Abstract
BACKGROUND & AIMS
Genome-wide association studies (GWAS) have identified 200 inflammatory bowel disease (IBD) loci, but the genetic architecture of Crohn’s disease (CD) and ulcerative colitis (UC) remains incompletely defined. Here we aimed to identify novel associations between IBD and functional genetic variants using the Illumina ExomeChip.
METHODS
Genotyping was performed in 10,523 IBD cases and 5,726 non-IBD controls. 91,713 functional single nucleotide polymorphism (SNP) loci in coding regions were analyzed. A novel identified association was further replicated in two independent cohorts. We further examined the association of the identified SNP with microbiota from 338 mucosal lavage samples in the Mucosal Luminal Interface (MLI) cohort measured using 16S sequencing.
RESULTS
We identified an association between CD and a missense variant encoding alanine (Ala) or threonine (Thr) at position 391 in the zinc transporter solute carrier family 39, member 8 protein (SLC39A8 Ala391Thr, rs13107325) and replicated the association with CD in two replication cohorts (combined meta-analysis p=5.55×10−13). This variant has previously been associated with distinct phenotypes including obesity, lipid levels, blood pressure and schizophrenia. We subsequently determined that the CD-risk allele was associated with altered colonic mucosal microbiome composition in both healthy controls (p=0.009) and CD cases (p=0.0009). Moreover, microbes depleted in healthy carriers strongly overlap with those reduced in CD patients (p=9.24×10−16) and overweight individuals (p=6.73×10−16).
CONCLUSIONS
Our results suggest that an SLC39A8-dependent shift in the gut microbiome could explain its pleiotropic effects on multiple complex diseases including CD.
Keywords: Genetics, Inflammatory Bowel Diseases, Microbiota
Introduction
The inflammatory bowel diseases (IBD), Crohn’s disease (CD) and ulcerative colitis (UC), are chronic relapsing inflammatory conditions of the gastrointestinal tract.1 These diseases are a significant cause of morbidity and have estimated direct and indirect costs of $6 billion annually in the United States.2
Currently, the etiology and pathogenesis of IBD are not fully understood, but it is widely accepted that genetic factors play an important role. Common variant genome-wide association studies have identified 200 IBD-associated loci.3, 4 However, these loci explain only part of the variance and genetic architecture of CD and UC.3, 4
Changes in the gut microbiota have also been associated with IBD.5–11 IBD patients have reduced bacterial diversity, and complex compositional changes in CD or UC patients include increased Enterobacteriaceae (such as E. coli) and Veillonellacae, and reduced Ruminococcaceae (such as F. prauznitzii), Roseburia, and Clostridials. Moreover, many of the known IBD susceptibility genes are associated with recognition and processing of bacteria.3, 4, 12–14 A ‘gardening’ effect of known IBD genetic variants on gut microbiome has also been reported, suggesting a role of the gut microbiota in the pathogenesis of IBD.15
In this study, we aimed to identify novel associations between IBD and functional genetic variants using the Illumina ExomeChip array in a large European ancestry cohort. We also examined the microbiome shift associated with an identified novel locus to elucidate its functional role and understand how it contributes to disease pathogenesis.
Materials and Methods
Overview
A collaborative group with a shared goal of conducting cost-effective genotyping of their case samples and shared control samples using the Illumina Infinium HumanExome BeadChip was formed. The HumanExome BeadChip was designed to complement common variant genotyping arrays by enabling cost-effective genotyping of putative functional exonic variants that were selected from over 12,000 individual exome and whole-genome sequences from diverse populations. Its content includes non-synonymous variants, splice variants, and stop altering variants, observed at least two times across two or more of the sequencing datasets. It also includes: tags for previously described GWAS hits; African American vs. European and Native American vs. European ancestry informative markers; a scaffold grid of markers designed for identity by descent analyses; a random set of synonymous variants; fingerprint SNPs shared among several major genotyping platforms; mitochondrial SNPs; chromosome Y SNPs; and HLA tag SNPs. Some of the collaborating groups designed custom content that was added to the HumanExome base content to address individual project-specific aims. The resultant Illumina Infinium HumanExome+ BeadChip was used to genotype all cases and shared control samples. Written, informed consent was obtained from all study participants and the institutional ethical review committees of the participating centers approved all protocols.
The data for all samples were pooled together in order to optimize accurate genotype calling and quality control filtering. In this manuscript, we report results from our analyses of predicted functional SNPs (missense, nonsense or splice variants) in non-Jewish European ancestry IBD case and control samples.
Illumina Infinium HumanExome+ BeadChip genotyping and quality control
DNA samples from 23,789 human peripheral blood or B-lymphoblastoid cell line specimens were processed using an Illumina Infinium HumanExome+ BeadChip at Cedars-Sinai Medical Center in Los Angeles, California; The Children’s Hospital of Philadelphia in Philadelphia, Pennsylvania; The Feinstein Institute for Medical Research in Manhasset, New York; and the University of Pittsburgh in Pittsburgh, Pennsylvania. A single compiled genotyping project was created (GenomeStudio v2011.1) and intensity data for 21,233 samples deemed to be the highest quality samples based on preliminary genotype call rate and p10GC statistics were used to recluster all SNPs, and then the resultant cluster file was applied to all samples. Variants were then systematically reviewed based on several marker statistic parameters including cluster separation, theta mean and deviation, heterozygous excess and frequency, call frequency, minor allele frequency, R intensity mean, and replicate error rate, in addition to review of mitochondrial and Y chromosome markers and indels.16 Following these quality control metrics, 6,849 SNPs were excluded without further manual review and 48,962 SNPs were manually reviewed and when possible, cluster locations adjusted to achieve optimal allele-calling. There was 99.9963% concordance for genotypes in 273 replicate control samples.
After genotype calling was complete, 1,161 samples were excluded based on the following criteria: p10GC and call rate statistics, gender discrepancies between reported and genotype-determined gender or ambiguous genotype-determined gender, misidentified samples, outlier samples consistently clustering outside the three distinct genotype clusters as identified by manual review of intensity data plots, high heterozygosity, and genetic relatedness. After the genotype calling and quality control filtering steps, data for 207,625 polymorphic SNP assays in 22,628 individuals remained.
We focused our subsequent analyses on 10,523 IBD cases (5,742 CD, 4,583 UC and 198 IBD unclassified) and 5,726 controls that formed a major European ancestry cluster based on principal components analyses, and on 153,486 autosomal and chromosome X SNPs predicted to be functional (missense, nonsense or splice variants) and available in the HumanExome base content with ≤0.5% missing data and Hardy-Weinberg equilibrium p-value in controls ≥1×10−5.
Statistical analyses
We adopted strategies previously utilized for ExomeChip single SNP analysis17. SNPs with at least 6 copies of minor alleles observed in the sample set were included in the single SNP analysis, and 61,773 SNPs with less than 6 copies were excluded. For the 91,713 variants included in the single SNPs analysis, the significance threshold was 5.45×10−7 after Bonferroni correction. To account for the rare variants in single SNP analysis, statistical inference on trait-SNP association was performed using linear regression assuming an additive genetic model, following examples in a previous study.17 In single SNP analysis, we also utilized logistic regression to estimate the Odds ratios (OR) and 95% confidence intervals (95% CI) when applicable. The first four principle components were included in the model as covariates to control for potential confounding effects due to population stratification. In addition to the standard genotyping quality control measures (listed above), genotype clusters for key SNPs listed in main tables were manually reviewed by two independent research personnel to ensure accurate allele-calling.
Replication cohorts
To validate novel association findings, we used two additional cohorts, including non-overlapping samples from a pediatric IBD GWAS cohort (1,096 CD cases and 6,088 non-IBD controls)18 and the Prospective Registry in IBD Study at Massachusetts General Hospital (PRISM) exome chip cohort (551 CD cases and 2,344 non-IBD controls).19 In both cohorts, association was tested using logistic regression with adjustment for principal components. We also performed an inverse-variance meta-analysis to combine results from all three cohorts, leading to a total sample size of 7,389 cases and 14,158 controls.
Microbiome analysis
The MLI cohort consists of 338 mucosal lavage samples from the cecum and sigmoid colon (i.e. 2 samples per person) of healthy individuals (22 SLC39A8 Thr391 allele carriers and 75 non-carriers) and CD patients in endoscopic remission (16 SLC39A8 Thr391 allele carriers and 58 non-carriers).20 Genomic DNA extraction, V4 region 16S ribosomal RNA gene amplification, data-preprocessing, and 97% operational taxonomic unit (OTU) picking were performed as previously described,20 yielding a median sampling sequencing depth of 606,105. Alpha diversity was assessed using the number of observed species, Chao1, phylogenetic diversity, and Shannon index on rarefied data. 16S rRNA abundances underwent normalization by a scaling factor (median of ratios of OTU counts to geometric mean across all samples).21 Distance matrices were calculated using root square Jensen-Shannon divergence, and then principal coordinates analysis was performed in QIIME. Additional beta diversity metrics including unweighted UniFrac, weighted UniFrac, and Bray–Curtis were measured using rarefied data in QIIME. P-values were calculated using Adonis.
Analysis of association between novel IBD-associated genetic variants and OTUs or genera was performed using Phyloseq22 and the DESeq2 algorithm (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html).23 OTUs present in less than 10% of samples were removed prior to analysis. An empirical Bayesian approach was used to shrink dispersion of normalized count data. Log fold changes for each OTU were fitted to a general linear model (fixed effects only) under a negative binomial model. Multivariate models included gender, lavage site, disease status, body mass index (BMI) (25< or >25), and SLC39A8 carrier status. OTUs or genera were filtered out by choosing a mean count threshold maximizing the number of OTUs returned at a given false discovery rate. Outliers were replaced by trimmed means, and p-values for the coefficients for carrier status in the linear models were calculated using the Wald test, then converted to q-values (http://www.bioconductor.org/packages/release/bioc/html/qvalue.html). Associations were considered significant if they were below a q-value threshold of 0.05. Hypothesis inference on the overlap of OTUs associated with CD, obesity and the SLC39A8 Thr391 allele was performed using the log-linear model.
Results
We analyzed 91,713 rare and common functional (missense, nonsense or splice variant) polymorphic SNPs that passed quality control (Table S1). Complete results for all SNPs in the single SNP analysis can be found in Table S2. QQ plots show modest genomic inflation (λGC=1.074, 1.093 and 1.094 for CD, UC and IBD, respectively). Functional variants in previously reported IBD loci such as NOD2, IL23R, and CARD93, 4 were significantly associated with CD, UC or both forms of IBD after Bonferroni correction (p<5.45×10−7) (Table S3). Furthermore, an additional common missense variant, rs13107325, which encodes SLC39A8 Ala391Thr, was significantly associated with CD (p=3.77×10−8; OR=1.31, 95% CI 1.19–1.44) (Table 1). We also examined the effects of different genotypes of the Ala391Thr variant and observed an OR of 1.32, 95% CI 1.19–1.47, for the heterozygous Ala/Thr genotype (genotype frequency 16.64% in CD and 13.10% in controls, p=1.66×10−7) and an OR of 1.53, 95% CI 0.96–2.36, for the homozygous Thr/Thr genotype (genotype frequency 0.92% in CD and 0.63% in controls, p=0.054), suggesting an additive heredity model. We checked the conservation of the SLC39A8 Ala391Thr variant across different species and observed a PolyPhen24 score of 0.782, indicating that this variant is “possibly damaging.” SLC39A8 Ala391Thr has a PHRED score of 18.44 in the Combined Annotation Dependent Depletion (CADD) database,25 suggesting that it is among the top 1.43% most deleterious substitutions possible in the human genome.
Table 1.
Meta-analysis of the association of SLC39A8 Ala391Thr variant with CD in discovery and replication cohorts
| Gene | SNP | Cohort | N cases | N controls | FA* | FU^ | P | OR(95% CI) |
|---|---|---|---|---|---|---|---|---|
| Discovery cohort | 5742 | 5726 | 0.092 | 0.072 | 3.77×10−8 | 1.31(1.19–1.44) | ||
| SLC39A8 | Ala391Thr | Pediatric CD GWAS | 1096 | 6088 | 0.108 | 0.088 | 2.35×10−3 | 1.26(1.09–1.46) |
| PRISM | 551 | 2344 | 0.109 | 0.072 | 1.20×10−4 | 1.56(1.24–1.95) | ||
| Meta-analysis | 7389 | 14158 | - | - | 5.55×10−13 | 1.33(1.21–1.45) |
FA: Frequencies of Thr allele in CD
FU: Frequencies of Thr allele in non-IBD
SLC39A8 is located approximately 200 Kb centromeric from NFKB1, a gene in a known UC-associated locus, and 250 Kb downstream of BANK1, a gene in a known CD-associated locus.3 Conditional analyses were performed by including both index SNPs (rs13126505 in BANK1 and rs1598859 in NFKB1, which has a linkage disequilibrium (LD) r2=1 with the reported rs37749593) in the model (Table 1). Effect estimates for SLC39A8 Ala391Thr did not vary much (ORcond=1.28, 95% CI 1.12–1.47), although the p-value dropped to 3.00×10−4 conditional on the two index variants in NFKB1 and BANK1. Interestingly, the effect of the known CD-associated BANK1 SNP rs13126505, which is in moderate LD with the SNP encoding SLC39A8 Ala391Thr (r2=0.50), greatly diminished in the joint model (in single SNP analysis: p=4.43×10−5, OR=1.24; in joint model: p=0.021, OR=1.07). Furthermore, the SLC39A8 SNP remained associated with CD in an analysis that was stratified by the BANK1 SNP (rs13126505), but the BANK1 SNP was no longer significant in the reciprocal stratification analysis (Table S4), indicating that the previously identified association signal at the BANK1 locus3 is at least partially driven by the SLC39A8 missense variant. The UC-associated NFKB1 SNP is not in LD with the SNP encoding the CD-associated SLC39A8 Ala391Thr variant (r2=0.004), strongly suggesting that its effect is independent of the observed association with SLC39A8. We also examined the interaction between SLC39A8 Ala391Thr and known CD SNPs genotyped on the HumanExome+ BeadChip and observed no statistically significant interactions after controlling for multiple testing (Table S5).
We then replicated the SLC39A8 Ala391Thr association in two independent cohorts (Table 1). We observed similar association in both replication cohorts (pediatric CD GWAS cohort: p=2.35×10−3; OR=1.26, 95% CI 1.09–1.46; PRISM cohort: p=1.20×10−4; OR=1.56, 95% CI 1.24–1.95). The evidence for association between CD and SLC39A8 Ala391Thr was highly significant (p=5.55×10−13; OR=1.33, 95% CI 1.21–1.45) in a meta-analysis of the combined discovery and replication cohorts.
A genotype-subphenotype correlation analysis (Table 2) for SLC39A8 Ala391Thr found significant evidence for association with ileal CD (p=0.002 compared to non-ileal CD; OR=1.36, 95% CI 1.12–1.66) and with complicated disease behavior (p=0.006 for stricturing (B2) or penetrating (B3) vs. non-stricturing, non-penetrating (B1) behavior; OR=1.22, 95% CI 1.06–1.40). Joint analysis indicates that the associations of SLC39A8 with ileal CD and B2/B3 are independent signals (data not shown).
Table 2.
Association of SLC39A8 Ala391Thr with clinical phenotypes of CD
| Gene | SNP | Outcome* | N | P | OR(95% CI) |
|---|---|---|---|---|---|
| B2 vs. B1 | 3415 | 0.030 | 1.20(1.02–1.42) | ||
| B3 vs. B1 | 3602 | 0.012 | 1.23(1.05–1.45) | ||
| B3 or B2 vs. B1 | 4852 | 0.006 | 1.22(1.06–1.40) | ||
| SLC39A8 | Ala391Thr | Colonic location | 4942 | 0.966 | 1.00(0.86–1.17) |
| Ileal location | 4823 | 0.002 | 1.36(1.12–1.66) | ||
| Jejunal location | 4460 | 0.205 | 1.17(0.92–1.50) | ||
| Perianal disease | 4876 | 0.868 | 0.99(0.85–1.15) | ||
| Surgery | 4841 | 0.268 | 1.08(0.94–1.24) |
Disease behavior classified according to Montreal classification: B1 – inflammatory or non-stricturing/non-internal penetrating phenotype; B2 – Stricturing phenotype; B3 – internal penetrating or fistulizing phenotype. Surgery is defined as abdominal surgery for complication of Crohn’s disease.
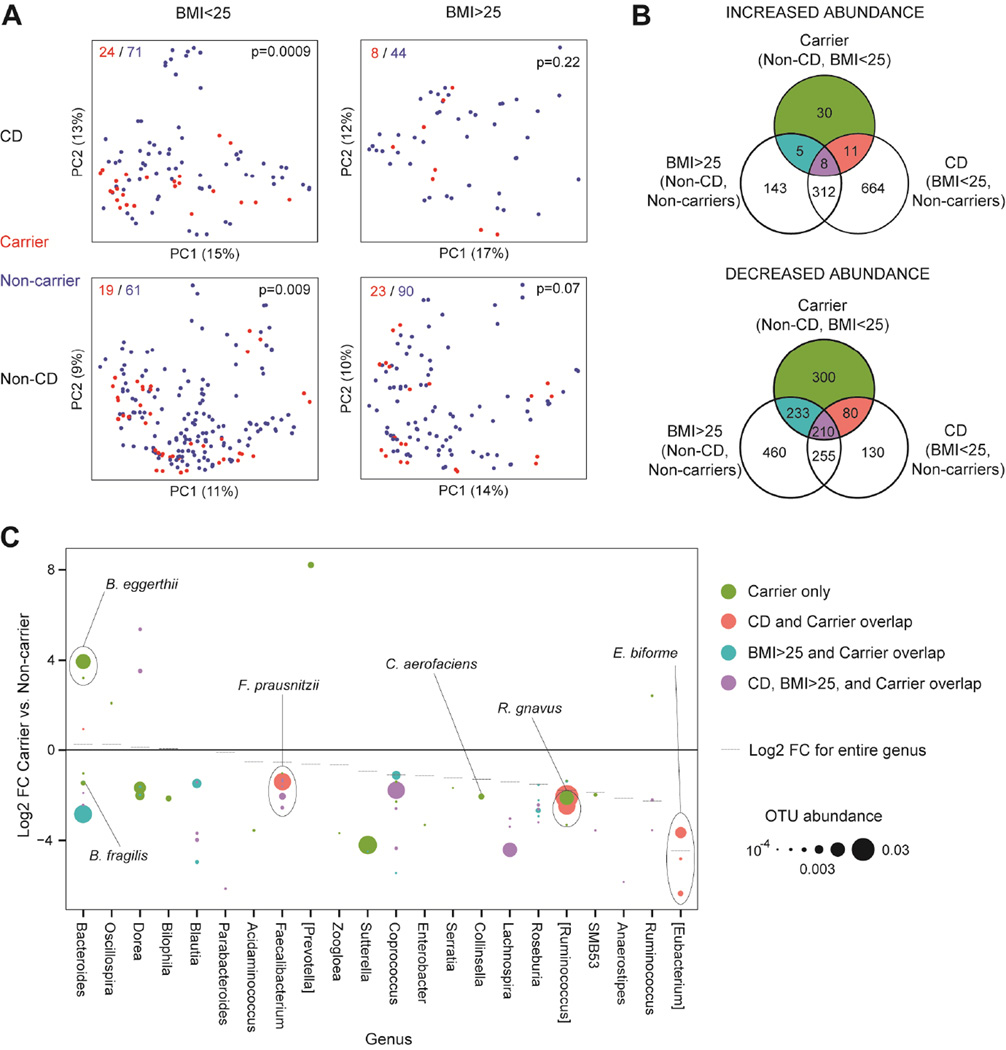
Recognizing the role that zinc plays in innate immunity and the pleitropic nature of this polymorphism with association with other microbiome associated traits including obesity, we hypothesized that the SLC39A8 genetic variant may affect CD susceptibility by altering the microbiome. Therefore, we tested the microbial composition of lavage samples from the cecum and sigmoid colon of SLC39A8 Thr391 allele carriers and non-carriers in the MLI15, 20, 26 cohort. CD patients and healthy controls were analyzed separately and were further subdivided by BMI (greater or less than 25) given previous associations of both CD and obesity with the microbiome. Carriage of the CD-associated Thr allele was associated with altered microbial composition as determined by 16S ribosomal RNA sequencing in both healthy controls and CD patients with BMI<25 (p=0.009 and 0.0009, respectively) (Figure 1A). We focused on carrier-associated changes in the microbiome of non-overweight healthy controls to capture pre-disease perturbations that may predispose to CD and obesity. A multivariate analysis of the non-overweight healthy controls incorporating carrier status, sampling site, and gender revealed statistically significant enrichment of 54 OTUs and depletion of 823 OTUs in the Thr allele carriers (Table S6). We further investigated whether the microbial signature of non-overweight healthy SLC39A8 variant carriers resembled that of CD patients. Indeed, 290 of the 823 depleted OTUs reduced in healthy Thr allele carriers were also reduced in non-carrier, non-overweight CD patients compared to non-carrier, non-overweight healthy controls, indicating a strongly statistically significant overlap (p=9.24×10−16) of the shift in gut microbiota (Figure 1B, Table S7). There was similar overlap of the SLC39A8 microbial signature with that of individuals with BMI>25: 443 OTUs depleted in non-overweight healthy Thr allele carriers were also reduced in overweight, non-IBD, non-carriers (p=6.73×10−16) (Table S8). This overlap was not observed in the enriched OTUs (19 and 13 of the 54 enriched OTUs also associated with CD and overweight status, with p=0.88 and 0.10, respectively).
Figure 1. SLC39A8 polymorphism is associated with shifts in the colonic microbiome paralleling those of Crohn’s disease and obesity.
A) Principal coordinates analysis plots of the MLI cohort, divided into subsets by CD status and BMI. Individuals are colored by SLC39A8 Thr391 allele carrier status. Each PCoA plot shows the number of samples from carriers (red) and non-carriers (blue) in the upper left corner and p-values for the difference in microbial composition between carriers and non-carriers in the upper right corner. B) Venn diagrams depicting the overlap of enriched and depleted OTUs in three comparisons: SLC39A8 Thr391 allele carriers vs. non-carriers in the non-CD subset with BMI<25, CD vs. non-CD individuals in the non-carrier subset with BMI<25, and BMI>25 vs. BMI<25 in the non-carrier, non-CD subset. OTUs were identified as enriched or depleted if they had q<0.05 in multivariate models with sample site (cecum, sigmoid) and gender as additional covariates. C) Log2 fold changes (FC) for OTUs with a statistically significant difference in abundance between carriers and non-carriers from multivariate models. Only OTUs identified to the genus level with mean normalized abundance > 10−4 in the non-IBD, BMI<25 subset are shown. Colors correspond to the Venn diagrams in panel B. Genera are ordered by the log2 fold changes of entire genera between carriers and non-carriers.
Among relatively abundant bacterial taxa, five genera depleted in SLC39A8 Thr391 allele carriers were also significantly (q<0.05) depleted in CD patients and overweight controls (Anaerostipes, Coprococcus, Roseburia, Lachnospira, and SMB53) (Figure 1C). Additional genera depleted in carriers and CD included Faecalibacterium prausnitzii and Ruminococcus gnavus. The most strongly depleted genus in Thr allele carriers was [Eubacterium] (q=9×10−10), attributable to reduced abundance of three OTUs identified as E. biforme (Figure 1C). Other changes specific to Thr allele carriers included specific species of the Bacteroides (e.g., B. eggerthii and B. fragilis), Dorea genera Sutterella, and Collinsella aerofaciens.
Discussion
In the current study, we utilized the Illumina ExomeChip to identify functional variants associated with IBD. We found that a missense variant in SLC39A8, a pleiotropic locus with effects in several phenotypes including hypertension, blood lipid, obesity as well as schizophrenia, is associated with CD. We replicated this finding in two independent cohorts and illustrated that this locus is also associated with disease location and behavior in CD. We further demonstrated that this locus is associated with shifts in the composition of the gut microbiota in both CD and controls. Moreover, these SLC39A8-associated shifts in the composition of the gut microbiota strongly overlap with gut microbiota changes associated with CD and obesity.
SLC39A8 is part of a family of zinc transporters that is localized to the plasma membrane and early endosomes27 and mediates zinc influx into the cytosol.28, 29 Zinc has concentration-dependent effects on immune function, and zinc deficiency has been shown to affect T cell counts and function.30 SLC39A8 is highly expressed in T cells, stimulated monocytes, and differentiated macrophages,28, 31 and a recent study demonstrated that continuous stimulation of pattern recognition receptors in macrophage derived monocytes increased intracellular zinc concentrations and enhanced macrophage clearance of bacteria via autophagy.32 Upregulated expression of SLC39A8 during T cell activation leads to increased intracellular zinc in the cytoplasm and increased IFN-γ expression.28 In addition, SLC39A8 is upregulated in response to tumor necrosis factor-alpha and lipopolysaccharide and serves as a negative regulator of innate immune function and of NF-κB signaling via zinc-mediated suppression of IκB kinase in monocytes and macrophages.33, 34 Such effects on T cell function and innate immunity are of potential key pathophysiologic relevance to IBD, and our findings highlight the importance of further defining the role of zinc in IBD. In a study of treatment naïve pediatric IBD patients, SLC39A8 was one of 1,281 genes that were differentially expressed in ileal biopsies from patients with CD.35 SLC39A8 Ala391Thr has also been associated with lipid levels, blood pressure, obesity and schizophrenia, emphasizing the pleiotropic and critical role of this variant in health and disease as well as further highlighting the intriguing relationship among metabolic features, psychiatric illness, and chronic inflammation.36–41
CD, dyslipidemia and obesity have all been linked to alterations in the intestinal microbiome,6, 42 and a recent study linked the same gut microbiome change (decrease in Akkermansia muciniphila) to both IBD and type II diabetes.43 Furthermore, in a study of the microbiome in new-onset CD, an exploratory analysis of genomes of organisms associated with CD identified a contribution to pathway components that included a zinc-dependent enzyme.6 We therefore hypothesized that the SLC39A8 genetic variant identified in this study may affect CD susceptibility by altering the microbiome. In the microbiome analysis, 5 genera (Anaerostipes, Coprococcus, Roseburia, Lachnospira, and SMB53) are depleted in SLC39A8 Thr391 allele carriers, CD patients and overweight controls. Faecalibacterium prausnitzii and Ruminococcus gnavus are depleted in both Thr allele carriers and CD. These genera (with the exception of SMB53) are notable for being producers of short-chain fatty acids, which can ameliorate colitis through induction of regulatory T cells.44, 45 Prior studies also found lower levels of Faecalibacterium prausnitzii in IBD,6, 11, 46, 47 and higher levels of this bacterium are associated with lower risk of postoperative CD recurrence and maintenance of remission in UC.47, 48 An early small Sanger sequencing study of ileal biopsies had suggested increased Ruminococcus gnavus in CD,49 possibly reflecting differences in ileal vs. colonic location or differences among Ruminococcus gnavus strains in their association with CD. This organism was also depleted in CD patients compared to healthy controls, an association not previously reported but perhaps related to the unique mucosal lavage sampling protocol used for this cohort. Thus, SLC39A8 is part of an emerging set of loci associated with taxonomically-restricted microbial composition.48
We investigated the effect of SLC39A8 Ala391Thr on protein structure. The closest transporters to SLC39A8 are SLC39A14 and SLC39A12 with 50% sequence identity. There is no known X-ray 3D structure for SLC39A8, and since this region is not homologous to other zinc transporters with X-ray 3D structures, the effect of the Ala391Thr variant on the structure of SLC39A8 cannot be inferred. Thus the effect of this variant on transporter structurr variant is located in a transmembrane domain (based on Uniprot) and mutations involving polar residues are particularly associated with protein malfunction.50 Loss of function of SLC39A8, a zinc solute transporter, may perturb zinc uptake by epithelial and immune cells and cause zinc deficiency as has been observed in SLC39A8 hypomorphic mice.51 Animal studies have indicated that zinc deficiency alters microbial function and composition.52 Moreover, some ion channels, such as KCNN4, which share membrane transporter properties and roles with solute transporters, are activated by intracellular calcium and regulate Paneth cell secretion53 with an impact on immune cell and epithelial cell function.54 Therefore, we speculate that by disturbing the transmembrane domain of SLC39A8, Ala391Thr may alter zinc metabolism in functionally relevant cells, which might in turn affect innate and adaptive immunity, as well as the gut microbiota.
In summary, we identified and replicated a novel association between CD and SLC39A8, a zinc transporter linked to multiple metabolic traits. The SLC39A8 Ala391Thr variant identified in our study was associated with altered microbiome composition in both healthy controls and CD patients. Moreover, the observed microbe signature in healthy SLC39A8 Thr391 allele carriers strongly overlaps with that in CD patients and overweight individuals, suggesting a pre-disease microbial susceptibility state originating due to genetic influence on the microbial ecosystem that contributes to the pleiotropic effect of SLC39A8 in CD and metabolic traits.
Acknowledgments
Grant Support: The following support for this work is acknowledged by the authors whose initials or names are in parentheses following each source of support. The NIDDK IBD Genetics Consortium is supported by National Institutes of Health (NIH) grants U01DK062413 (DPBM, J Braun), U01DK062420 (RHD, MDR), U01DK062422 (JHC, KYH, DDP), U01DK062429 (JHC, LPS, YS), U01DK062423 (MSS, RM, JMS), U01DK062431 (SRB), U01DK062432 (JDR). This work was also supported by NIH grants F30DK098927 (KYH), P01DK046763 (SRT, J Braun, DPBM), P30CA016042 (J Braun), R01CA141743 (RHD), R01DK087694 (SK), R01DK092235 (JHC, KYH), R01DK098231 (SK), R01HS021747 (DPBM), T32DK007180 (JPJ), T32GM007205 (KYH), U01AI067068 (DPBM), U54DE023798 (J Braun), and UL1TR000124 (J Braun). Additional sources of support included the Crohn’s and Colitis Foundation of America (J Braun, HH), Deutsche Forschungsgemeinschaft (DFG BR 1912/6-1) (SRB), Deutsche Forschungsgemeinschaft (DFG; projects Ni575/7-1 and Ni 575/4-1) (J-HN), Else Kröner-Fresenius-Stiftung (Else Kröner Exzellenzstipendium 2010; 2010_EKES.32) (SRB), Inflammatory Bowel Disease Genetic Research Chair at the University of Pittsburgh (RHD), Institutional Development Fund The Children’s Hospital of Philadelphia (HH), Örebro University Hospital Research Foundation (JH), Royal Brisbane and Women’s Hospital Research Foundation (GR-S), Sanford J Grossman Charitable Trust (JHC), SUCCESS (JHC), Swedish Research Council (521-2011-2764) (JH), Swedish Research Council (VR 2010–2976) (M D’Amato), Swedish Research Council (VR 2013–3862) (M D’Amato), Swiss National Science Foundation (SNF) 146290 (J-HN), The Eli and Edythe Broad Foundation, Proposal No. IBD-0164R (CB), The European Union (DPBM), The Kenneth Rainin Chair for IBD Research (J-PA), The Leona M and Harry B Helmsley Charitable Trust (DPBM), and The National Health and Medical Research Council (APP498405) (GR-S). The authors thank the following investigators for providing additional control samples: M Ilyas Kamboh with support from NIH grants R01AG030653, R01AG041718, P50AG005133, R01AG007562, and R01AG023651; David C Whitcomb with support from NIH grant R01DK061451; Todd Lencz; and Peter K Gregersen.
Abbreviations used in this paper
- Ala
alanine
- BMI
body mass index
- CI
confidence intervals
- GWAS
genome-wide association studies
- LD
linkage disequilibrium
- MLI
Mucosal Luminal Interface
- NIH
National Institutes of Health
- OR
odds ratio
- OTU
operational taxonomic unit
- PRISM
Prospective Registry in IBD Study at Massachusetts General Hospital
- SNP
single nucleotide polymorphism
- Thr
threonine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: Nothing to disclose.
Author Contributions: Overall project supervision and management: RHD, DPBM, J Braun. Genotype calling: TH, RHD. Genotype data cleaning and quality control: TH, RHD, LK, BD, LPS. Population stratification analysis: DL, KYH, LK, BD, RHD. Genetic association analysis: DL, LK, BD, DPBM, RHD. SNP annotation: KYH. Microbiome analysis: JPJ, J Braun, J Borneman. Primary drafting of the manuscript: DL, J-PA, TH, JPJ, J Braun, DPBM, RHD. Major contribution to drafting of the manuscript: M D’Amato, SB, JH, M Daly, JDR, JHC. The remaining authors contributed to the study conception, design, subject recruitment, subject phenotyping, genotyping, microbial 16S ribosomal RNA sequencing, and/or data management. All authors saw, had the opportunity to comment on, and approved the final draft.
References
- 1.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12:720–727. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- 3.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellaguarda E, Chang EB. IBD and the gut microbiota--from bench to personalized medicine. Curr Gastroenterol Rep. 2015;17:15. doi: 10.1007/s11894-015-0439-z. [DOI] [PubMed] [Google Scholar]
- 6.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manichanh C, Borruel N, Casellas F, et al. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 8.Ray KIBD. Understanding gut microbiota in new-onset Crohn’s disease. Nat Rev Gastroenterol Hepatol. 2014;11:268. doi: 10.1038/nrgastro.2014.45. [DOI] [PubMed] [Google Scholar]
- 9.Wu GD, Bushmanc FD, Lewis JD. Diet, the human gut microbiota, and IBD. Anaerobe. 2013;24:117–120. doi: 10.1016/j.anaerobe.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Swidsinski A, Weber J, Loening-Baucke V, et al. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salucci V, Rimoldi M, Penati C, et al. Monocyte-derived dendritic cells from Crohn patients show differential NOD2/CARD15-dependent immune responses to bacteria. Inflamm Bowel Dis. 2008;14:812–818. doi: 10.1002/ibd.20390. [DOI] [PubMed] [Google Scholar]
- 13.Stockinger S, Reutterer B, Schaljo B, et al. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J Immunol. 2004;173:7416–7425. doi: 10.4049/jimmunol.173.12.7416. [DOI] [PubMed] [Google Scholar]
- 14.Couturier-Maillard A, Secher T, Rehman A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013;123:700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong M, McHardy I, Ruegger P, et al. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn’s disease risk polymorphism. ISME J. 2014;8:2193–2206. doi: 10.1038/ismej.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grove ML, Yu B, Cochran BJ, et al. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS One. 2013;8:e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmen OL, Zhang H, Fan Y, et al. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat Genet. 2014;46:345–351. doi: 10.1038/ng.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imielinski M, Baldassano RN, Griffiths A, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fowler SA, Ananthakrishnan AN, Gardet A, et al. SMAD3 gene variant is a risk factor for recurrent surgery in patients with Crohn’s disease. J Crohns Colitis. 2014;8:845–851. doi: 10.1016/j.crohns.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McHardy IH, Goudarzi M, Tong M, et al. Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite interrelationships. Microbiome. 2013;1:17. doi: 10.1186/2049-2618-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kircher M, Witten DM, Jain P, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong M, Li X, Wegener Parfrey L, et al. A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS One. 2013;8:e80702. doi: 10.1371/journal.pone.0080702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang CY, Jenkitkasemwong S, Duarte S, et al. ZIP8 is an iron and zinc transporter whose cell-surface expression is up-regulated by cellular iron loading. J Biol Chem. 2012;287:34032–34043. doi: 10.1074/jbc.M112.367284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aydemir TB, Liuzzi JP, McClellan S, et al. Zinc transporter ZIP8 (SLC39A8) and zinc influence IFN-gamma expression in activated human T cells. J Leukoc Biol. 2009;86:337–348. doi: 10.1189/jlb.1208759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lichten LA, Cousins RJ. Mammalian zinc transporters: nutritional and physiologic regulation. Annu Rev Nutr. 2009;29:153–176. doi: 10.1146/annurev-nutr-033009-083312. [DOI] [PubMed] [Google Scholar]
- 30.Rink L, Haase H. Zinc homeostasis and immunity. Trends Immunol. 2007;28:1–4. doi: 10.1016/j.it.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Begum NA, Kobayashi M, Moriwaki Y, et al. Mycobacterium bovis BCG cell wall and lipopolysaccharide induce a novel gene, BIGM103, encoding a 7-TM protein: identification of a new protein family having Zn-transporter and Zn-metalloprotease signatures. Genomics. 2002;80:630–645. doi: 10.1006/geno.2002.7000. [DOI] [PubMed] [Google Scholar]
- 32.Lahiri A, Abraham C. Activation of pattern recognition receptors up-regulates metallothioneins, thereby increasing intracellular accumulation of zinc, autophagy, and bacterial clearance by macrophages. Gastroenterology. 2014;147:835–846. doi: 10.1053/j.gastro.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Besecker B, Bao S, Bohacova B, et al. The human zinc transporter SLC39A8 (Zip8) is critical in zinc-mediated cytoprotection in lung epithelia. Am J Physiol Lung Cell Mol Physiol. 2008;294:L1127–L1136. doi: 10.1152/ajplung.00057.2008. [DOI] [PubMed] [Google Scholar]
- 34.Liu MJ, Bao S, Galvez-Peralta M, et al. ZIP8 regulates host defense through zinc-mediated inhibition of NF-kappaB. Cell Rep. 2013;3:386–400. doi: 10.1016/j.celrep.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haberman Y, Tickle TL, Dexheimer PJ, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrera N, Arrojo M, Sanjuan J, et al. Association study of nonsynonymous single nucleotide polymorphisms in schizophrenia. Biol Psychiatry. 2012;71:169–177. doi: 10.1016/j.biopsych.2011.09.032. [DOI] [PubMed] [Google Scholar]
- 37.Ehret GB, Munroe PB, et al. International Consortium for Blood Pressure Genome-Wide Association Studies. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraja AT, Chasman DI, North KE, et al. Pleiotropic genes for metabolic syndrome and inflammation. Mol Genet Metab. 2014;112:317–338. doi: 10.1016/j.ymgme.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Vliet-Ostaptchouk JV, den Hoed M, Luan J, et al. Pleiotropic effects of obesity-susceptibility loci on metabolic traits: a meta-analysis of up to 37,874 individuals. Diabetologia. 2013;56:2134–2146. doi: 10.1007/s00125-013-2985-y. [DOI] [PubMed] [Google Scholar]
- 41.Waterworth DM, Ricketts SL, Song K, et al. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol. 2010;30:2264–2276. doi: 10.1161/ATVBAHA.109.201020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 43.Yassour M, Lim MY, Yun HS, et al. Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med. 2016;8:17. doi: 10.1186/s13073-016-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masui R, Sasaki M, Funaki Y, et al. G protein-coupled receptor 43 moderates gut inflammation through cytokine regulation from mononuclear cells. Inflamm Bowel Dis. 2013;19:2848–2856. doi: 10.1097/01.MIB.0000435444.14860.ea. [DOI] [PubMed] [Google Scholar]
- 45.Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Png CW, Linden SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 47.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an antiinflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knights D, Silverberg MS, Weersma RK, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6:107. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joossens M, Huys G, Cnockaert M, et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 50.Partridge AW, Therien AG, Deber CM. Missense mutations in transmembrane domains of proteins: phenotypic propensity of polar residues for human disease. Proteins. 2004;54:648–656. doi: 10.1002/prot.10611. [DOI] [PubMed] [Google Scholar]
- 51.Galvez-Peralta M, He L, Jorge-Nebert LF, et al. ZIP8 zinc transporter: indispensable role for both multiple-organ organogenesis and hematopoiesis in utero. PLoS One. 2012;7:e36055. doi: 10.1371/journal.pone.0036055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed S, Neuman H, Moscovich S, et al. Chronic Zinc Deficiency Alters Chick Gut Microbiota Composition and Function. Nutrients. 2015;7:9768–9784. doi: 10.3390/nu7125497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ayabe T, Satchell DP, Pesendorfer P, et al. Activation of Paneth cell alpha-defensins in mouse small intestine. J Biol Chem. 2002;277:5219–5228. doi: 10.1074/jbc.M109410200. [DOI] [PubMed] [Google Scholar]
- 54.Ayabe T, Wulff H, Darmoul D, et al. Modulation of mouse Paneth cell alpha-defensin secretion by mIKCa1, a Ca2+-activated, intermediate conductance potassium channel. J Biol Chem. 2002;277:3793–3800. doi: 10.1074/jbc.M107507200. [DOI] [PubMed] [Google Scholar]