Abstract
Yeast surface display has proven to be an effective tool in the discovery and evolution of ligands with new or improved binding activity. Selections for binding activity are generally carried out using immobilized or fluorescently labeled soluble domains of target molecules such as recombinant ectodomain fragments. While this method typically provides ligands with high affinity and specificity for the soluble molecular target, translation to binding true membrane-bound cellular target is commonly problematic. Direct selections against mammalian cell surfaces can be carried out either exclusively or in combination with soluble target-based selections to further direct towards ligands for genuine cellular target. Using a series of fibronectin domain, affibody, and Gp2 ligands and human cell lines expressing a range of their targets, epidermal growth factor receptor and carcinoembryonic antigen, this study quantitatively identifies the elements that dictate ligand enrichment and yield. Most notably, extended flexible linkers between ligand and yeast enhances enrichment ratios from 1.4±0.8 to 62±57 for a low-affinity (>600 nM) binder on cells with high target expression and from 14±13 to 74±25 for a high-affinity binder (2 nM) on cells with medium valency. Inversion of the yeast display fusion from C-terminal display to N-terminal display still enables enrichment albeit with 40% to 97% reduced efficacy. Collectively, this study further enlightens the conditions – while highlighting new approaches – that yield successful enrichment of yeast-displayed binding ligands via panning on mammalian cells.
Keywords: ligand engineering, yeast display, panning, protein engineering
Introduction
Protein-based therapeutics have shown effectiveness in alleviating disease states as inhibitors, targeting agents for drug delivery, radioisotope carriers, and immune system engagers (Leader et al., 2008). Engineered ligands have also been used for diagnostic purposes, such as targeted molecular imaging for patient stratification and treatment monitoring (James and Gambhir, 2012) and ex vivo molecular analysis of blood and urine (Dijkstra et al., 2014; Husseinzadeh, 2011; Yotsukura and Mamitsuka, 2015). The landscape of clinical targets continues to grow with new genomic and proteomic discovery methods (Mäbert et al., 2014). Moreover, biophysical constraints placed on the ligand for ‘developability’ require further ligand engineering (Drake and Papalia, 2012). To meet the demand for engineered ligands, numerous robust, high-throughput methods for selection of ligands with unique or improved specific binding activity have been developed.
Yet, selections for ligands targeting cell surface receptors are often directed by the use of recombinantly produced soluble extracellular domains for previously characterized biomarkers. These target molecules are often immobilized on a solid support (Ackerman et al., 2009; McCafferty et al., 1990) or labeled by a fluorescent or affinity tag for efficient screening (Boder and Wittrup, 1997). While this selection strategy has yielded success in various campaigns, it has two major shortcomings. First, for known antigens, ligands with a binding phenotype to the soluble extracellular domain of the target of interest may not necessarily translate to binding effectively to target expressed on an intact cell. Potential causes include: 1) improper folding of the soluble target due to instability introduced by lack of transmembrane domain, storage conditions, or purification steps, 2) differential post-translational modification between the production host and the cell type of interest, 3) binding to a non-natural epitope resulting from the biological or chemical addition of tags to the target molecule to aid in purification or selection, or 4) lack of accessibility of the bound epitope in the presence of the transmembrane domain, cell membrane, and extracellular molecules. Second, these soluble target-based strategies are limited to targets that have been identified previously. Direct selection of ligands binding to mammalian cell surfaces overcomes these shortcomings. Target molecules are presented in their normal conformation with appropriate post-translational modification and no additional tags. Further, due to the wide array of cell surface proteins, cell-based selections can simultaneously be used as a proteomic strategy, allowing for discovery of previously uncharacterized protein expression while also evolving a ligand for the new target.
The use of a genotype-phenotype linkage strategy allows for the screening of large combinatorial libraries of affinity proteins. One such genotype-phenotype linkage strategy is yeast surface display (Boder and Wittrup, 1997; Gera et al., 2013). In yeast surface display, proteins of interest encoded by expression plasmids are produced as fusions with the yeast mating protein agglutinin 2 (Aga2p) and secreted. The fusion is tethered to the yeast cell surface in quantities of approximately 104–105 per cell by disulfide linkage with yeast protein agglutinin 1, which is anchored to the cell wall (Lu et al., 1995). Yeast surface display has been successfully applied (Pepper et al., 2008) in screening for numerous ligands including, but not limited to, peptides (VanAntwerp and Wittrup, 2000), antibody fragments (Chao et al., 2006), and fibronectin domains (Hackel et al., 2008). Yeast surface display has been previously applied to cell-based selections. A protocol for these selections against cell monolayers has been optimized using fluorescein-labeled rat brain endothelial (RBE4) cells and anti-fluorescein single-chain antibody variable fragments (scFvs) (Wang and Shusta, 2005). Non-immune scFv libraries were effectively applied in ligand-biomarker co-discovery experiments using this optimized protocol to isolate ligands for receptors expressed by RBE4 cells (Wang et al., 2007) and androgen-dependent prostate cancer cells (Williams et al., 2014). Other applications of this protocol include combination with soluble target-based screening to ensure that isolated anti-B7-H4 scFvs would translate to binding true cellular B7-H4 (Dangaj et al., 2013) and isolation of mutants of the I domain of integrin Mac-1 that achieve high affinity conformations (Hu et al., 2010). An alternative method for direct cellular selections utilizing disadhered mammalian cells and Ficoll density centrifugation for isolation of yeast-mammalian cell complexes has also been optimized (Richman et al., 2006).
Although not evaluated in the current study, it should be noted that alternative display approaches have also been used for cellular selections. Phage display approaches have been employed successfully to generate cancer-specific scFvs (Sanchez-Martin et al., 2015) and peptides (Newton and Deutscher, 2008) by cellular selection. The small size of phage particles does afford the advantage of multiple different selection methods (Sanchez-Martin et al., 2015) including direct cellular selection against either adhered or disadhered cells (Barry et al., 1996), microdissection methods (Yujing et al., 2009), and in vivo selections (Pasqualini and Ruoslahti, 1996). Although successful, phage display approaches can suffer from weak enrichment (often less than 10-fold per selection round) (Fu et al., 2014; Mutuberria et al., 2004). Phage display systems also do not have access to the eukaryotic translation machinery that yeast surface display employs, leading to lower functional diversity in phage display libraries relative to their yeast surface displayed counterparts (Bowley et al., 2007). Aptamers for several targets have also been developed (Dua et al., 2011) by selections against whole cells (Shangguan et al., 2006), selections against membrane preparations (Huang et al., 2007), and in vivo selections (Cheng et al., 2013).
This study aims to optimize and better understand the parameters for successful direct cellular selection using yeast surface display and cell monolayers. Incubation conditions, stringency of washing, target expression on mammalian cells, ligand expression on yeast cells, ligand binding affinity, ligand linker length and display orientation, location of bound epitope, and ligand protein scaffold were systematically varied to understand how each parameter affects enrichment ratio and yield of binding yeast in an EGFR-expressing model system. A strong expression dependence from the standpoint of the mammalian cell is shown. Further, the orientation of these interactions is significantly important. The addition of longer peptide linkers extending the distance between the yeast cell surface and binding ligand aids recovery of binding ligands. Understanding and applying these parameters will aid in the application of this protocol for ligand selection experiments using a wide array of affinity protein scaffolds.
Materials and Methods
Cells and Cell Culture
MDA-MB-468 and MDA-MB-231 were kind gifts from Professor Jayanth Panyam (Department of Pharmaceutics, University of Minnesota – Twin Cities). MCF7 and SKOV-3 were kind gifts from Professor Deepali Sachdev (Department of Medicine, University of Minnesota – Twin Cities). LS174T and MDA-MB-435 were kind gifts from Professor Tim Starr (Department of Obstetrics, Gynecology & Women’s Health, University of Minnesota – Twin Cities). A431 was a kind gift from Professor Daniel Vallera (Department of Therapeutic Radiology, University of Minnesota – Twin Cities). All cell lines were grown in DMEM with 4.5 g/L glucose, sodium pyruvate, and glutamine supplemented with 10% (v/v) fetal bovine serum. All cell lines were grown at 37 °C in a humidified atmosphere with 5% CO2.
Yeast surface display was performed essentially as described (Chen et al., 2013). Expression plasmids were transformed into EBY100 Saccharomyces cerevisiae yeast by EZ-Yeast Transformation (Zymo Research, Irvine, CA). Yeast harboring expression plasmids were grown in SD-CAA medium (16.8 g sodium citrate dihydrate, 3.9 g citric acid, 20.0 g dextrose, 6.7 g yeast nitrogen base, 5.0 casamino acids in 1 L deionized H2O) at 30 °C with shaking. Protein expression was induced by transferring yeast cells in logarithmic phase (OD600nm<6) into SG-CAA medium (10.2 g sodium phosphate dibasic heptahydrate, 8.6 g sodium phosphate monobasic monohydrate, 19.0 g galactose, 1.0 g dextrose, 6.7 g yeast nitrogen base, 5.0 g casamino acids in 1 L deionized H2O) and growing at 30 °C with shaking for at least 8 hours, unless otherwise stated. EBY100 not harboring plasmid were grown in YPD medium (10.0 g yeast extract, 20.0 g peptone, 20.0 g dextrose in 1 L deionized H2O) at 30 °C with shaking.
Expression Plasmids
The pCT plasmid (Boder and Wittrup, 1997) was used as the expression vector for yeast surface display on the C-terminus of Aga2p. The vector encodes for Aga2p followed by a 40-amino acid linker – including a Factor Xa cleavage site, an HA epitope, and a glycine-rich peptide – followed by the ligand with a C-terminal MYC epitope peptide (Table I). Fibronectin clones E6.2.6′ (Hackel et al., 2010), E6.2.6′ N78S, E6.2.6′ AASV, EI4.4.2 (Hackel et al., 2010), EI3.4.3 (Hackel et al., 2010), WT′ (Hackel et al., 2012b), and C7.4.3 (Pirie et al., 2011), Gp2 clones GαEGFR2.2.3 (Kruziki et al., 2015) and GαRIgG3.2.3 (Kruziki et al., 2015), and affibody clones EA68 (Case and Hackel, 2016) and A5 (Woldring and BJH, unpublished) were cloned into pCT vector by NheI and BamHI restriction sites.
Table 1.
Relevant Ligand and Linker Sequences
| Format | Ligand Domain | Name | Sequence |
|---|---|---|---|
| C-terminal | Fibronectin | E6.2.6′ | [Aga2p] – linker – VSDVPRDLEVVAATPTSLLISWFDYAVTYYRITYGETGGNSPVQEFTVPGWISTATISGLKPGVDYTITVYAVTDNSRWPFRSTPISTNYRTEIDKPPQ |
| E6.2.6′ N78S | [Aga2p] – linker – VSDVPRDLEVVAATPTSLLISWFDYAVTYYRITYGETGGNSPVQEFTVPGWISTATISGLKPGVDYTITVYAVTDSSRWPFRSTPISTNYRTEIDKPPQ | ||
| E6.2.6′ AASV | [Aga2p] – linker – VSDVPRDLEVVAAAPTSLLISWFDYAVTYYRITYGETGGNSPAQEFTVPGWISTATISGLKPGVDYTITVYAVTDSSRWPFRSTPVSTNYRTEIDKPPQ | ||
| EI4.4.2 | [Aga2p] – linker – VSDVPRDLEVVAATPTSLLISWYFRDPRYVDYYRITYGETGGNSPAQEFTVPWYLPEATISGLKPGVDYTITVYAVTGDDQNAGLPISINYRTEIDKPSQ | ||
| EI3.4.3 | [Aga2p] – linker – VSDVPRDLEVVAATPTSLLISWLHHRSDVRSYRITYGETGGNSPVQKFTVPGSRSLATISGLKPGVDYTITVYAVTWGSYCCSNPISINYRTEIDKPSQ | ||
| WT′ | [Aga2p] – linker – VSDVPRDLEVVAATPTSLLISWDAPAVTVRYYRITYGETGGNSPVQEFTVPGSRSTATISGLRPGVDYTITVYAVTGRDGSPASSRPISINYRT | ||
| C7.4.3 | [Aga2p] – linker – VSDGTLSRDLEVVAATPTSLLISWYYSYSHHYSSYRITYGETGGNSPVQEFTVPRYRAFATISGLKPGVDYTITVYAVTSSSSYSYPISINYRTEIDKPSQ | ||
|
| |||
| Affibody | EA68 | [Aga2p] – linker – AEAKYAKEMWAAWEEIRNLPNLTGWQMTAFIAALVDDPSQSANLLAEAKKLNDAQAPK | |
| A5 | [Aga2p] – linker – AEAKYAKENFNATSEIYYLPNLTHFQRSAFSNALFDDPSQSSELLSEAKKLNDSQAPK | ||
|
| |||
| Gp2 | GαEGFR2.2.3 | [Aga2p] – linker – KFWATVSRGDSYWFEVPVYAETLDEALELAERQYPMYHIYYVTRVRP | |
| GαRIgG3.2.3 | [Aga2p] – linker – KFWATVHSVHGYFEVPVYAETLDEALELAFGQYGNALGYVTRVRP | ||
|
| |||
| N-terminal | Fibronectin | E6.2.6′ | VSDVPRDLEVVAATPTSLLISWFDYAVTYYRITYGETGGNSPVQEFTVPGWISTATISGLKPGVDYTITVYAVTDNSRWPFRSTPISTNYRTEIDKPPQ – linker – [Aga2p] |
| E6.2.6′ N78S | VSDVPRDLEVVAATPTSLLISWFDYAVTYYRITYGETGGNSPVQEFTVPGWISTATISGLKPGVDYTITVYAVTDSSRWPFRSTPISTNYRTEIDKPPQ – linker – [Aga2p] | ||
| E6.2.6′ AASV | VSDVPRDLEVVAAAPTSLLISWFDYAVTYYRITYGETGGNSPAQEFTVPGWISTATISGLKPGVDYTITVYAVTDSSRWPFRSTPVSTNYRTEIDKPPQ – linker – [Aga2p] | ||
| EI3.4.3 | VSDVPRDLEVVAATPTSLLISWLHHRSDVRSYRITYGETGGNSPVQKFTVPGSRSLATISGLKPGVDYTITVYAVTWGSYCCSNPISINYRTEIDKPSQ – linker – [Aga2p] | ||
| WT′ | VSDVPRDLEVVAATPTSLLISWDAPAVTVRYYRITYGETGGNSPVQEFTVPGSRSTATISGLRPGVDYTITVYAVTGRDGSPASSRPISINYRT – linker – [Aga2p] | ||
|
| |||
| Affibody | EA68 | AEAKYAKEMWAAWEEIRNLPNLTGWQMTAFIAALVDDPSQSANLLAEAKKLNDAQAPK – linker – [Aga2p] | |
|
| |||
| Gp2 | GαEGFR2.2.3 | KFWATVSRGDSYWFEVPVYAETLDEALELAERQYPMYHIYYVTRVRP – linker – [Aga2p] | |
|
| |||
| Linkers | - | pCT | KDNSSTIEGRYPYDVPDYALQASGGGGSGGGGSGGGGSAS |
| pCT-20 | KDNSSTIEGRYPYDVPDYALQASASPAAPAPASPAAPAPSAPAGGGGSGGGGSGGGGSAS | ||
| pCT-40 | KDNSSTIEGRYPYDVPDYALQASASPAAPAPASPAAPAPSAPAASPAAPAPASPAAPAPSAPAGGGGSGGGGSGGGGSAS | ||
| pCTN | GSEQKLISEEDLGGGGSGGGGSGGGGSLQ | ||
Codons encoding for 20 or 40 additional amino acid residues, based on the PAS#1 peptide (Schlapschy et al., 2013), within the linker between Aga2p and ligand were constructed by DNA assembly and ligated into pCT vector by PstI and NheI restriction sites, generating pCT-20 and pCT-40 vectors (Table I). The construct for yeast surface display on the N-terminus of Aga2p was purchased as a full cassette (Integrated DNA Technologies, Coralville, IA) and ligated into pCT vector by EcoRI and XhoI restriction sites, generating pCTN vector.
Expression Level of Target Receptors
EGFR and CEA expression on cell lines was quantified by flow cytometry in comparison to a calibration curve from anti-mouse IgG beads (Bangs Laboratories, Inc., Fishers, IN). Polystyrene beads with known quantities of immobilized monoclonal anti-mouse IgG were labeled with mouse anti-EGFR clone ab30 (10 μg/mL) or mouse anti-CEA clone ab4451 (20 μg/mL, Abcam, Cambridge, MA) for 30 minutes at room temperature. Beads were washed once with phosphate-buffered saline with 0.1% bovine serum albumin (PBSA) and pelleted at 2,500g for 2.5 minutes. The beads were then labeled by goat anti-mouse Alexa Fluor 647 conjugate (Life Technologies, Carlsbad, CA) for 30 minutes at 4°C, washed once with PBSA, and again pelleted at 2,500g for 2.5 minutes. Fluorescence was analyzed by flow cytometry using an Accuri C6 (BD Biosciences, San Jose, CA).
Mammalian cells were allowed to grow to 70–90% confluence. Culture medium was removed and the cells were washed once with 5 mL PBS. Cells were disadhered by trypsin-EDTA treatment for 3–7 minutes, and then quenched by the addition of serum-containing culture medium and centrifuged at 500g for 3 minutes. Culture medium was removed and cells were resuspended in PBSA and counted on a hemacytometer. Aliquots of 40,000 cells were pelleted at 500g for 3 minutes at 4°C, then labeled with 20 μL ab30 (10 μg/mL) or ab4451 (20 μg/mL) for 30 minutes at 4°C. Cells were washed once with PBSA and pelleted at 500g for 3 minutes. The cells were then labeled by goat anti-mouse Alexa Fluor 647 conjugate for 30 minutes at 4°C, washed once with PBSA, and again pelleted at 500g for 3 minutes at 4°C. Fluorescence was analyzed by flow cytometry using an Accuri C6.
Yeast Surface Display Cell Panning
Yeast surface display cell panning experiments were carried out following established protocols (Tillotson et al., 2013). Cell lines were grown in 6-well plates to approximately 80% confluence, washed three times with ice cold PBSA, and kept at 4°C for the duration of the experiments. Yeast mixtures – with 1×108 plasmidless EBY100 yeast and 1×105 ligand-displaying yeast – were washed in PBSA and added to each well in 1 mL ice cold PBSA. Cells were incubated without shaking for 2 hours at 4°C. Cells were washed three times as described (Tillotson et al., 2013) – tilted gently 25 times, and rotated 5 times until the final wash, during which plates were rotated 10 times – and detached by scraping into 1 mL PBSA. Dilutions of recovered material were plated on SD-CAA plates to measure recovery of clone-expressing yeast. Dilution plating on YPD plates measured recovery of all yeast. This set of conditions constitutes the baseline for optimization experiments.
Optimization of Incubation and Washing
To determine the optimal washing condition, the baseline conditions described above were repeated while varying the number of wash steps from one to five. For the one wash case, 1 mL ice cold PBSA was added to each well and plates were tilted gently 25 times and rotated 5 times. For the two wash case, a washing step involving only rotating the plate gently 10 times was added. For each subsequent case, steps of gently tilting 25 times and rotating five times were sequentially added. Recovery was quantified by dilution plating.
In separate experiments, yeast populations were added to mammalian cells in 0.5 mL, 1 mL, or 2 mL ice cold PBSA. Incubations were carried out according to the baseline conditions. After incubation, cells were washed three times and recovery was quantified by dilution plating.
Following the addition of yeast populations to mammalian cells, plates were either nutated, allowed to be static, or immediately centrifuged (300 g for 3 min) and allowed to be static for the duration of incubation. After incubation, cells were washed three times and recovery was quantified by dilution plating.
Influence of Ligand Expression on Selections
Expression levels of fibronectin clones pCT E6.2.6′ and pCT E6.2.6′ N78S were modulated by modifying the duration of induction. Yeast clones were grown to OD=0.25 in SD-CAA medium at 30°C with shaking. Yeast cells were then pelleted and changed to SG-CAA medium for induction. Aliquots of culture were removed 1, 2, 4, 8, and 24 hours post-induction. To quantify ligand expression, yeast were labeled with mouse anti-c-Myc primary antibody (9E10, BioLegend, San Diego, CA) followed by goat anti-mouse Alexa Fluor 647 conjugate and analyzed by flow cytometry. These samples were compared to a standard curve of anti-mouse conjugated beads similar to the EGFR expression experiments detailed previously. Upon choosing multiple distinct expression levels, the effects of ligand expression on recovery in selections was tested under the baseline conditions previously described.
Isolation of Dilute Binders by Multiple Round Selection
To determine the number of rounds of cell panning until a binder dominates a population, mixtures of 108 WT′-displaying yeast and 7×104 or 105 binder-displaying yeast were screened against MDA-MB-468 or MDA-MB-231 using the four or five wash conditions. Yeast were recovered after each round, amplified in SD-CAA medium, and protein expression was induced in SG-CAA medium. For quantification, plasmid was recovered from 1×108 amplified yeast by zymoprep and analyzed using qPCR with clone-specific primers: qRDGf: GTGAGCGACGTTCCAAGAGATC, qRDGr: GATAATTAATGCTGATCGGACGGCTG, q626′BCf: TACTGATCAGCTGGTTCGACTACG, q626′r: GAAATTGGAGTAGAGCGAAAAGGC.
Visualization of Binding Interactions by Phase Microscopy
MDA-MB-468 and A431 were grown in 6-well plates to 70–90% confluence, washed with ice cold PBSA, and kept at 4°C for the duration of the experiment. 2.5×107 yeast displaying fibronectin clone pCT E6.2.6′ were introduced to monolayers in 1 mL ice cold PBSA and allowed to bind for 1.5 hours. Cells were washed 5 times with ice cold PBSA and visualized in phase mode using an EVOS FL Cell Imaging System (Thermo Fisher Scientific, Waltham, MA) with a 10X objective.
Visualization of Binding Interactions by Variable Angle TIRF Microscopy
1×108 yeast displaying fibronectin clone E6.2.6′ were washed three times with PBS, then incubated with fluorescein isothiocyanate (FITC, 1 μg/mL) diluted in PBS for 45 minutes at room temperature in the dark. Excess FITC was quenched and removed by 3 washes with PBSA.
MDA-MB-468 and A431 were grown to 80% confluence in 35 mm uncoated glass bottom dishes (MatTek Corporation, Ashland, MN). Cells were washed three times with PBS, then incubated with wheat germ agglutinin – Alexa Fluor 594 conjugate (1 μg/mL, Thermo Fischer Scientific, Waltham, MA) diluted in Hank’s balanced salt solution for 10 minutes at 37°C in the dark. Cells were then washed twice with ice cold PBSA to remove excess wheat germ agglutinin.
FITC-labeled yeast were introduced to cells dropwise in 1 mL ice cold PBSA and allowed to bind for 1.5 hours. Cells were washed 5 times with ice cold PBSA. Cells were imaged using a Nikon Eclipse TiE microscope (Nikon, Melville, NY) using 488 nm and 561 nm lasers and variable angle TIRF (i.e., TIRF microscopy with the laser angle adjusted to view a deeper evanescent field). A Nikon CFI Apochromat 100X 1.49 NA oil objective and Andor iXon3 EM-CCD camera fitted with a 2.5X projection lens was used to capture images with a 64 nm pixel size in a 512×512 pixel field of view. Full 3D volumes were recorded at 500-nm Z-steps for each randomly selected field of view (32–57 planes depending on the total thickness of all of the yeast and human cells present in the field of view). Images were reconstructed using the FIJI software package. Maximal-intensity projections of the entire field of view volume were generated along the z-plane (axial view) and 3D projections were constructed with brightest point projection and rotation about the y-plane.
The relative z-axis position of yeast bound to mammalian cells was quantified using the straight line selection tool for measurement in the FIJI software package. Two measurements were taken: the distance from the center of each individual bound yeast to the bottom of the interacting portion of the mammalian cell and the distance from the top to the bottom of the interacting portion of the bound mammalian cell. These distances were used to determine a normalized position for bound yeast relative to the z-dimensional thickness of the interacting portion of each mammalian cell (Figure 10E).
Results
Yeast Surface Display Cell Panning to Enrich Fibronectin Domains
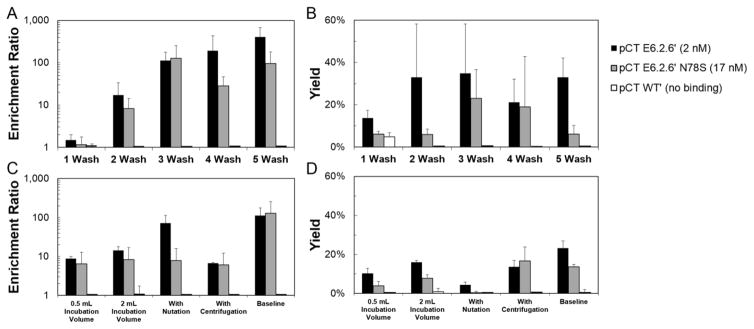
We sought to quantify the experimental elements that dictate success or failure in panning yeast-displayed ligands, particularly fibronectin domains, on mammalian cell monolayers. To determine if evolved fibronectin domains could be effectively enriched, the Shusta lab protocol (Tillotson et al., 2013) was used with highly expressing human cells (MDA-MB-468, 1.5±0.6×106 EGFR per cell) and a high-affinity ligand (fibronectin domain E6.2.6′, 2±2 nM affinity for EGFR). Indeed, the high-affinity ligand was effectively enriched 112±66-fold relative to non-displaying yeast with 35±23% yield (Figure 1A, B). The baseline condition significantly outperforms the less stringent two-wash and one-wash conditions by increasing enrichment (p=0.009 and p=0.02, respectively) and maintaining yield. Increasing the number of washes from three to five increases the enrichment ratio to 442±273 (p=0.003) while not significantly changing the binder yield (33±9%) in this context. Overall, enrichment follows a monotonic increasing correlation with number of washes (Spearman’s ρ=0.73, p<0.001). Notably, non-binding fibronectin mutant WT′ – wild-type human protein with the 77RGD79 tripeptide mutated to 77RDG79 to eliminate integrin binding – is not appreciably enriched under any of these conditions.
Figure 1. The effect of washing and incubation conditions on enrichment ratio and recovery of yeast displaying fibronectin domain ligands panned on EGFRhigh cells.
Yeast displaying E6.2.6′, E6.2.6′ N78S, and WT′ (affinities indicated) mixed 1:1,000 with non-displaying yeast were panned against EGFR-expressing MDA-MB-468. The enrichment and yield of binding ligands is presented as the mean ± standard deviation of 3–9 replicates. (A and B) Selections were performed under baseline conditions with the exception of varied number of washing steps. (C and D) Selections were performed with the indicated modulation of incubation conditions.
To study the impact of binding affinity on enrichment, a mutant ligand was engineered with reduced affinity. A yeast display library was created in which the entire fibronectin gene was mutated by error-prone PCR. Fluorescence-activated cell sorting was used to isolate mutants with reduced binding affinity to recombinant EGFR ectodomain. Several mutants were produced recombinantly in E. coli and titrated by flow cytometry for binding to A431 human epidermoid cancer cells. A single mutation in the FG loop, N78S, reduced binding strength to 17±4 nM. Using the mutant in yeast-mammalian cell panning experiments produced effective enrichment and a similar monotonic increase with an increasing number of washes (Spearman’s ρ=0.61, p<0.001) (Figure 1A, B). Notably, enrichment and yield were generally modestly reduced (median: 2.0-fold; mean: 3.1-fold for enrichment) relative to the high-affinity ligand. To further evaluate the limit of weak affinity, additional affinity reduction was performed. Adding T16A, V45A, and I88V mutations to E6.2.6′ N78S results in weak binding at high concentrations (>600 nM affinity) in detergent solubilized cell lysate experiments with yeast surface display (Cho and Shusta, 2010). The low-affinity mutant, E6.2.6′ AASV, was not able to be definitively enriched relative to non-displaying yeast (1.3±0.9-fold) using the five-wash condition.
While the observed enrichment ratios and yields enable practical use, consistent with the aforementioned scFv studies, we attempted to elevate selection efficiency though modification of incubation conditions. Decreased incubation volume or centrifugation of selections prior to incubation were explored for potentially improved yeast-mammalian cell contacting. Increased incubation volume or nutation during incubation were tested for increased stringency against non-specific yeast-mammalian cell interactions. For the high- and mid-affinity mutants, neither centrifugation nor reduction of incubation volume improved yields; moreover, enrichments relative to non-displaying yeast were decreased relative to baseline conditions (p=0.01 and p=0.01, respectively; Figure 1C, D). Increasing the incubation volume and adding nutation to incubation were also unable to increase enrichment ratio and yield for both clones tested (Figure 1C, D).
Multi-round Library Selections
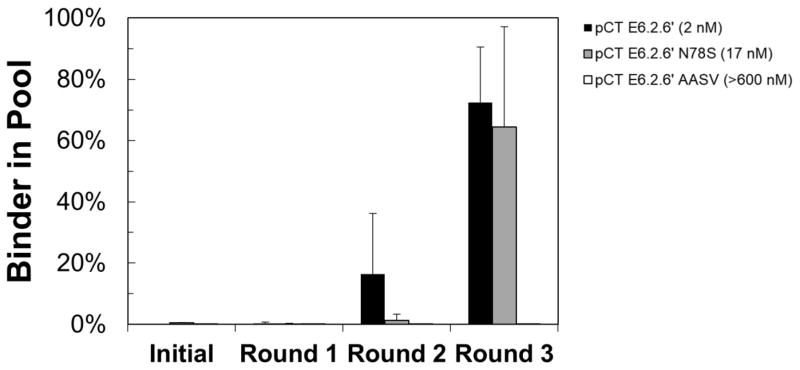
To further corroborate these data, we performed multi-round selections to evaluate the ability to isolate high-, mid-, or low-affinity ligands from a mock library of 105 binders and 108 non-binders. Enrichment to 16% frequency was observed within two rounds for the high-affinity mutant. Within three rounds of panning, the high- and mid-affinity binders were dominant relative to non-binders whereas the low-affinity mutant was not noticeably enriched (Figure 2).
Figure 2. Multi-round library selections.
EBY100 yeast displaying fibronectin clones E6.2.6′, E6.2.6′ N87S, or E6.2.6′ AASV (affinities indicated) mixed 1:1,000 with yeast displaying WT′ were panned against MDA-MB-468 monolayers for multiple rounds in triplicate using the four wash condition. After each round, the fraction of yeast harboring plasmid encoding for the binding ligand was quantified by quantitative PCR.
Effect of Target Expression on Cell Panning Efficiency
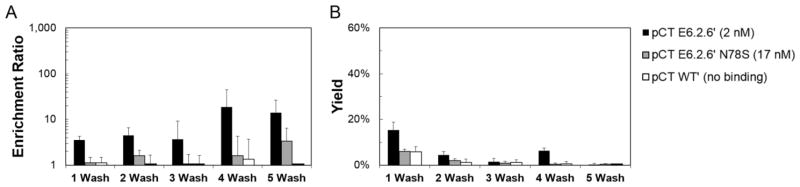
To assess the impact of EGFR expression on panning performance, yeast displaying high- or mid-affinity fibronectin domains were panned on MDA-MB-231 breast cancer cells, which express 1.9±0.6×105 EGFR per cell (8-fold lower than MDA-MB-468). Enrichment was observed for high-affinity E6.2.6′ albeit with a lower ratio (14±13-fold enrichment over non-displaying (Figure 3A) as compared to 112±66 on highly expressing MDA-MB-468; p<0.001) and lower yield (2.2±1.9% (Figure 3B) as compared to 35±23%; p<0.001). Yield improved with reduced washing but at the expense of enrichment. Enrichment and yield for mid-affinity E6.2.6′ N78S were essentially non-functional.
Figure 3. The effect of washing on enrichment ratio and recovery of yeast displaying fibronectin domain ligands panned on EGFRmid cells.
Yeast displaying E6.2.6′, E6.2.6′ N78S and WT′ (affinities indicated) mixed 1:1,000 with non-displaying yeast were panned against MDA-MB-231. The enrichment (A) and yield (B) of binding ligands is presented as the mean ± standard deviation of 3–9 replicates.
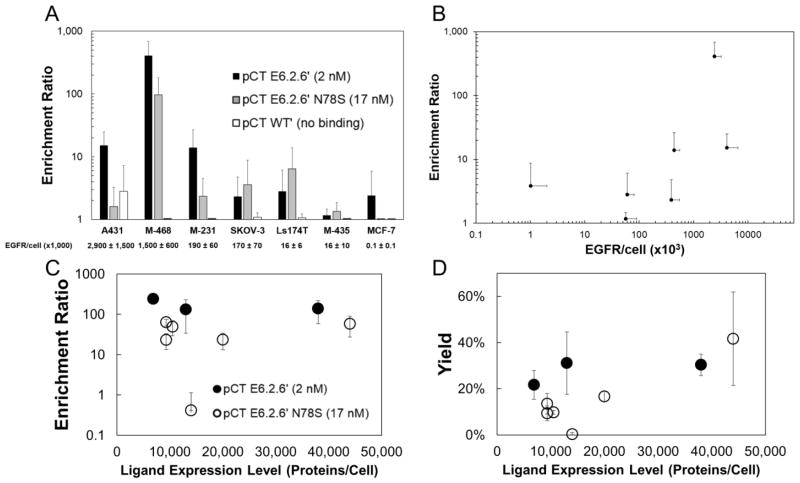
As MDA-MB-468 and MDA-MB-231 cells differ beyond simply EGFR expression levels, additional cell lines were evaluated (Figure 4A). In the context of the E6.2.6′ fibronectin/EGFR system, enrichment shows a strong positive monotonic correlation with target expression (Spearman’s ρ=0.72, p<0.001) (Figure 4B). Using the stringent five-wash protocol, yeast displaying E6.2.6′ are enriched 410±273-fold versus non-displaying yeast when panned on MDA-MB-468 (1.5±0.6×106 EGFR/cell) and 15±10-fold on A431 (2.9±1.4×106 EGFR/cell) but only 14±13 and 2.4±2.5 on mid-expressing cells (MDA-MB-231 (1.9±0.6×105 EGFR/cell) and SKOV-3 (1.7±0.7×105 EGFR/cell), respectively), and only 2.8±3.3, 1.2±0.3, and 2.4±3.5 on low-expressing MDA-MB-435 (1.5±1.1×104 EGFR/cell), LS174T (1.7±0.6×104 EGFR/cell), and MCF7 (1.2±1.5×102 EGFR/cell) cells (Figure 4). As a control, yeast displaying non-binding WT′ did not appreciably enrich on any cells. These results are consistent with the concept of multivalency in which numerous ligand-receptor interactions are needed to maintain durable cell-cell binding. Yet the superior performance on MDA-MB-468 versus A431 is unclear.
Figure 4. Enrichment across cell lines with varying EGFR expression and yeast cells with varying ligand expression.
Yeast displaying E6.2.6′, E6.2.6′ N78S, and WT′ mixed 1:1,000 with non-displaying yeast were panned using the five-wash condition against seven cell lines with varying EGFR expression. (A) Enrichment is presented as the mean ± standard deviation of 6–9 replicates. EGFR expression per cell, quantified by flow cytometry is indicated. M-468: MDA-MB-468. M-231: MDA-MB-231. M-435: MDA-MB-435. (B) Data from (A) for E6.2.6′ are plotted as enrichment vs. EGFR expression. Yeast displaying E6.2.6′ and E6.2.6′ N78S with varying ligand expression mixed 1:1,000 with non-displaying yeast were panned against MDA-MB-468. The enrichment (C) and yield (D) of binding ligands is presented as the mean ± standard deviation of 3 replicates. Ligand expression, which varies because of different induction times, was quantified by flow cytometry.
Effect of Yeast-Displayed Ligand Expression on Cell Panning Efficiency
Given the impact of EGFR expression level on panning performance, we examined the effect of ligand expression at the previously described baseline conditions (Figure 4C, D). A 2.9-fold reduction from 38,000 to 6,800 ligands/cell changes neither the enrichment ratio relative to non-displaying yeast nor yield of E6.2.6′ when panned against MDA-MB-468. A slight decrease in enrichment and yield of weaker affinity mutant E6.2.6′ N78S is observed with decreasing ligand expression, but these changes are not significant with the exception of one instance of a culture expressing 14,000 ligands/cell. Regrowth of this clone with a similar ligand expression level (9,300 ligands/cell) shows similar enrichment (64±11 and 23±10) and yield (9±3% and 14±4%) to yeast expressing 10,500 (50±20 and 10±1%) and 20,000 (24±10 and 17±2) ligands/cell, suggesting that the single trial showing weak enrichment and yield was an aberration. Enrichment ratio and yield of E6.2.6′ N78S at an expression level of 44,000 ligands/cell is statistically indistinguishable from E6.2.6′ at an expression level of 38,000 ligands/cell (p=0.2 and p=0.4, respectively). Overall, ligand expression does not drastically effect the enrichment and yield of binding clones in this context.
Impact of Paratope
While the strong performance on high-expressing cells – even for mid-affinity ligands – is encouraging, several applications would benefit from improved performance on mammalian cells expressing 104–105 targets. Notably, the fluorescein-binding antibody fragment system evaluated in the Shusta lab exhibited good yields (~20%) and enrichment (~60-fold) with 104 targets per cell (Wang and Shusta, 2005). Numerous experimental differences could explain the discrepancy including ligand type (antibody fragment versus fibronectin domain), target (fluorescein conjugated to cell surface molecules versus membrane-integrated EGFR), and geometric considerations: orientation of ligand paratope relative to yeast and orientation of target epitope relative to mammalian cell.
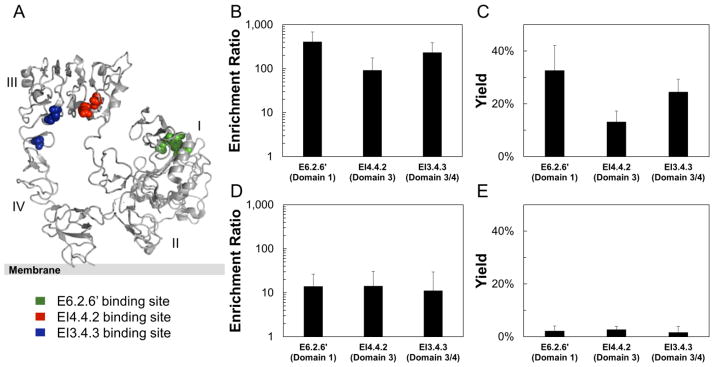
To evaluate the impact of the receptor epitope and its orientation with regards to the mammalian cell or the presentation of the ligand paratope on yeast, three high-affinity fibronectin domains that bind various locations on EGFR (Hackel et al., 2012a) were evaluated. E6.2.6′ binds EGFR in domain 1, EI4.4.2 binds EGFR domain 3, and EI3.4.3 binds EGFR at the domain 3/4 interface (Figure 5A). When panned against high-expressing MDA-MB-468, EI4.4.2 showed significantly lower enrichment (92±83) than E6.2.6′ (410±273) (p = 0.002), suggesting some epitope dependence may exist. EI3.4.3 enriched strongly (233±157) at an intermediate level between E6.2.6′ (p=0.10) and EI4.4.2 (p=0.08) (Figure 5B, C). Yeast displaying each ligand were all similarly enriched on mid-expressing MDA-MB-231, though to a lesser extent relative to MDA-MB-468 (Figure 5D, E).
Figure 5. The effect of epitope:paratope dependence on enrichment and yield.
The epitopes for fibronectin domains E6.2.6′ (green), EI4.4.2 (red), and EI3.4.3 (blue) (Hackel et al., 2012a) occur at different locations on the EGFR extracellular domain (1NQL) (Ferguson et al., 2003) (A). Yeast displaying the indicated clones mixed 1:1,000 with non-displaying yeast were panned against MDA-MB-468 (B, C) and MDA-MB-231 (D, E). The enrichment (B, D) and yield (C, E) of binding ligands is presented as the mean ± standard deviation of 6–12 replicates.
Effect of Linker Length and Ligand Orientation
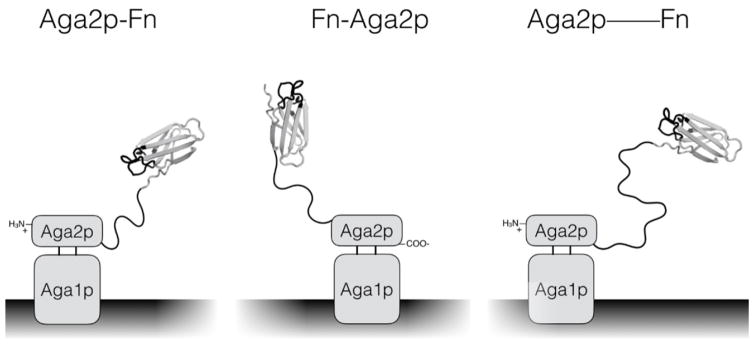
The fibronectin domains used in the previous experiments were displayed in the conventional yeast display format in which the C-terminus of Aga2p is fused to the N-terminus of fibronectin via polypeptide linker. This format permits binding to recombinant EGFR ectodomain immobilized on magnetic beads – which was used for the discovery of these ligands (Hackel et al., 2010) – and cellular EGFR as evidenced by the effective panning results shown up to this point. However, the N-terminus of fibronectin is proximal to the three engineered loops that comprise the presumed paratope (Figure 6). Additionally, the orientation of the N-terminus seemingly favors display of the fibronectin domain in an orientation in which the engineered loops are more frequently oriented towards the yeast cell surface. A longer linker aids separation of the fibronectin domain from the yeast surface and adds flexibility to reorient the ligand to prevent cell-cell steric hindrance. Fusion to the C-terminus of fibronectin moves the linker distal to the engineered loops and seemingly favors display in which the engineered loops are more frequently oriented away from the yeast cell surface.
Figure 6. Schematic of yeast surface display formats used in this study.
The original pCT vector encodes a protein of interest expressed as a fusion to the C-terminus of Aga2p with a 40 amino acid linker (Aga2p-Fn). The designed pCTN vector encodes for a protein of interest as a fusion to the N-terminus of Aga2p with a 29 amino acid linker (Fn-Aga2p). The pCT-20 and pCT-40 constructs follow the same expression scheme as the original pCT, except 20- and 40-mer versions of the PAS#1 linker are added to the aforementioned 40 amino acid linker (Aga2p—Fn).
Modified yeast display vectors were constructed to insert an additional 20 or 40 amino acids – using the proline-alanine-serine (PAS#1) design (Schlapschy et al., 2013) – into the polypeptide linker between Aga2p and the fibronectin domain.
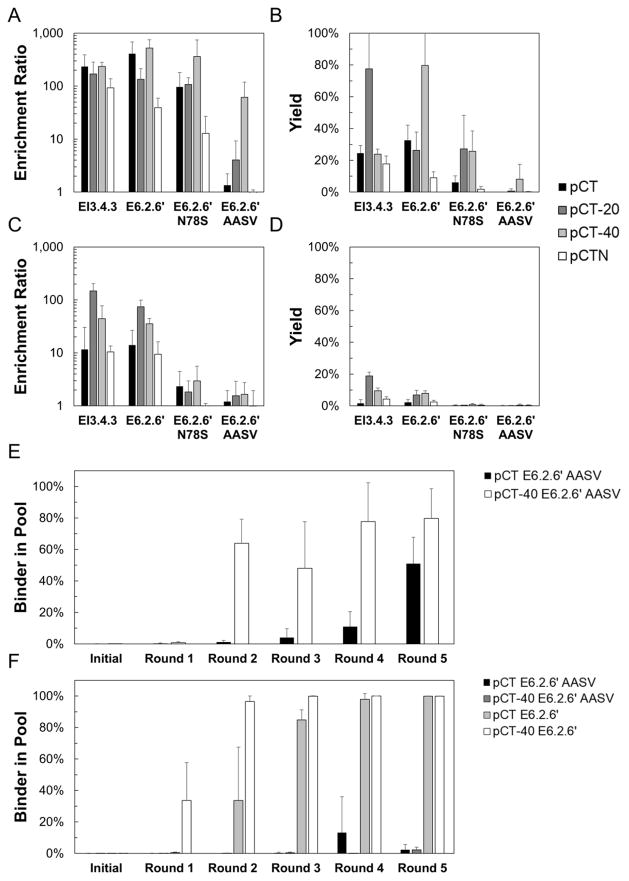
As high-affinity binders already achieve strong enrichment and yield when panned against highly-expressing cells, the extended linker has minimal further benefit (Figure 7A, B). However, when panned against mid-expressing MDA-MB-231 cells, the extended linker improves enrichment for clone E6.2.6′ from 14±13 using the standard linker to 74±24 with pCT-20 (p=0.001) and 35±9 with pCT-40 (p=0.002) and yield from 2.2±1.9% to 6.9±2.9% (p=0.008) and 7.8±1.7% (p<0.001) respectively (Figure 7C, D). A similar trend is seen using clone EI3.4.3, with the extended linker improving enrichment from 11±19 to 148±55 (p=0.001) and 73±13 (p=0.002) and yield from 2±2% to 19±2% (p<0.001) and 30±21% (p=0.02) for standard linker, pCT-20, and pCT-40, respectively. Further, the low-affinity ligand – which lacks observable single-round enrichment with the standard linker even on highly expressing cells – can be effectively enriched on highly expressing cells when displayed with an extended linker: 62±57 for pCT-40 versus 1.3±0.9 for standard linker (p=0.02). Unfortunately, the extended linker is not sufficient to enhance enrichment on mid-affinity ligands on mid-expressing cells.
Figure 7. The effect of varying linker length and display orientation on enrichment and yield.
Yeast displaying EI3.4.3, E6.2.6′, E6.2.6′ N78S, and E6.2.6′ AASV mixed 1:1,000 with non-displaying yeast were panned against MDA-MB-468 (A, B) or MDA-MB-231 (C, D). C-terminal fusions (Aga2p-Fn) with the native linker, +20 linker, and +40 linker, as well as the N-terminal fusion (Fn-Aga2p) were tested. The enrichment (A, C) and yield (B, D) of binding ligands is presented as the mean ± standard deviation of 6–12 replicates. Yeast displaying E6.2.6′ or E6.2.6′ AASV as C-terminal fusions with the native linker or +40 linker were mixed 1:1,500 with WT′ displaying yeast in the native linker or +40 linker. Mixtures were panned in triplicate against MDA-MB-468 (E) or MDA-MB-231 (F) monolayers for five rounds. After each round, the fraction of yeast harboring the binding ligand was quantified using quantitative PCR.
To further corroborate these data, multi-round selection experiments were conducted using the pCT and pCT-40 versions of E6.2.6′ and E6.2.6′ AASV in 1:1,500 starting ratio with the equivalent pCT and pCT-40 versions of WT′. For selections against MDA-MB-468, pCT-40 E6.2.6′ AASV enriched to 64±15% of the pool after just two selection rounds, while pCT E6.2.6′ AASV required five rounds to reach 51±17% of the pool (Figure 7E). For selections against MDA-MB-231, pCT-40 E6.2.6′ enriched to 96±24% of the pool after just two selection rounds, while pCT E6.2.6′ required four selection rounds to reach a comparable 98±4% (Figure 7F). Unfortunately, neither pCT E6.2.6′ AASV nor pCT-40 E6.2.6′ AASV were robustly enriched after five rounds.
Inverting the orientation from C-terminal fusion (Aga2p—Fn) to N-terminal fusion (Fn—Aga2p) maintains or hinders enrichment and yield on high- and mid-expressing cell lines for both high- and mid-affinity ligands (Figure 7). The enrichment ratio for high affinity clone E6.2.6′ decreases from 410±273 for the C-terminal fusion to 39±20 (p=0.001) for the N-terminal fusion when panned against highly expressing MDA-MB-468 cells. Enrichment for mid-affinity clone E6.2.6′ N78S decreases from 96±83 to 13±14 (p = 0.057) when using the same cells. This trend holds for panning against mid-expressing MDA-MB-231 cells as well. For high-affinity EI3.4.3, enrichment is 233±157 for C-terminal and 93±45 for N-terminal (p = 0.08). Notably, the expression level of multiple ligands tested on the yeast surface is comparable for N-terminal display and C-terminal display (Supplemental Figure 1).
Further exploration of the impact of paratope was conducted using the high-affinity CEA-binding fibronectin domain C7.4.3 to examine if the observed results are generalizable beyond fibronectin domain-EGFR complexes. When expressed as a C-terminal fusion to Aga2p with a 40-amino acid PAS linker and panned against LS174T human colorectal adenocarcinoma, the enrichment ratio (126±66) and yield (20±8%) remain quite strong, validating that fibronectin domains can bind other proteins on the mammalian cell surface in the yeast surface display context. Notably, LS174T cells expressed 4.4±4.5×104 CEA/cell on average although the distribution of CEA expression on a given day was much broader than EGFR expression on any cell line (Supplemental Figure 2). In fact, the top 10% of highest expressers averaged 2.9±0.8×105 CEA/cell.
Effect of Ligand Protein Scaffold
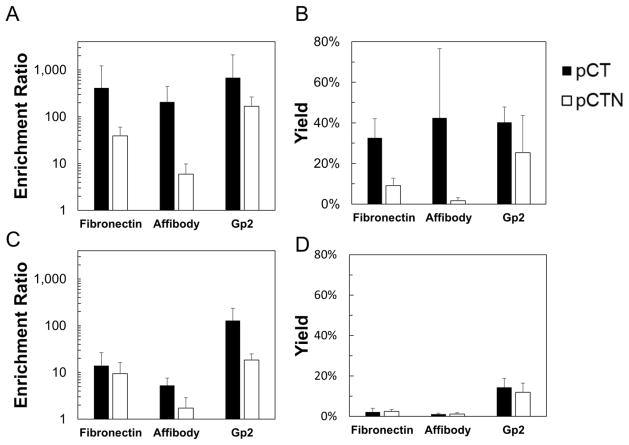
Spatial consideration can be further evaluated by testing alternative ligand scaffolds, which provide different paratope:epitope interactions as well as alternative display geometries. EGFR-binding fibronectin clone E6.2.6′, affibody clone EA68, and Gp2 clone GαEGFR2.2.3 were tested. In the C-terminal fusion construct, all three scaffolds effectively enriched against highly expressing MDA-MB-468 (Figure 8A, B). Non-binding fibronectin clone WT′, affibody clone A5, and Gp2 clone GαRIgG3.2.3 showed no appreciable enrichment (data not shown). However, on mid-expressing MDA-MB-231, Gp2 clone GαEGFR2.2.3 exhibits significantly higher enrichment than either fibronectin clone E6.2.6′ (p=0.04) or affibody clone EA68 (p=0.035) (Figure 8C, D). The same holds true for yield (p<0.001 in both cases). Notably, the Gp2 scaffold has its N-terminus distal to the presumed paratope whereas the fibronectin and affibody do not. All three ligands show significant decreases in enrichment and yield when displayed as N-terminal fusions to Aga2p relative to their C-terminal fusion counterparts. Interestingly, the superior enrichment and yield of GαEGFR2.2.3, relative to the fibronectin domain and affibody, are retained in the N-terminal fusion. The affibody variant exhibited lower enrichment than the other scaffolds on both cell types in both display orientations (p<0.05 for all scenarios (high- and mid-expression with N- and C-terminal display compared to fibronectin and Gp2) except p=0.07 for C-terminal display panned on mid-expressing MDA-MB-231 cells as compared to fibronectin)
Figure 8. The effect of protein scaffold on enrichment and yield.
Yeast displaying fibronectin domain E6.2.6′, affibody domain EA68, and Gp2 domain GαE2.2.3 mixed 1:1,000 with non-displaying yeast were panned against MDA-MB-468 (A, B) and MDA-MB-231 (C, D). Both N-terminal fusions and C-terminal fusions were tested. The enrichment (A, C) and yield (B, D) of binding ligands is presented as the mean ± standard deviation of 6–12 replicates.
Microscopic Visualization of Mammalian Cell – Yeast Interaction
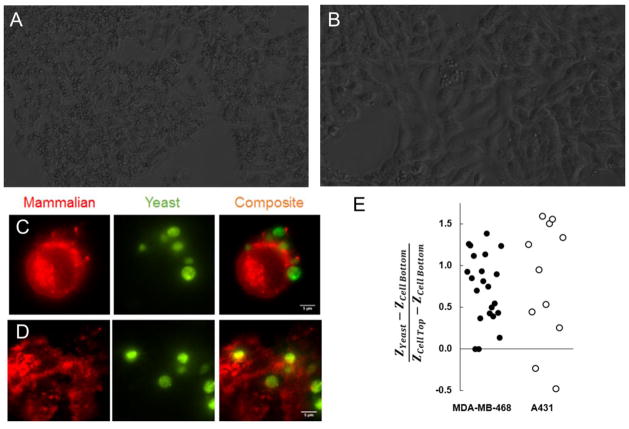
MDA-MB-468 and A431 express similar levels of EGFR per cell, but panning against these cell lines using fibronectin clone E6.2.6′ shows much stronger performance when using MDA-MB-468. Microscopic visualization was employed to help better understand cellular interactions in this system. Phase microscopy was initially used to visualize a macroscopic view of the mammalian cell – yeast binding landscape (Figure 9A, B). Visual inspection of the images corroborates panning data; many more yeast are seen binding to MDA-MB-468 than A431. In both cases, yeast are most often seen binding at the interfaces between mammalian cells or the interface of a mammalian cell and the plate surface. Yeast are rarely seen binding to the middle of a mammalian cell unless the mammalian cell is particularly crowded with yeast.
Figure 9. Visualization of mammalian cell – yeast binding interactions.
(A, B) Yeast displaying fibronectin domain E6.2.6′ were introduced directly to monolayers of MDA-MB-468 (A) or A431 (B) for imaging by phase microscopy. (C–E) Yeast displaying fibronectin domain E6.2.6′ were labeled by fluorescein isothiocyante conjugation (green) and introduced directly to monolayers of Alexa Fluor 594 wheat germ agglutinin-labeled (red) MDA-MB-468 (C) or A431 (D) for imaging by variable angle TIRF microscopy. Mammalian cells are shown as a single z-slice through the center. Yeast are shown by z-integration. Yeast binding interface position relative to the total height of the mammalian cell was quantified for both cell lines (E).
Variable angle TIRF microscopy was employed to gain a higher resolution, three-dimensional view of these binding interactions (Figure 9C, D, Supplemental Movies 1,2). Quantification of nine images using each cell line shows that the distribution of yeast binding locations are not appreciably different between both cell types (Figure 9E). The majority of yeast cells bound to the top portions of mammalian cells; very few yeast cells were found underneath mammalian cells. Variable angle TIRF images generally corroborated the phase microscopy images wherein the majority of yeast are found at mammalian cell – mammalian cell interfaces rather than in central portions of mammalian cells.
Discussion
Yeast surface display methods for direct cellular selections have shown success in the past, but applications have thus far been limited. We sought to gain a better understanding of how several parameters including washing stringency, incubation conditions, ligand and receptor stoichiometry, paratope location, yeast-displayed linker length and orientation, and ligand scaffold affect the ability to recover binding clones using these methods. The receptor expression levels, increased yeast-displayed linker length, and ligand scaffold had the most profound effects on the ability to enrich binding clones.
Multiple small alternative protein scaffolds – fibronectin domain, affibody, and Gp2 – can be enriched by panning ligand-displaying yeast on target-expressing human cell monolayers as described for antibody fragment systems (Wang et al., 2007). Enrichment is highly functional for mid-nanomolar affinity ligands when panned on human cells with millions of EGFR per cell (Figures 1 and 2). Slightly improved enrichment is observed for a point mutant with low nanomolar affinity. Conversely, a mutant engineered via four point mutations for >600 nM affinity required five rounds of selection in the standard system for moderate enrichment. Though limiting to binder discovery, the reduced enrichment of lower affinity mutants can be advantageous for affinity discrimination to empower evolution of stronger binders.
In addition to affinity, receptor density on the human cell surface strongly impacts ligand enrichment (Figures 3 and 4). The strong, monotonic correlation between enrichment of binding clones and receptor expression is explained by an increase in avid interaction between yeast and mammalian cells. Without sufficient avidity, yeast are unable to remain bound during washing and are lost. While limiting for enrichment of binders to some targets, selective enrichment based on receptor expression should aid the ability to isolate receptor-specific binders rather than binders to molecules ubiquitous, but lowly expressed on many cell types.
However, the correlation between receptor density and enrichment is not perfect; enrichment using the A431 cell line was significantly lower than when using the MDA-MB-468 cell line despite similar EGFR-expression levels. In order to better understand what causes this difference, TIRF microscopy was used to visualize yeast-mammalian cell binding interactions for both cell lines. In both cases, yeast tend to bind above the mammalian cells; there is no appreciable difference in the location of the binding interfaces despite a significant difference in morphology. Thus, the differences in enrichment do not appear to be differences in cellular-level physical characteristics. Although it is beyond the scope of this study, differences on a molecular level such as disparate receptor clustering could explain these differences.
Differing from the receptor expression results, decreased avidity with respect to yeast-displayed ligand expression does not show a significant impact on the ability to enrich binding clones (Figure 4). Notably, even with substantial variance in culture induction times (1 – 24 hours), the display of fibronectin domains only varied 3.5-fold. This expression level appears to still allow for the minimum functional avidity for recovery in the context of this experiment. Thus, the lack of impact on enrichment was likely partially driven by the small valency range evaluated. It remains to be determined if a yeast system engineered for ultra-high valency would provide improved enrichment. Moreover, we could not readily obtain very poor expression with our model system to evaluate lower valency conditions that may result from certain proteins or alternative display methods.
Fibronectin enrichment was broadly effective as evolved variants targeting three different EGFR epitopes were robustly enriched on highly expressing MDA-MB-468 cells (Figure 5). Moreover, a CEA-binding fibronectin was enriched 130±70-fold on LS174T cells. Breadth was further evaluated by testing three alternative small protein scaffolds: 10 kDa β-sandwich fibronectin domain, 7 kDa αhelical bundle affibody, and 5 kDa αβ Gp2 domain (Figure 8). EGFR-binding variants of all three were strongly enriched (210±50 to 680±410-fold) on highly expressing MDA-MB-468 cells and at least modestly enriched (5±2 to 130±110-fold) on mid-expressing MDA-MB-231 cells. Despite the fact that the Gp2 variant tested has modestly weaker affinity than the affibody and the fibronectin, GαEGFR2.2.3 outperformed these molecules on both cell lines. Moreover, the affibody variant EA68 had the lowest enrichment of the three tested scaffolds. These differences may be caused by paratope orientation or epitope accessibility. The presumed paratope of Gp2 is distal to its termini, which may reduce steric hindrance from the linker and/or the yeast surface to allow better accessibility in binding. Conversely, the affibody and fibronectin scaffolds have N-termini proximal to the evolved regions. It is important to note that general conclusions cannot be drawn from these scaffold comparisons because the observations relate to only one variant for each. In order to make stronger generalizations, additional variants from each scaffold type should be tested. Moreover, while numerous scaffolds, ligand mutants, and cell lines were used throughout to aid generalizability, experimental specifics should be carefully considered when analyzing data to differentiate between general principles and case-specific results.
Though advantageous for affinity discrimination and target specificity, the limited enrichment when using low-affinity ligands or mid-expressing cell monolayers can limit some applications. We hypothesized that an extended linker between the Aga2p tethering domain and the ligand could aid the ability for ligand to engage with receptor. The addition of a 40-mer PAS linker, based on the PAS#1 peptide (Table I) (Schlapschy et al., 2013), allowed for successful enrichment of E6.2.6′ AASV, the>600 nM affinity clone, using MDA-MB-468 (Figure 7). The ability to enrich molecules of such modest affinities is important for many applications, such as mimicking immune cell interactions and isolating binders from naïve combinatorial libraries to initiate directed evolution of new activity. Further, longer linkers allow for 2.5- to 13-fold improved enrichment of strong affinity clones E6.2.6′ and EI3.4.3 against the MDA-MB-231 cell line, which has 8-fold lower EGFR expression than MDA-MB-468. This broadens the reach of yeast panning for target-ligand co-discovery by strengthening enrichment on cells with upregulated target expression, albeit only to a ~200,000 target/cell level, which is the case for many important biomarkers. The enhanced enrichment also allows for differential affinity discrimination dependent on target density and affinity. Further experiments would be needed to identify the optimal linker length that balances gains in accessibility with potential detriments.
While extended linkers aided enrichment, changing the yeast surface display construct from Aga2p—ligand to ligand—Aga2p (Figure 6) did not; rather performance was generally decreased when ligands were fused to the N-terminus of Aga2p. These results were counter to the hypothesis that this orientation would enhance accessibility for fibronectin and affibody paratopes. While the polypeptide linker lengths were not equal (29 amino acids in ligand—Aga2p and 40 amino acids in Aga2p—ligand), this was designed to match distance between the yeast surface and the evolved fibronectin loops; i.e. to account for the length of the conserved fibronectin framework in the ligand—Aga2p construct (Figure 6). Notably, many of the clones tested were matured as C-terminal fusions to Aga2p. Changing the anchor point of these ligands may contribute to this decrease in performance and is not recommended for future ligand evolution campaigns.
Conclusions
In conclusion, yeast displaying small ligands of at least 20 nM affinity were readily enriched by panning on human cell monolayers expressing millions of targets per cell. Extension of the polypeptide linker by 20 or 40 amino acids enabled enrichment of a μM-affinity binder and improved enrichment of nM-affinity binders on cells expressing 105 targets per cell. Yeast display as C-terminal fusion to Aga2p yielded broadly superior enrichment relative to N-terminal fusion for the molecules tested; thus, orientation should be carefully considered in future use. These considerations can be applied to direct cellular selections both for ligand evolution against known biomarkers and ligand-biomarker co-discovery experiments using a variety of ligand scaffolds. Ultimately, the findings of this study will increase the efficiency of ligand discovery by removing the challenge of translatability presented by current soluble target-based selection methods.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Minnesota Futures program, the National Institutes of Health (R21EB019518), and the University of Minnesota.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Ackerman M, Levary D, Tobon G, Hackel BJ, Orcutt KD, Wittrup KD. Highly avid magnetic bead capture: an efficient selection method for de novo protein engineering utilizing yeast surface display. Biotechnol Prog. 2009;25:774–783. doi: 10.1002/btpr.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry M, Dower W, Johnston S. Toward cell–targeting gene therapy vectors: Selection of cell–binding peptides from random peptide–presenting phage libraries. Nat Med. 1996;2:299–305. doi: 10.1038/nm0396-299. [DOI] [PubMed] [Google Scholar]
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Bowley DR, Labrijn aF, Zwick MB, Burton DR. Antigen selection from an HIV-1 immune antibody library displayed on yeast yields many novel antibodies compared to selection from the same library displayed on phage. Protein Eng Des Sel. 2007;20:81–90. doi: 10.1093/protein/gzl057. [DOI] [PubMed] [Google Scholar]
- Case BA, Hackel BJ. Synthetic and natural consensus design for engineering charge within an affibody targeting epidermal growth factor receptor. Biotechnol Bioeng. 2016;9999 doi: 10.1002/bit.25931. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–68. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- Chen TF, De Picciotto S, Hackel BJ, Wittrup KD. Engineering fibronectin-based binding proteins by yeast surface display. Methods Enzymol. (1) 2013;523:303–326. doi: 10.1016/B978-0-12-394292-0.00014-X. [DOI] [PubMed] [Google Scholar]
- Cheng C, Chen YH, Lennox Ka, Behlke Ma, Davidson BL. In vivo SELEX for identification of brain-penetrating aptamers. Mol Ther — Nucleic Acids. 2013;2:e67. doi: 10.1038/mtna.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YK, Shusta EV. Antibody library screens using detergent-solubilized mammalian cell lysates as antigen sources. Protein Eng Des Sel. 2010;23:567–577. doi: 10.1093/protein/gzq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangaj D, Lanitis E, Zhao A, Joshi S, Cheng Y, Sandaltzopoulos R, Ra HJ, Danet-Desnoyers G, Powell DJ, Scholler N. Novel recombinant human B7-H4 antibodies overcome tumoral immune escape to potentiate T-cell antitumor responses. Cancer Res. 2013;73:4820–4829. doi: 10.1158/0008-5472.CAN-12-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra S, Mulders PFa, Schalken Ja. Clinical use of novel urine and blood based prostate cancer biomarkers: A review. Clin Biochem. 2014;47:889–896. doi: 10.1016/j.clinbiochem.2013.10.023. [DOI] [PubMed] [Google Scholar]
- Drake AW, Papalia GA. Biophysical condiserations for development of antibody-based therapeutics. Dev Antibody-Based Ther. 2012:95–139. [Google Scholar]
- Dua P, Kim S, Lee D. Nucleic acid aptamers targeting cell-surface proteins. Methods. 2011;54:215–225. doi: 10.1016/j.ymeth.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- Fu B, Zhang Y, Long W, Zhang A, Zhang Y, An Y, Miao F, Nie F, Li M, He Y, Zhang J, Teng G. Identification and characterization of a novel phage display-derived peptide with affinity for human brain metastatic breast cancer. Biotechnol Lett. 2014;36:2291–2301. doi: 10.1007/s10529-014-1608-0. [DOI] [PubMed] [Google Scholar]
- Gera N, Hussain M, Rao BM. Protein selection using yeast surface display. Methods. 2013;60:15–26. doi: 10.1016/j.ymeth.2012.03.014. [DOI] [PubMed] [Google Scholar]
- Hackel BJ, Ackermn ME, Howland SW, Wittrup KD. Stability and CDR composition biases enrich binder functionality landscapes. J Mol Biol. 2010;401:84–96. doi: 10.1016/j.jmb.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel BJ, Kapila A, Wittrup KD. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J Mol Biol. 2008;381:1238–52. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel BJ, Neil JR, White FM, Wittrup KD. Epidermal growth factor receptor downregulation by small heterodimeric binding proteins. Protein Eng Des Sel. 2012a;25:47–57. doi: 10.1093/protein/gzr056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel B, Kimura R, Gambhir S. Use of 64Cu-labeled fibronectin domain with EGFR-overexpressing tumor xenograft: molecular imaging. Radiology. 2012b;263:179–188. doi: 10.1148/radiol.12111504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Kang S, Lefort C, Kim M, Jin MM. Combinatorial libraries against libraries for selecting neoepitope activation-specific antibodies. Proc Natl Acad Sci U S A. 2010;107:6252–7. doi: 10.1073/pnas.0914358107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Pei W, Jayaseelan S, Shi H, Niu L. RNA aptamers selected against the GluR2 glutamate receptor channel. Biochemistry. 2007;46:12648–55. doi: 10.1021/bi701036p. [DOI] [PubMed] [Google Scholar]
- Husseinzadeh N. Status of tumor markers in epithelial ovarian cancer has there been any progress? A review. Gynecol Oncol. 2011;120:152–157. doi: 10.1016/j.ygyno.2010.09.002. [DOI] [PubMed] [Google Scholar]
- James ML, Gambhir SS. A molecular imaging primer: modalities, imaging agents, and applications. Physiol Rev. 2012;92:897–965. doi: 10.1152/physrev.00049.2010. [DOI] [PubMed] [Google Scholar]
- Kruziki MA, Bhatnagar S, Woldring DR, Duong VT, Hackel BJ. A 45-amino-acid scaffold mined from the PDB for high-affinity ligand engineering. Chem Biol. 2015;22:946–956. doi: 10.1016/j.chembiol.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nat Rev Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- Lu CF, Montijn RC, Brown JL, Klis F, Kurjan J, Bussey H, Lipke PN. Glycosyl phosphatidylinositol-dependent cross-linking of alpha-agglutinin and beta 1,6-glucan in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1995;128:333–340. doi: 10.1083/jcb.128.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäbert K, Cojoc M, Peitzsch C, Kurth I, Souchelnytskyi S, Dubrovska A. Cancer biomarker discovery: current status and future perspectives. Int J Radiat Biol. 2014;90:659–77. doi: 10.3109/09553002.2014.892229. [DOI] [PubMed] [Google Scholar]
- McCafferty J, Griffiths aD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- Mutuberria R, Satijn S, Angelique H, van der Linden E, Lichtenbeld H, Chames P, Arends J, Hoogenboom H. Isolation of human antibodies to tumor-associated endothelial cell markers by in vitro human endothelial cell selection with phage display libraries. J Immunol Methods. 2004;287:31–47. doi: 10.1016/j.jim.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Newton J, Deutscher SL. Phage peptide display. Handb Exp Pharmacol. 2008;185:145–163. doi: 10.1007/978-3-540-77496-9_7. [DOI] [PubMed] [Google Scholar]
- Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- Pepper LR, Cho YK, Boder ET, Shusta EV. A decade of yeast surface display technology: where are we now? Comb Chem High Throughput Screen. 2008;11:127–134. doi: 10.2174/138620708783744516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirie CM, Hackel BJ, Rosenblum MG, Wittrup KD. Convergent potency of internalized gelonin immunotoxins across varied cell lines, antigens, and targeting moieties. J Biol Chem. 2011;286:4165–72. doi: 10.1074/jbc.M110.186973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman Sa, Healan SJ, Weber KS, Donermeyer DL, Dossett ML, Greenberg PD, Allen PM, Kranz DM. Development of a novel strategy for engineering high-affinity proteins by yeast display. Protein Eng Des Sel. 2006;19:255–64. doi: 10.1093/protein/gzl008. [DOI] [PubMed] [Google Scholar]
- Sanchez-Martin D, Sorensen MD, Lykkemark S, Sanz L, Kristensen P, Ruoslahti E, Alvarez-Vallina L. Selection strategies for anticancer antibody discovery: searching off the beaten path. Trends Biotechnol. 2015;33:292–301. doi: 10.1016/j.tibtech.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlapschy M, Binder U, Börger C, Theobald I, Wachinger K, Kisling S, Haller D, Skerra A. PASylation: A biological alternative to PEGylation for extending the plasma half-life of pharmaceutically active proteins. Protein Eng Des Sel. 2013;26:489–501. doi: 10.1093/protein/gzt023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillotson BJ, Cho YK, Shusta EV. Cells and cell lysates: a direct approach for engineering antibodies against membrane proteins using yeast surface display. Methods. 2013;60:27–37. doi: 10.1016/j.ymeth.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanAntwerp JJ, Wittrup KD. Fine affinity discrimination by yeast surface display and flow cytometry. Biotechnol Prog. 2000;16:31–7. doi: 10.1021/bp990133s. [DOI] [PubMed] [Google Scholar]
- Wang XX, Cho YK, Shusta EV. Mining a yeast library for brain endothelial cell-binding antibodies. Nat Methods. 2007;4:2006–2008. doi: 10.1038/nmeth993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XX, Shusta EV. The use of scFv-displaying yeast in mammalian cell surface selections. J Immunol Methods. 2005;304:30–42. doi: 10.1016/j.jim.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Williams RM, Hajiran CJ, Nayeem S, Sooter LJ. Identification of an antibody fragment specific for androgen-dependent prostate cancer cells. BMC Biotechnol. 2014;14:81. doi: 10.1186/1472-6750-14-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yotsukura S, Mamitsuka H. Evaluation of serum-based cancer biomarkers: A brief review from a clinical and computational viewpoint. Crit Rev Oncol Hematol. 2015;93:103–115. doi: 10.1016/j.critrevonc.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Yujing S, Shukla GS, Kennedy GG, Warshaw DM, Weaver DL, Pero SC, Floyd L, Krag DN. Biopanning phage-display libraries on small tissue sections captured by laser capture microdissection. Jounral Biotechnol Res. 2009;1:55–63. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.