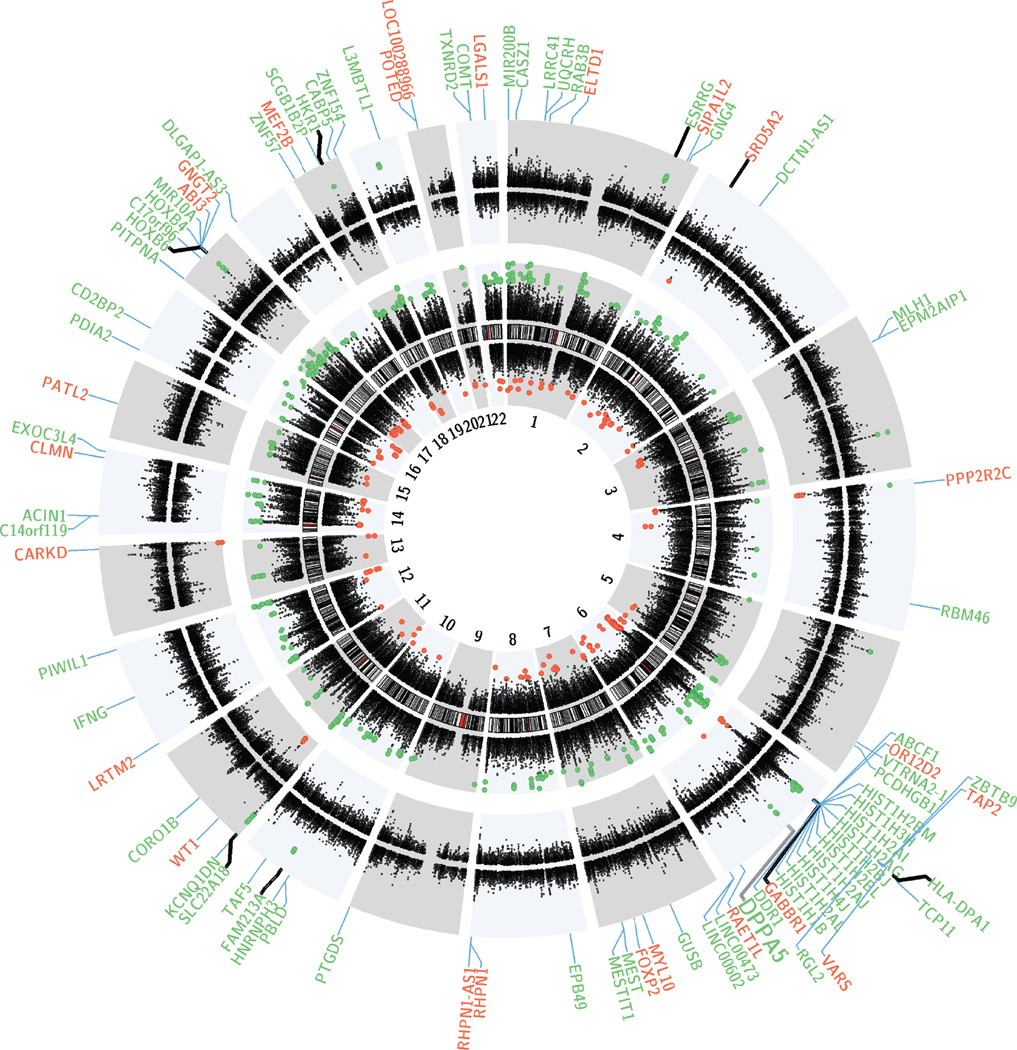
Figure 3. Genome-wide DNA methylation changes in previously malnourished G1 subjects and their G2 offspring.
Circos plot depicting the entire autosomal complement, green (red) genes with transcription start sites within ±2kb of significantly hyper-(hypo-)methylated DMRs. Genes with thin connector (Gene abbreviation to plot) line =G1; thick connector =G2. The black innermost ring (with vertical lines) represents autosome ideograms (annotated is the chromosomal number), with the pter-qter orientation in a clockwise direction. The dots on the inner side (negative) and outer side (positive) of the ideogram represent the Fisher’s method –log10 (P value) for each 1kb window analyzed in G1. Each dot marks the location of Illumina 450K probe along the genome. The next outermost black circle (with dots on the inner/negative and outer/positive side) represents the Fisher’s method –log10 (P value) for each 1kb window analyzed in G2. DPPA5 (green, larger font) was significantly hypermethylated both in G1 and G2 cohorts. Only genes where the DMR overlap its promoter i.e. ±2kb of TSS are shown.
Nutrition-sensitive DNA methylation sites correlated with attention and cognition
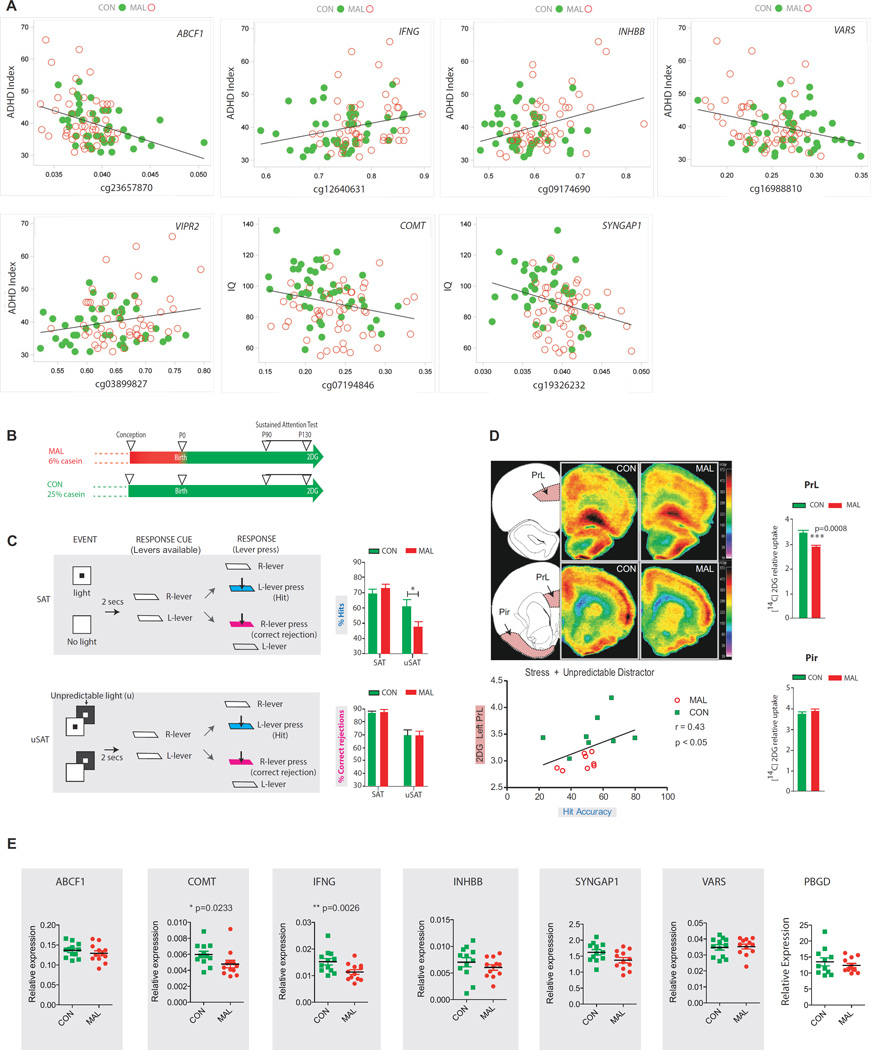
Malnutrition in the first year of life causes long-term impairments in cognition and, independent of the lower IQ, attention deficits (primarily inattention), lasting into adulthood(2, 23). Given the strong link between childhood malnutrition and adult attention-deficit hyperactivity disorder (ADHD)(23), together with the cognitive deficits and lower IQ(2), we then asked whether such neuropsychiatric sequels are associated with specific DNA methylation signatures. Here we focus on the G1 cohort, the generation directly exposed to malnutrition. Virtually all (≥95%) of G1 subjects contributed to the psychometric data. Partial correlations after adjusting for age and gender as key demographic factors(24) revealed methylated CpGs in at least 13 DMRs moderately correlated with the ADHD index on the CAARS questionaire (R2>0.04, FDR <0.1) (Supplemental Table S10A,B). These included two genes (IFNG, VIPR2) known to play a role in neuronal function and neuropsychiatric disease (Figure 4A, Supplemental Table S9). Furthermore, at least three sites were significantly correlated with IQ scores, including a DMR associated with ZBTB9 and SYNGAP1; SYNGAP1 is a gene critical for excitatory signaling in cerebral cortex and other brain regions(25) (Figure 4A, Supplemental Table S9,S11). However, with a more stringent cut-off (R2>0.13, FDR <0.05), a single gene, ABCF1, was significantly associated with the ADHD index (Figure 4A, Supplemental Table S9,S10). ABCF1, which encodes an ATP-binding cassette, was recently described as a major ‘hub’ gene in the blood transcriptome of subjects diagnosed with schizophrenia, a neurodevelopmental disorder(26). Additional neuropsychiatric risk genes (COMT, DCTN1_AS1, MIR200B, WT1) emerge with less stringent correlational filter criteria (Figure 4A, Supplemental TablesS 9-S11). Importantly, adjustment for potential confounds including childhood socioeconomic standards of living (see Supplemental Methods), and age, gender or white blood cell count, overall resulted only in very minor changes in the associations between CpG methylation and ADHD index or IQ (Supplemental Tables S10, S11).
Figure 4. Epigenetic and transcriptional dysregulation in adult blood and brain associated with impaired attention and cognition after infant malnutrition.
(A) Correlation graphs, (y-axis) ADHD index/CAARS questionnaire or WASI IQ and (x-axis) H values for top significance scoring Illumina CpG probe showing association between methylation and attention and cognition in G1 BNS subjects for seven DMR-associated genes, as indicated (see Supplemental Table s10 ,s11 for complete list of correlations). (B) Time line for rat malnourishment study, to illustrate exposure to 6% casein/low protein diet (MAL) or 25% casein normal diet (CON) from birth to conception, followed by 25% casein/normal protein diet in both groups onwards from birth. Behavioral training (SAT) occurred between P90-P130, followed by SAT testing, 2DG injection and brain harvest. (C) Overview of the sustained attention task (SAT), include light signal-based event and unpredictable distractor, and response scheme. Adult rats previously exposed to prenatal malnutrition (red bars) were more susceptible to the effects on an unpredictable, visual distractor (uSAT) than controls (green bars) and detected fewer correct targets in the presence of the distractor than controls (% hits). (D) Region-specific hypometabolism in adult rat prefrontal cortex after prenatal malnutrition. Coronal sections at level of rostromedial prelimibic cortex (PrL) from control (CON) and malnourished (MAL) animal, showing specific decrease in 2DG signal in rostro-medial cortex, including PrL, while lateral somatomotor cortex and other corticolimbic structures including piriform cortex (Pir) show normal glucose metabolism. Bar graphs summarizing 2DG uptake (mean ±S.E.M., normalized to white matter) in PrL and Pir, n=8 MAL, white and n=8 CON animals, black bars. *** P=0.0008, two-tailed t-test. Correlation graph shows strong association between glucose metabolism in left area PrL vs. hit accuracy in (unpredictable distractor) uSAT. (E) Left PrL RNA quantification for Abcf1, Comt, Ifng, Inhbb, Syngap1, and Vars, genes associated with nutrition-sensitive DMRs in BNS cohort, and Pbdg control gene. All data normalized to Hprt housekeeping gene. N=12/group. P, two-tailed t-test. After Bonferroni correction, only Ifng remained significant.
Nutrition-sensitive DNA methylation sites are enriched in imprinted genes
Imprinted genes play an important role in neurodevelopment and remain sensitive to environmentally-induced epigenomic disruption(27). We examined whether early life malnutrition was associated with DNA methylation changes in regions that show parent-of-origin specific DNA methylation patterns. We observed that several nutrition-associated DMR loci in our study occurred at known or putative imprinted genes, including L3MBTL1, MEST/MESTIT1, JAKMIP1, and VTRNA2-1(28–30). In total, 73 of the 1000 autosomal probes that showed a significant difference in methylation levels between MAL and CON G1 individuals correspond to regions that exhibit a parent-of-origin bias in DNA methylation levels, representing a highly significant 40.6 fold enrichment when compared to the non-imprinted genomic regions (p=3.5×10−91) (Supplemental Table S12). Importantly, only 3 out of 102 G1 DMRs, or less than 3%, showed concordant alterations in the G2 cohort, which would suggest that the very large majority of DNA methylation alterations after early childhood malnutrition, including the aforementioned imprinted genes, are not subject to intergenerational transmission. This could be explained by remethylation of imprinted genes in primordial germ cells—which give rise to the next generation—before birth (at least in males) (31), therefore predating the malnutrition period. Interestingly, all 3 ‘intergenerational’ DMRs were located on chromosome 6, including a ~700bp CpG island (hg18 chr 6: 74120243-74121316) next to the DPPA5 gene promoter, and CpG rich sequences surrounding the 5’ends of ZFP57 (hg18 chr6: 29756140-29757064), and PSORS1C3 (hg18 chr6: 31256311-31256592), all three significantly hypermethylated both in MAL1 G1 and MAL G2 (Supplemental Tables S2, S3). Unsurprisingly, because CpG’s within proximity of common SNPs had been removed from all analyses (see Methods), 81 of 102 autosomal G1 DMRs—including all neuropsychiatric risk genes listed in Supplemental Table S7—did not harbor any CpGs linked to known mQTLs (a genetic variant, usually a SNP, correlated with methylation levels at a nearby CpG site)(32), and the remaining 22 DMRs mostly harbored a single CpG match (Supplemental Table S13). One exception was the DMR of the aforementioned ZFP57 ‘intergenerational’ gene, with six CpGs matching to an mQTL (Supplemental Table S13). We then assessed the genotype of the mQTL driver SNP (rs396660) at the ZFP57 DMR in MAL and CON subjects, and observed strong genotype effects for the majority of the CpGs, with methylation levels consistently highest in C/C, intermediate in C/T and lowest in T/T allele carriers (Supplemental Figure S1). Of note, mQTL SNP rs396660 is positioned ~2kb distal to the cluster of probes that form the DMR at ZFP57. The SNP does not create or destroy a CpG, suggesting that methylation state of the SNP position itself is unlikely to be determinant for the methylation state of the DMR. Instead, the SNP is located centrally within a putative regulatory element, marked by DNAse I hypersensitivity and multiple transcription factor and enhancer protein binding sites (Supplemental Figure S2). Therefore, local DNA polymorphisms, affecting transcription factor and enhancer protein binding, are likely to explain at least some of the epigenetic alterations at ZFP57 locus reported here, and in other nutrition-focused studies(33). To explore this, we re-analyzed the ZFP57 DMR using linear regression, with nutrition and genotype as covariates. Indeed, both in G1 and G2, genotype consistently showed the strongest p-values and had the largest impact on the variation of methylation, with a much smaller (but still significant) effect by nutrition (Supplemental Table S14).
To technically verify DNA methylation changes detected by the Illumina array, we selected sequences within DPPA5, PSCORS1C and COMT (encoding catechol-O-methyltransferase critical for monoamine signaling at the site of cortical synapses) from the Illumina array and quantified methylation levels of several individual CpG sites within these regions in MAL and CON samples by bisulfite-pyrosequencing. Specific CpGs with significant MAL versus CON differences on the array showed similar changes by pyrosequencing. Table 2 shows the summary of Pearson’s correlation of CpG sites measured by Illumina array and pyrosequencing assays (n=10 G1 MAL and 10 G1 CON). Furthermore, there were concordant changes for neighboring CpGs not included in the Illumina array (Supplemental Figure S3). These additional studies provide verification and extension of the array data.
| Illumina probe ID |
Gene | Methylation status |
Correlation (r) | P |
|---|---|---|---|---|
| cg09071762 | DPPA5 | ↑ | 0.57 | 0.008 |
| cg18052665 | DPPA5 | ↑ | 0.54 | 0.015 |
| cg09357589 | PSORS1C3 | ↑ | 0.64 | 0.003 |
| cg27547543 | PSORS1C3 | ↑ | 0.55 | 0.015 |
| cg06860277 | COMT | ↑ | 0.66 | 0.010 |
| cg07194846 | COMT | ↑ | 0.58 | 0.030 |
Malnutrition-sensitive genes are dysregulated in cerebral cortex of rats with attention deficits
Here we showed that long-lasting epigenetic dysregulation in subjects with histories of protein-energy malnutrition limited to the first year of life is correlated with impairments in attention and cognition. This would imply that at least a subset of such genes could impact brain function and behavior. To explore whether animals with defects in attention and cognition show altered gene expression in brain, we exposed rats from (pre)conception to birth to a low protein diet(34) (Figure 4B). Note that this model is not congruent with the exposure timing in our Barbadian subject who were affected by malnutrition after birth and in early childhood. Nonetheless, our prenatally malnourished rats show attention deficits as adults, which is of interest given that some subjects exposed to early life malnutrition show a similar phenotype (23, 24, 35). Furthermore, according to our findings presented here, a subset of nutrition-sensitive blood methyl-CpGs show a significant correlation with attentional scores. Note that postnatal malnourishment is extremely difficult to model in rodent because of altered maternal behaviors, which may not be dissociated from the nutritional deficit as a cause of later brain/ behavioral changes. To confirm that our early life malnourished rats are affected by attentional and cognitive defects as previously reported for this model (23, 24, 35), we trained prenatally malnourished MAL and well-nourished control CON rats, starting postnatal day 90, first to make the correct choice between two lever press systems to discriminate brief, temporally asynchronous visual targets from non-targets. Subsequently, the animals performed the same task in the context of an unpredictable distractor (salient light flashing at unpredictable rates) (Figure 4C). Indeed, there was a significant main effect of nutrition on target discrimination (F(1,16)=5.54, p=0.032). Post-hoc analyses showed that MAL rats (N=10 males, 1/litter) performed significantly worse in the sustained attention task in presence of the unpredictable distractor (uSAT), in comparison to control (N=8 males, 1 /litter) (t(16)=2.4; P=0.03) (Figure 4C). This attention deficit in MAL, as evidenced by the decrease in the number of correct target choices, was highly specific, because nutritional history had no effect on sustained attention in the absence of the distractor. Furthermore, all animals were less able to correctly reject non-targets in presence of the distractor (F(1,16)=69.59). None of these effects differed on the basis of prior nutrition (all p>0.10). All animals maintained approximately neutral side biases for lever pressings (MAL=0.41±0.01; CON=0.42±0.02). Of note, adult rats exposed to malnourishment during development show region-specific glucose hypometabolism in rostro-medial cerebral cortex(36), considered the homolog of frontal association cortex in primates (37). We tested whether these neural are affected by altered expression of differentially methylated genes associated with attention and cognition in our human cohort. To this end, we injected MAL and CON animals (N=18, see above) with radiolabeled dexoxyglucose (2DG) prior to the behaviorally assay (uSAT), and brains harvested 45 min thereafter. Densitometry on coronal sections cut through the level of the rostro-medial cortex of the left hemisphere showed that 2DG uptake was significantly decreased, specifically in the prelimibic area (PrL) and surrounding prefrontal cortex in MAL rats relative to CON (Figure 4D). 2DG uptake in PrL correlated with correct lever press choices in the uSAT paradigm (r=0.43, P<0.05) (Figure 4D). We then quantified RNA in adult PFC of MAL and CON rats for six genes (Ifng, Inhbb, Abcf1, Comt, Syngap1, Vars, all moderately expressed at baseline see Supplemental Figure S4) associated with nutrition-sensitive DMRs and showing moderate correlation between CpG methylation and attentional/IQ scores in our human cohort (Figure 4A,E). Ifng and Inhbb genes show a significant degree of conservation of regulatory sequences between human and rat within the nutrition-sensitive DMR (Supplemental Table S15). Indeed, Ifng (gamma interferon, implicated in the regulation of cognition and emotion via cytokine signaling pathways(38, 39)) transcript was significantly decreased in MAL PFC. These changes were specific, because expression of the remaining genes (with the possible exception of Comt ), and Pbdg ‘housekeeping’ gene remained unaltered from control (Figure 4E).
DISCUSSION
The BNS is a longitudinal study following for 48 years individuals who had moderate to severe protein-energy malnutrition limited to the first year of life after normal prenatal development. Of note, the primary selection criteria, birth weight >2500 g and normal APGAR scores and physical exam at time of birth, with no evidence of wasting or other clinical signs of malnutrition, essentially excludes the likelihood of fetal malnourishment. We now provide the first evidence for long-lasting DNA methylation changes in humans exposed to malnutrition in early infancy. Genome-wide DNA methylation scans from 168 subjects revealed, after stringent statistical analyses, 134 nutrition-sensitive genomic loci that showed significant differential methylation in previously malnourished subjects and their offspring after correction for age, gender and cell type composition. Among the nutrition-sensitive genomic loci, there was a significant overrepresentation of CpG islands and other cis-regulatory sequences including promoters, suggesting long-lasting epigenetic dysregulation of gene expression. Brain is among the organs affected, because nutrition-sensitive CpG methylation at various genomic loci showed weak to moderate correlations with measures of attention and cognition in BNS subjects that could not be explained by differences in childhood socioeconomic status or other confounds. Prefrontal expression of Ifng, a gene associated with a hypermethylated DMR in our human subjects with a history of infant malnutrition, was significantly decreased in a rat model for attention deficits.
Our studies draw a strong link between protein malnutrition during early childhood and epigenetic dysregulation associated with attentional and cognitive deficits in the adult. These findings significantly extend previous work reporting epigenetic dysregulation after prenatal malnutrition(36, 40–42). Given that DNA and histone methylation landscapes show highly dynamic developmental regulation of neuronal and to some extent also non-neuronal chromatin of human cerebral cortex during infancy and early childhood(43–47), the blood DNA methylation changes as reported here could reflect on epigenetic maladaptations in brain, in analogy to epigenetic changes in the brain and behavioral dysregulation in response to early life adversity(48, 49).
In the present study, the number of autosomal DMRs in G2 offspring was ~10-fold lower compared to G1 parents (16 vs. 102). Fewer than 3% (3/102) of epigenetically altered loci showed evidence for intergenerational transmission, or concordant methylation changes in G2 offspring of G1 parents malnourished in their childhood. Therefore, the very large majority of malnutrition-induced DNA methylation changes after early childhood malnutrition are not transmitted into the next generation. Alternatively, our study may have lacked the power to detect more subtle methylation changes at some of the nutrition-sensitive loci in G2. We note that in the present study, regulatory sequences at DPPA5 were hypermethylated in MAL G1 and in their G2 offspring. It is not unreasonable to speculate that DPPA5, a gene extremely highly expressed in primordial germ cells (PGC)(50), with its CpG island heavily methylated in blood and other somatic tissues(51) could transduce nutrition-related epigenome remodeling from the G1 parent to the G2 offspring. Finally, as G3 was not included in this study, it remains unclear whether or not epigenomic alterations after childhood malnutrition carry transgenerational potential as reported for nutritional exposures during the fetal period (52).
We note that at least seven of our nutrition-sensitive G1 DMRs are in close spatial proximity to imprinted genes, including: IGF1R insulin growth factor1 receptor 1(53); KCNQ1DN and WT1 important for Wilms tumor biology (54) and (WT1) implicated in neurodegenerative disease(55); the MEST and MESTIT1 genes as maternal stress-sensitive genes regulating parental behaviors and body mass in offspring(56); and SLC22A18AS and VTRNA2-1 which have not yet been explored in the nervous system. Such epigenetic dysregulation—with an impressive >40-fold overrepresentation of methyl-CpG changes for imprinted loci in the present study— could affect not only the risk exposed G1 subjects but also the earliest stages of development in their offspring, even in the absence of an intergenerational transmission of the G1-specific DNA methylation changes.
Our study was the first to assess epigenetic alterations after childhood malnutrition in the first year of postnatal life. However, epigenomic sensitivity to nutritional restriction is likely to exist onward from the earliest phases of prenatal period. For example, a recent study reported alterations in six metastable epialleles (CpG sequences with high inter-individual variation presumably due to stochastic methylation in early development) in 84 infants conceived during the protein-energy- and methyl-donor limited Gambian rainy season, in comparison to infants born in the dry season(57). Strikingly, one of the six nutrition-dependent epilalleles in the Gambian cohort(57), RBM46, encoding a regulator of early blastocyst and trophoectoderm differentiation(58), completely matched one of the G1 DMRs in our study (Gambia DMR hg18, chr4:155,922,422-155,922,489, Barbados DMR: hg18, chr4: 155,921,670-155,922,589). Furthermore, DNA methylomics on 24 infants exposed to the war-related 1944/45 Dutch hunger winter at the time of conception, in comparison to their unaffected siblings, identified 181 DMRs, many of which related to growth and metabolism(59). The Dutch study and our BNS datasets intersects at a single DMR (GNGT2/ABI3 promoter, hg18, chr17:44,641,718-44,642,919). These three human studies (incl. ours presented here), taken together, point to epigenome shaping by nutritional history across a wide age range, while DNA methylation signatures differ by exposure timing. For example, malnourishment around the time of conception would affect a set of genes and loci largely different from those epigenetically dysregulated when exposure occurs after birth. Furthermore, DNA methylation at a subset of genes, including the metastable RBM46 epiallele and GNGT2/ABI3, remain sensitive to nutrition from conception onwards at least until early childhood. Finally, nutrition-related DNA methylation changes are subtle. For example, in our study, we did not observe all-or-none methylation changes but rather subtle concordant changes (either higher or lower β-value average) in multiple CpGs distributed at specific gene loci. It is likely therefore that the combined effect of genome-wide DNA methylation changes, with a limited contribution from each specific site, amount to an epigenetic risk architecture, that could in conjunction with additional factors impact long term physical and mental health after childhood malnutrition.
Supplementary Material
Acknowledgments
This research was supported by grants from the NIH (HD060986 and MH074811 to JRG, NS076958 and MH086509 to SA, and DA033660, HG006696, HD073731 and MH097018 to AJS), the Brain & Behavior Research Foundation (NARSAD Distinguished Investigator Grant to SA and NARSAD Young Investigator Grant to MK), and research grant 6-FY13-92 from the March of Dimes to AJS. The authors would like to thank the dedicated staff of the Barbados Nutrition Study and the participants without whom this work could not have been done. We also acknowledge the contribution of the Barbados Reference Laboratory, Ltd, and the Icahn School of Medicine at Mount Sinai Genomics Core, incl. Dr. Milind Mahajan and Dr. Yumi Kasai.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: Evaluated BNS subjects, analyzed neuropsychological data: J.R.G., L.K.F., C.P.B., G.M. Designed and performed molecular assays and designed experiments : C.J.P., M.K., C.R., M.G., E.I.G., SA. Designed and performed behavioral and 2DG experiments (animals): A.C.A, D.L.R., J.A.M. Bioinformatical and genomic analyses: A.J.S., P.G, A.D., M.K., C.J.P. Wrote the paper: S.A., J.R.G., A.J.P., M.K., C.J.P.
Competing Financial Interests: The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Walker SP, Wachs TD, Gardner JM, Lozoff B, Wasserman GA, Pollitt E, et al. Child development: risk factors for adverse outcomes in developing countries. Lancet. 2007;369:145–157. doi: 10.1016/S0140-6736(07)60076-2. [DOI] [PubMed] [Google Scholar]
- 2.Waber DP, Bryce CP, Girard JM, Zichlin M, Fitzmaurice GM, Galler JR. Impaired IQ and academic skills in adults who experienced moderate to severe infantile malnutrition: a 40-year study. Nutritional neuroscience. 2014;17:58–64. doi: 10.1179/1476830513Y.0000000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galler JR, Bryce CP, Zichlin ML, Waber DP, Exner N, Fitzmaurice GM, et al. Malnutrition in the first year of life and personality at age 40. Journal of child psychology and psychiatry, and allied disciplines. 2013;54:911–919. doi: 10.1111/jcpp.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galler JR, Bryce CP, Zichlin ML, Fitzmaurice G, Eaglesfield GD, Waber DP. Infant malnutrition is associated with persisting attention deficits in middle adulthood. J Nutr. 2012;142:788–794. doi: 10.3945/jn.111.145441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, Isganaitis E, et al. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 2014;345:1255903. doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franklin TB, Russig H, Weiss IC, Graff J, Linder N, Michalon A, et al. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68:408–415. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 9.Gapp K, von Ziegler L, Tweedie-Cullen RY, Mansuy IM. Early life epigenetic programming and transmission of stress-induced traits in mammals: how and when can environmental factors influence traits and their transgenerational inheritance? Bioessays. 2014;36:491–502. doi: 10.1002/bies.201300116. [DOI] [PubMed] [Google Scholar]
- 10.Galler JR, Bryce CP, Waber DP, Zichlin ML, Fitzmaurice GM, Eaglesfield GD. Socioeconomic outcomes in adults malnourished in the first year of life: A 40-year study. Pediatrics. 2012a;130:e1–e7. doi: 10.1542/peds.2012-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conners CK, Erhardt D, Sparrow E. Conners' Adult ADHD Rating Scales (CAARS) North Tonawanda, NY: Multi-Health Systems Inc; 1999. [Google Scholar]
- 12.Wechsler D. Wechsler Abbreviated Scale of Intelligence (WASI) New York: Psychological Corporation; 1999. [Google Scholar]
- 13.Galler JR, Bryce C, Waber DP, Zichlin ML, Fitzmaurice GM, Eaglesfield D. Socioeconomic outcomes in adults malnourished in the first year of life: a 40-year study. Pediatrics. 2012;130:e1–e7. doi: 10.1542/peds.2012-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics : official journal of the DNA Methylation Society. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 15.Hernando-Herraez I, Prado-Martinez J, Garg P, Fernandez-Callejo M, Heyn H, Hvilsom C, et al. Dynamics of DNA methylation in recent human and great ape evolution. PLoS Genet. 2013;9:e1003763. doi: 10.1371/journal.pgen.1003763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huynh JL, Garg P, Thin TH, Yoo S, Dutta R, Trapp BD, et al. Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat Neurosci. 2014;17:121–130. doi: 10.1038/nn.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Li X, Aryee MJ, Ekstrom TJ, Padyukov L, Klareskog L, et al. GeMes, clusters of DNA methylation under genetic control, can inform genetic and epigenetic analysis of disease. Am J Hum Genet. 2014;94:485–495. doi: 10.1016/j.ajhg.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharp AJ, Stathaki E, Migliavacca E, Brahmachary M, Montgomery SB, Dupre Y, et al. DNA methylation profiles of human active and inactive X chromosomes. Genome research. 2011;21:1592–1600. doi: 10.1101/gr.112680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes & development. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature genetics. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nature genetics. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galler JR, Bryce CP, Zichlin ML, Fitzmaurice G, Eaglesfield GD, Waber DP. Infant malnutrition is associated with persisting attention deficits in middle adulthood. The Journal of nutrition. 2012;142:788–794. doi: 10.3945/jn.111.145441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stein AD, Wang M, DiGirolamo A, Grajeda R, Ramakrishnan U, Ramirez-Zea M, et al. Nutritional supplementation in early childhood, schooling, and intellectual functioning in adulthood: a prospective study in Guatemala. Archives of pediatrics & adolescent medicine. 2008;162:612–618. doi: 10.1001/archpedi.162.7.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozkan ED, Creson TK, Kramar EA, Rojas C, Seese RR, Babyan AH, et al. Reduced cognition in Syngap1 mutants is caused by isolated damage within developing forebrain excitatory neurons. Neuron. 2014;82:1317–1333. doi: 10.1016/j.neuron.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Jong S, Boks MP, Fuller TF, Strengman E, Janson E, de Kovel CG, et al. A gene co-expression network in whole blood of schizophrenia patients is independent of antipsychotic-use and enriched for brain-expressed genes. PloS one. 2012;7:e39498. doi: 10.1371/journal.pone.0039498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaSalle JM, Powell WT, Yasui DH. Epigenetic layers and players underlying neurodevelopment. Trends in neurosciences. 2013;36:460–470. doi: 10.1016/j.tins.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paliwal A, Temkin AM, Kerkel K, Yale A, Yotova I, Drost N, et al. Comparative anatomy of chromosomal domains with imprinted and non-imprinted allele-specific DNA methylation. PLoS genetics. 2013;9:e1003622. doi: 10.1371/journal.pgen.1003622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Docherty LE, Rezwan FI, Poole RL, Jagoe H, Lake H, Lockett GA, et al. Genome-wide DNA methylation analysis of patients with imprinting disorders identifies differentially methylated regions associated with novel candidate imprinted genes. Journal of medical genetics. 2014;51:229–238. doi: 10.1136/jmedgenet-2013-102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Court F, Tayama C, Romanelli V, Martin-Trujillo A, Iglesias-Platas I, Okamura K, et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome research. 2014;24:554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bale TL. Epigenetic and transgenerational reprogramming of brain development. Nat Rev Neurosci. 2015;16:332–344. doi: 10.1038/nrn3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. elife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amarasekera M, Martino D, Ashley S, Harb H, Kesper D, Strickland D, et al. Genome-wide DNA methylation profiling identifies a folate-sensitive region of differential methylation upstream of ZFP57-imprinting regulator in humans. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2014;28:4068–4076. doi: 10.1096/fj.13-249029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGaughy JA, Amaral AC, Rushmore RJ, Mokler DJ, Morgane PJ, Rosene DL, et al. Prenatal malnutrition leads to deficits in attentional set shifting and decreases metabolic activity in prefrontal subregions that control executive function. Developmental neuroscience. 2014;36:532–541. doi: 10.1159/000366057. [DOI] [PubMed] [Google Scholar]
- 35.Walker SP, Chang SM, Powell CA, Simonoff E, Grantham-McGregor SM. Early childhood stunting is associated with poor psychological functioning in late adolescence and effects are reduced by psychosocial stimulation. The Journal of nutrition. 2007;137:2464–2469. doi: 10.1093/jn/137.11.2464. [DOI] [PubMed] [Google Scholar]
- 36.Amaral AC, Jakovcevski M, McGaughy JA, Calderwood SK, Mokler DJ, Rushmore RJ, et al. Prenatal protein malnutrition decreases KCNJ3 and 2DG activity in rat prefrontal cortex. Neuroscience. 2015;286:79–86. doi: 10.1016/j.neuroscience.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uylings HB, Groenewegen HJ, Kolb B. Do rats have a prefrontal cortex? Behavioural brain research. 2003;146:3–17. doi: 10.1016/j.bbr.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 38.McAfoose J, Baune BT. Evidence for a cytokine model of cognitive function. Neuroscience and biobehavioral reviews. 2009;33:355–366. doi: 10.1016/j.neubiorev.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 39.Campos AC, Vaz GN, Saito VM, Teixeira AL. Further evidence for the role of interferon-gamma on anxiety- and depressive-like behaviors: involvement of hippocampal neurogenesis and NGF production. Neuroscience letters. 2014;578:100–105. doi: 10.1016/j.neulet.2014.06.039. [DOI] [PubMed] [Google Scholar]
- 40.Begum G, Davies A, Stevens A, Oliver M, Jaquiery A, Challis J, et al. Maternal undernutrition programs tissue-specific epigenetic changes in the glucocorticoid receptor in adult offspring. Endocrinology. 2013;154:4560–4569. doi: 10.1210/en.2013-1693. [DOI] [PubMed] [Google Scholar]
- 41.Xu J, He G, Zhu J, Zhou X, St Clair D, Wang T, et al. Prenatal nutritional deficiency reprogrammed postnatal gene expression in mammal brains: implications for schizophrenia. Int J Neuropsychopharmacol. 2015;18 doi: 10.1093/ijnp/pyu054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grissom NM, Herdt CT, Desilets J, Lidsky-Everson J, Reyes TM. Dissociable deficits of executive function caused by gestational adversity are linked to specific transcriptional changes in the prefrontal cortex. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2015;40:1353–1363. doi: 10.1038/npp.2014.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siegmund KD, Connor CM, Campan M, Long Tl, Weisenberger DJ, Biniszkiewicz D, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PloS one. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. American journal of human genetics. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shulha HP, Cheung I, Guo Y, Akbarian S, Weng Z. Coordinated cell type-specific epigenetic remodeling in prefrontal cortex begins before birth and continues into early adulthood. PLoS genetics. 2013;9:e1003433. doi: 10.1371/journal.pgen.1003433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mill J, Petronis A. Pre- and peri-natal environmental risks for attention-deficit hyperactivity disorder (ADHD): the potential role of epigenetic processes in mediating susceptibility. Journal of child psychology and psychiatry, and allied disciplines. 2008;49:1020–1030. doi: 10.1111/j.1469-7610.2008.01909.x. [DOI] [PubMed] [Google Scholar]
- 48.Kundakovic M, Gudsnuk K, Herbstman JB, Tang D, Perera FP, Champagne FA. DNA methylation of BDNF as a biomarker of early-life adversity. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1408355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Provencal N, Suderman MJ, Guillemin C, Massart R, Ruggiero A, Wang D, et al. The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:15626–15642. doi: 10.1523/JNEUROSCI.1470-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim SK, Suh MR, Yoon HS, Lee JB, Oh SK, Moon SY, et al. Identification of developmental pluripotency associated 5 expression in human pluripotent stem cells. Stem Cells. 2005;23:458–462. doi: 10.1634/stemcells.2004-0245. [DOI] [PubMed] [Google Scholar]
- 51.Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS genetics. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, et al. Sex-specific, male-line transgenerational responses in humans. EurJ Hum Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 53.Sun J, Li W, Sun Y, Yu D, Wen X, Wang H, et al. A novel antisense long noncoding RNA within the IGF1R gene locus is imprinted in hematopoietic malignancies. Nucleic acids research. 2014;42:9588–9601. doi: 10.1093/nar/gku549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xin Z, Soejima H, Higashimoto K, Yatsuki H, Zhu X, Satoh Y, et al. A novel imprinted gene, KCNQ1DN, within the WT2 critical region of human chromosome 11p15.5 and its reduced expression in Wilms' tumors. Journal of biochemistry. 2000;128:847–853. doi: 10.1093/oxfordjournals.jbchem.a022823. [DOI] [PubMed] [Google Scholar]
- 55.Chaudhry M, Wang X, Bamne MN, Hasnain S, Demirci FY, Lopez OL, et al. Genetic variation in imprinted genes is associated with risk of late-onset Alzheimer's disease. Journal of Alzheimer's disease : JAD. 2015;44:989–994. doi: 10.3233/JAD-142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vidal AC, Benjamin Neelon SE, Liu Y, Tuli AM, Fuemmeler BF, Hoyo C, et al. Maternal stress, preterm birth, and DNA methylation at imprint regulatory sequences in humans. Genetics & epigenetics. 2014;6:37–44. doi: 10.4137/GEG.S18067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dominguez-Salas P, Moore SE, Baker MS, Bergen AW, Cox SE, Dyer RA, et al. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nature communications. 2014;5:3746. doi: 10.1038/ncomms4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang C, Chen Y, Deng H, Gao S, Li L. Rbm46 regulates trophectoderm differentiation by stabilizing Cdx2 mRNA in early mouse embryos. Stem cells and development. 2015;24:904–915. doi: 10.1089/scd.2014.0323. [DOI] [PubMed] [Google Scholar]
- 59.Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nature communications. 2014;5:5592. doi: 10.1038/ncomms6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.