Abstract
Besides being the carrier of genetic information, DNA is also an excellent biological organizer to establish well-designed nanostructures in the fields of material engineering, nanotechnology, and biomedicine. DNA-based materials represent a diverse nanoscale system primarily due to their predictable base pairing and highly regulated conformations, which greatly facilitate the construction of DNA nanostructures with distinct shapes and sizes. Integrating the emerging advancements in bioconjugation techniques, DNA nanostructures can be readily functionalized with high precision for many purposes ranging from biosensors to imaging to drug delivery. Recent progress in the field of DNA nanotechnology has exhibited collective efforts to employ DNA nanostructures as smart imaging agents or delivery platforms within living organisms. Despite significant improvements in the development of DNA nanostructures, there is limited knowledge regarding the in vivo biological fate of these intriguing nanomaterials. In this review, we summarize the current strategies for designing and purifying highly-versatile DNA nanostructures for biological applications, including molecular imaging and drug delivery. Since DNA nanostructures may elicit an immune response in vivo, we also present a short discussion of their potential toxicities in biomedical applications. Lastly, we discuss future perspectives and potential challenges that may limit the effective preclinical and clinical employment of DNA nanostructures. Due to their unique properties, we predict that DNA nanomaterials will make excellent agents for effective diagnostic imaging and drug delivery, improving patient outcome in cancer and other related diseases in the near future.
Keywords: DNA nanotechnology, DNA nanoparticle, DNA nanostructure, DNA, molecular imaging, drug delivery
Graphical Abstract
1. Introduction
There are many bio-functional molecules, such as small molecules [1–3], peptides [4–6], and proteins [7–9], that have proven practical as imaging tracers or therapeutic agents. By virtue of the rapid development of nanotechnology, enhanced imaging contrast and drug delivery is among several promising futures based on the assumption that nanoplatforms may maintain or optimize biological properties of drugs in vivo, while helping to decrease necessary pharmaceutical dosages and minimize potential side effects [10, 11]. Advancements in nanotechnology have allowed scientists to develop highly sophisticated DNA-based drug delivery systems capable of transporting a variety of bio-functional molecules directly to sites of interest through active and passive targeting mechanisms [12–14]. For example, most traditional chemotherapeutics may be loaded into micelles [15], polymers [16, 17], carbon-based nanomaterials [18–20], quantum dots [21–23], metallic nanoparticles [24], and others for in vivo tumor-targeted imaging and therapy. In addition, some inorganic nanomaterials, such as AuNPs [25–27] and mesoporous silica nanoparticle [28] may serve as both contrast agents for diagnostic imaging, therapeutic agents for various forms of cancer treatment (e.g. photothermal therapy, PTT; or photodynamic therapy, PDT), or a combination of both, also known as theranostic agents [29, 30].
Deoxyribonucleic acid (DNA) is commonly used for in vivo biomedical applications in the forms of single-stranded DNA (ssDNA), double-stranded DNA (dsDNA), and circulated plasmid DNA. Due to its diversity in sequences and consistency in structure, DNA is an ideal material for integrating versatile functional groups for targeting of diseased tissues in biomedical applications [31]. Based on the Watson-Crick base-pairing principle, complementary DNA strands can form DNA double helices under physiological environments with almost 100% predictability and reproducibility, which offers an excellent model for constructing DNA nanomaterials with high designability. The shape, size, and dimension of DNA nanomaterials could be readily controlled by sequence design and arrangement of oligonucleotides [32]. Besides the structural designability of DNA nanostructures, DNA possesses several unique properties that make it attractive for in vivo diagnostic and therapy [33]. For example, taking advantages of the rapid developments in the fields of nucleic acid synthesis and conjugation, we can easily construct multifunctional DNA nanomaterials with highly modified DNA strands. Moreover, highly sophisticated DNA tools, such as DNA ligases, polymerases, and nucleases allow scientists to design and edit DNA strands with even higher efficiency. Lastly, DNA and DNA nanocomposites are both biocompatible and biodegradable, which can ease most of the toxicity concerns associated with traditional nanoplatforms. Together, all these features make DNA nanomaterials a suitable and promising tool for molecular imaging and targeted drug delivery in vivo.
While DNA nanotechnology has been previously reviewed in literature focusing on the formation, design, and in vitro cellular delivery mechanisms of DNA nanostructures [34–38], this review extends beyond these basic concepts to investigate the applications of DNA nanotechnology for in vivo molecular imaging and drug delivery. First, the origin and development of DNA nanomaterials are discussed with an emphasis on designing DNA nanoparticles with and without DNA template strands. Next, we examine the methods currently employed for purifying and enhancing the stability of DNA nanomaterials. Later, we illustrated the in vivo applications of DNA nanomaterials regarding imaging and drug delivery before outlining the potential opportunities and challenges of DNA nanomaterials for in vivo applications.
2. Evolution of DNA Nanotechnology for Biomedical Applications
2.1. Initial Development of DNA Nanostructures
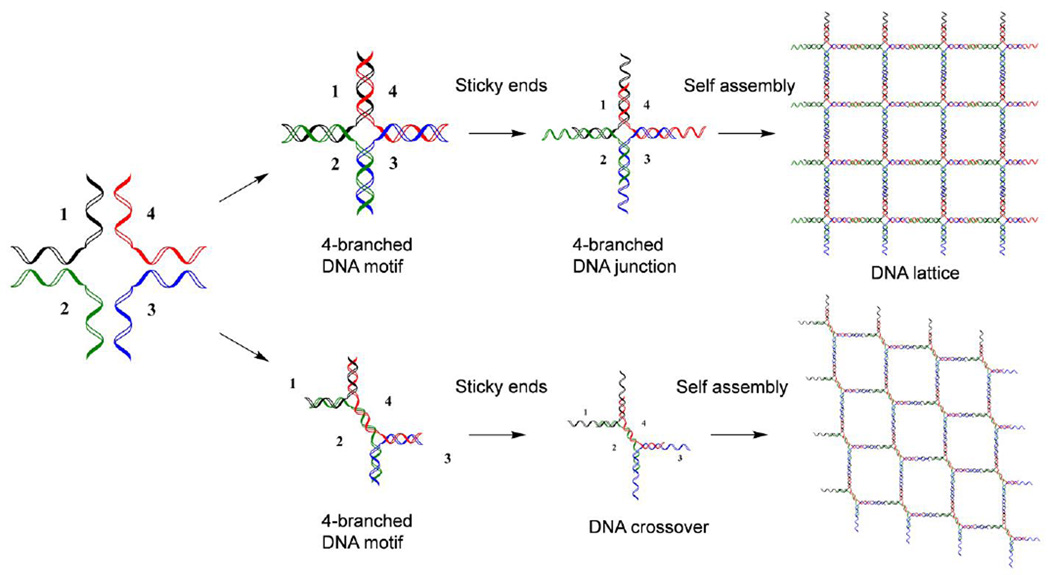
It was after the discovery of the double-helical structure of DNA and its deformational products, including G-quadruplexes (G4-DNA) and X- or Y-shaped DNA [39], when researchers recognized that the unique structural properties of DNA could be employed to develop 3-dimenisonal (3D) nanostructures. In the early 1980s, Nadrian C. Seeman was reportedly inspired by a piece of wood engraved artwork by M. C. Escher called the Depth. While examining this intrinsic artwork, he conceptualized the process constructing 3D lattices of DNA [40]. During this time, his laboratory exploited the Watson-Crick base-pairing principle of DNA to modify and compose linear DNA strands into branched DNA junctions via the addition of extended overhangs of nucleic acids, also known as sticky ends, at each arm of the DNA motif. These sticky ends act as toeholds for self-assembly processes and provide active building blocks for precise control over the formation of DNA nanostructures (Fig. 1) [41, 42]. However, DNA junctions with sticky ends could only offer two-dimensional DNA lattices with limited rigidity and undefined geometries [43]. To overcome these issues, researchers have provided modified DNA junctions, including double-crossovers [44, 45], triangle tensegrity DNA [46], and 4 × 4 DNA tiles [47]. The high designability of DNA nanostructures allows for the construction of highly-ordered 2D DNA lattices and 3D polyhedrons by hybridizing the sticky ends to form rigid DNA junctions (Fig. 1). While an effective approach, the lateral size of DNA nanostructures composed by such strategies were difficult to control; thus, a stoichiometric assembly method is required for massive and complex fabrication of DNA nanostructures. Since this discovery, the emerging role of DNA as structural nanomaterials has become well recognized and has led to the rapid development and growth of the field of DNA nanotechnology.
Fig. 1.
Formation of 2D DNA lattices with DNA junctions or DNA crossovers with the sticky end design. The colored lines represent different oligonucleotides.
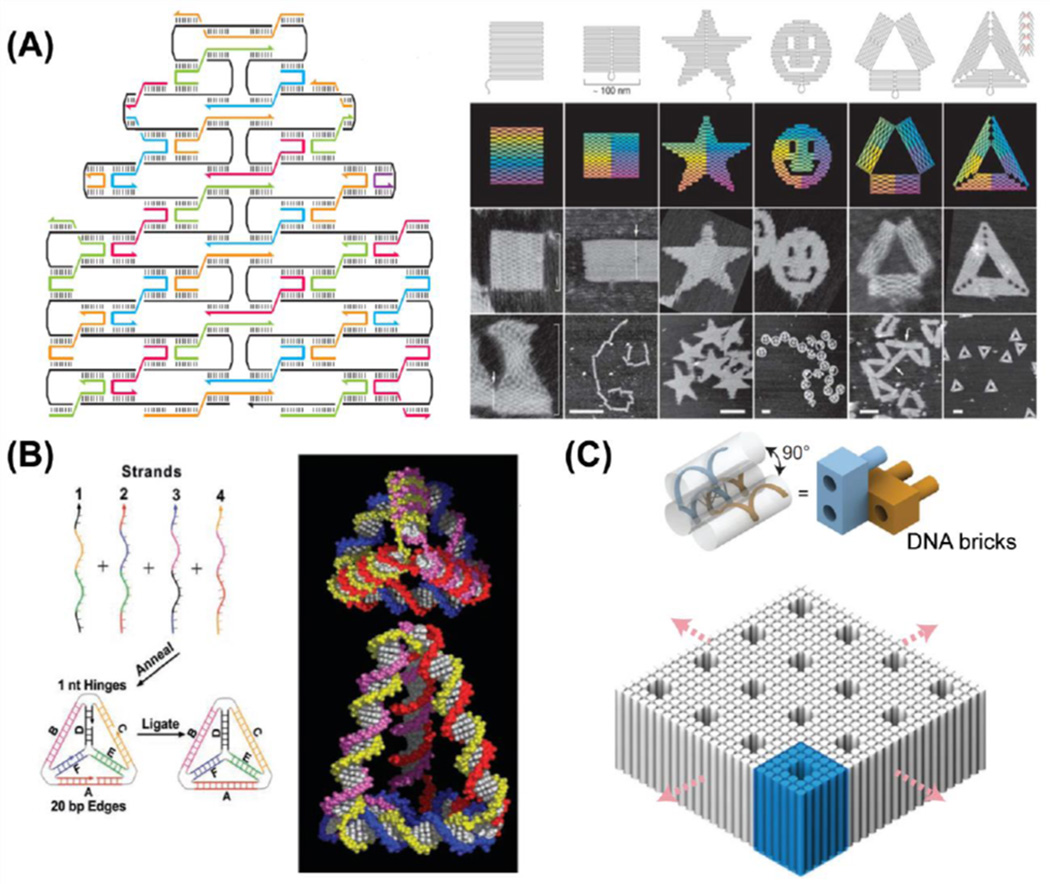
In 2006, Dr. Rothemund from California Institute of Technology developed a novel process of synthesizing DNA nanostructures from a template, which was later termed "scaffold DNA origami [48].” In the typical process of preparing DNA origami (Fig. 2A), circular genomic DNA from the virus M13mp18 (a 7249-nucleotide DNA that was screened for secondary structures) is utilized as the scaffold/template, and more than 200 short DNA stranded (termed as staple strands, all complementary to some parts of the scaffold DNA) are designed to fold the long piece of single-stranded DNA at specific sites [49–51]. The one-pot synthesis of DNA origami incorporates the DNA crossover design into the folding process while leaving no specific limitations on the molar ratio of staple strands to the DNA scaffold. The presence of scaffold DNA not only makes the assembly process stoichiometric, but also sets a stopping point for the building of complex DNA nanostructures. In some advanced techniques, DNA helices bumps could be designed on the surface of DNA origami to enable patterns or characters, which are useful for control of spatial positions of molecules or to serve as a matrix for versatile interaction studies.
Fig. 2.
Schematic display of formation processes of DNA origami and DNA nanocages (DNA tetrahedron and DNA bipyramid). Colored lines represent different oligonucleotides. (A) A long-circulated plasmid DNA could be folded by many short oligonucleotides (staple strands) to form DNA origami nanocomposites with specific patterns, such as rectangles, stars, smiling faces, and triangles. Adapted with permission from [48]. (B) Four oligonucleotides could hybridize with each other to form 3-dimensional DNA tetrahedron-shaped nanocages. Adapted with permission from [52]. (C) Formulation of 3-dimensional DNA nanostructures with multivalent DNA bricks. Adapted with permission from [56].
Meanwhile, Turberfield and co-workers paved another path for constructing three-dimensional DNA nanocages. Taking advantages of the base-pairing abilities of DNA, they developed a simple and rapid method to prepare small DNA tetrahedral nanocages around 7 nm, which exhibited tunable edge length and substantial structure rigidity [52]. Four single DNA strands with designated sequences have been designed to be four surfaces of the tetrahedral nanocage; and at each edge, every two strands hybridize with each other. The DNA polyhedral nanocages (tetrahedron [53], bipyramid [54], cube [40], and others) formed via hybridization of several DNA strands require no additional DNA templates, and the size of nanocages could be well controlled due to the self-enclosing structure design (Fig. 2B). Thus, these DNA nanocages could be well developed as small building blocks for complex higher-ordered DNA nanostructures. Besides the employment of short synthetic oligonucleotides for the creation of different DNA nanocages, highly-ordered DNA nanostructures can also be formulated with multi-valence DNA building blocks formed with short ssDNA (32 nt) using an expandable assembly strategy. Recently, Yin and colleagues described a methodology for synthesizing 3D nanostructures using short synthetic DNA strands, also called DNA bricks [55]. Each of these DNA bricks has the ability to bind four neighboring DNA bricks (Fig. 2C), thus allowing for accurate control of the shape, size, and sophisticated surface properties (tunnels and interior cavities) of the DNA nanostructures [56]. The high controllability and sophisticated geometric morphology of DNA brick-based nanostructures may allow for biological applications requiring the addition of target molecules at precise spatial locations. In addition, these DNA brick-based nanostructures may be synthesized using synthetic forms of DNA, like informational polymers, to reduce immunogenicity and increase nuclease resistance [55].
To date, a large variety of DNA nanostructures has been constructed with unprecedented speed, providing robust platforms for integrating other functional materials, such as small molecules, fluorescent dyes, tumor-targeting ligands, and inorganic nanoparticles. Taking advantages of DNA's unique properties including spontaneous self-recognition under physiological condition, stable forming of DNA helices, and the ability to locate any DNA bases within the 3D space; it is highly intriguing to explore the potential of DNA nanostructures for in vivo biological applications.
2.2. Purification of DNA Nanostructures
Despite continuous efforts in designing highly stable DNA nanostructures, the assembly yields using current protocols are suboptimal. It is essential for DNA nanostructures to be impurity-free and homogenous for potential preclinical and clinical utilization [57]. Thus, purification methods are included as a standard procedure during the preparation of DNA nanostructures. Among all purification methods, agarose gel electrophoresis (AGE) is the most widely-employed and cost-effective method for purifying DNA nanomaterials [58]. Assembled DNA nanostructures are divided into various bands in agarose gels, separating well-folded structures from misfolded DNA aggregates and unfolded DNA strands. Then, desired structures could be extracted from the gel and reconstituted in desired buffers with the desired concentration. However, traditional AGE methods always lead to low yields, structural damages, and/or agarose contaminations [59].
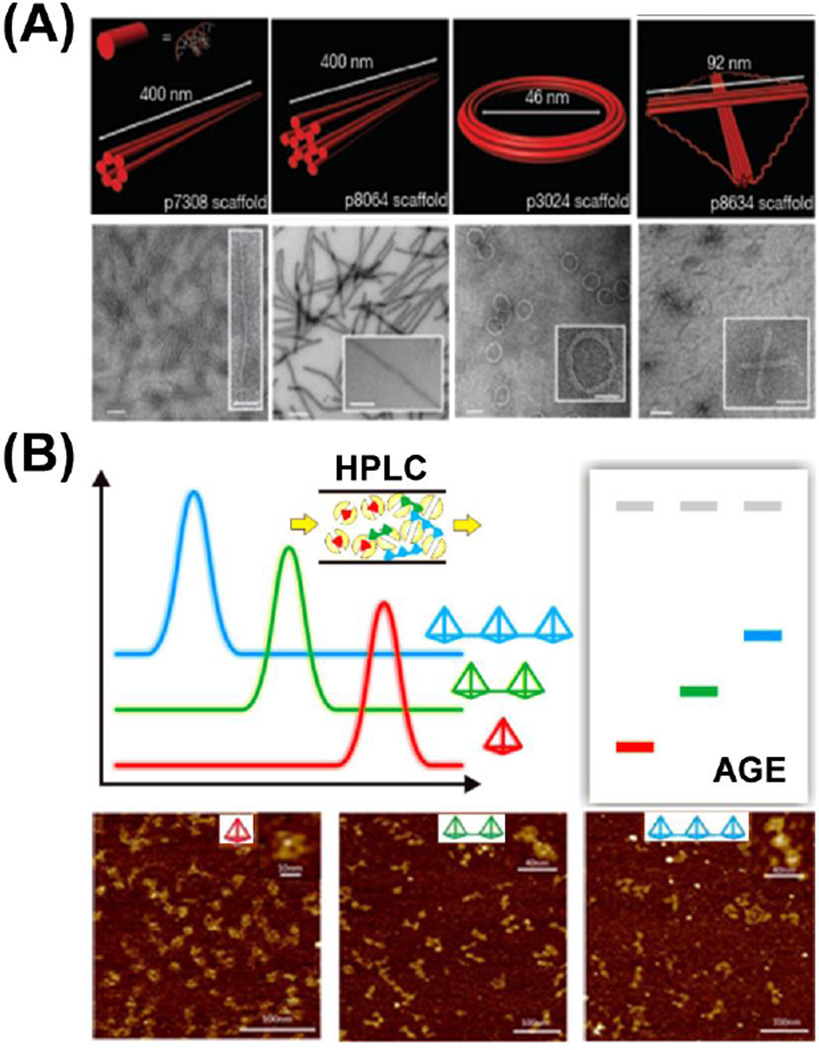
Bellot et al. modified traditional AGE purification methods for DNA origami, presenting an electro-elution way of AGE with increased efficiency, higher resolution, and large-scale reproducibility [59]. They prepared a dual-layer AGE gel with a layer of 1–2% agarose (sample resolving layer) on top of a thinner yet rigid 4% AGE gel (the basement layer). After loading DNA samples into the resolving layer and running the AGE gel to separate by-products, an elution well was cut in front of the band of interest. The well was filled with 30–50% sucrose buffer, and the band of interest could be eluted out by electrophoresis into the sucrose buffer and further collected by micro-pipetting (Fig. 3A). The electro-elution AGE could offer an enrichment efficiency of well-folded DNA origami within the range of 45–71%. With this method, DNA nanostructures could be obtained with high purity and minimal agarose contamination, thus more sophisticated 2D or 3D DNA nanostructures could be constructed with enhanced control over their designability.
Fig. 3.
Purification of different DNA origami nanocomposites and DNA nanocages. (A) Purification of DNA origami with double-layer agarose gel electrophoresis (AGE), the electro-elution AGE could minimize the possible contamination during the gel extraction process. Adapted with permission from [59]. (B) Purification of different DNA nanocages with high-performance liquid chromatography (HPLC). DNA nanocages with smaller sizes are retained longer in the size exclusion column. Adapted with permission from [60].
Furthermore, this group presented a scalable protocol for purifying DNA origami using rate-zonal centrifugation, which showed several benefits over conventional AGE methods with a comparable separation resolution [61]. A linear glycerol gradient (15–45%, v/v) was first prepared, and DNA origami containing 10% glycerol was loaded on top of the gradient. The solution was mixed and subjected to ultracentrifuge at a speed of 50,000 rpm (300,000 g) for approximately 2 h at 4°C before being divided into 12–16 fractions and characterized by both AGE and TEM to confirm the separation efficiency. Quantification analysis of gel electrophoresis showed that ~85% of desired DNA nanostructures were concentrated in 10–20% volume of the glycerol gradient. Rate-zonal centrifugation offers a universal gradient for purification of different DNA origami, and could be adapted to a versatile amount of DNA nanostructures (0.1–100 µg), thus enabling the preparation of large-scale DNA nanostructures.
The rate-zonal centrifugation method is suitable for large DNA origami, but was proved non-effective for small DNA nanocages composed by only several oligonucleotides. Thus, the construction of higher-ordered DNA complex using small building blocks is hampered by the lack of purified individual DNA nanocages. Recently, Xing et al. introduced size exclusion chromatography (SEC) for the large-scale preparation of versatile tetrahedral DNA nanocages (TDNs) [60]. Seven TDNs with different edge lengths (7, 10, 13, 17, 20, 26, 30 base pairs) and one bi-pyramidal DNA nanocage were purified by SEC-HPLC method with high purities. SEC-HPLC has several advantages over traditional gel methods. First, as for the construction of higher-order DNA complex, unpurified DNA nanocages would result in a poor yield of 14% for dimer and 12% for trimer structures. However, the SEC-HPLC purified aggregation-free DNA nanocages could propel the assembly efficiency to 98% for dimer and 95% for trimer (Fig. 3B). Thus, the SEC-HPLC method offers better quantification than normal gel electrophoresis (PAGE or AGE) for the precise fabrication of DNA nanocage complexes. Finally, this method can minimize possible re-aggregation and/or degradation that exist in traditional gel methods during the process of recovery and reconstitution.
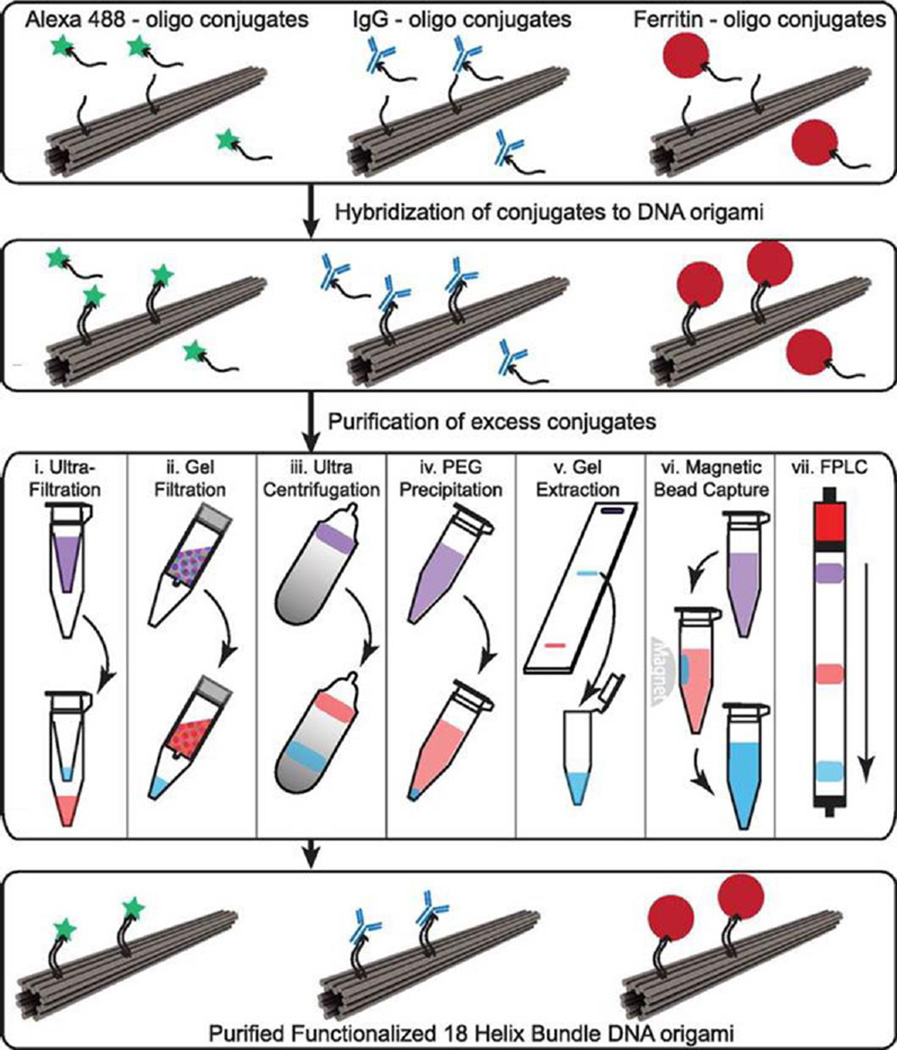
The high programmability of DNA offers precision with designed DNA nanostructures, which further opens up diverse possibilities to arrange functional groups for various applications with DNA nanostructures. However, when it comes to the actual manipulation, functionalized DNA nanocomposites suffer from the existence of unpurified functional groups and/or mismatched DNA nanostructures [62]. Shaw et al. provided a systematic study comparing seven purification methods for DNA origami nanostructures functionalized with either small molecules (Alexa 488 fluorophore), antibodies (human IgG) or large proteins (Ferritin), with the aim to provide a guideline for effectively fabricating and scalable production of modified DNA origami constructs (Fig. 4) [63]. Other than several traditional purification methods including ultrafiltration, gel filtration, glycerol gradient ultracentrifugation, PEG precipitation, gel extraction, the authors reported two new approaches for purification of DNA origami with function groups: magnetic bead capture and fast protein liquid chromatography (FPLC) using a Superose 6 column. As for the purification efficiencies for seven methods in terms of recovery yield as well as percentage of contaminations: ultrafiltration, magnetic beads and FPLC offered better results for Alexa488-modified DNA nanostructures (60–85% recovery yield, and almost no contaminations); PEG precipitation, magnetic beads and FPLC provided good outcomes for IgG1-modified DNA origami (over 50% recovery yield and <5% contamination left); then for Ferritin-DNA origami, gel filtration and PEG precipitation excelled other methods by providing a 60–70% recovery yield, though at least 10–20% contamination still existed in the modified DNA nanostructures. Overall, scalable production of designed DNA oligonucleotides, in addition to proper choices of purification methods, may boost the development of biomedical studies using highly modified and purified DNA nanostructures for biological applications in vivo.
Fig. 4.
Purification process of functionalized DNA origami nanocomposites. DNA origami functionalized with small molecules (Alexa 488), antibodies (IgG), and large proteins (Ferritin). Samples were purified using one of seven methods with violet indicating the starting solution with excess free functional groups and blue indicating purified DNA origami with functional groups. Adapted with permission from [63].
2.3. Stability of DNA Nanostructures
Before DNA nanostructures can be implemented for clinical use, their safety and stability must be thoroughly examined. As with other nanoplatforms, potential toxicity remains a critical barrier to successful translation of DNA nanomaterials. There are three essential requirements that must be met for successful employment of DNA-based nanoconstructs. For example, DNA nanostructures should remain intact within biological environments, especially in the plasma once injected into the bloodstream. Also, the nanomaterials should not elicit an immune response when injected into animals [64]. Until now, several groups have dedicated their intelligence to study and strengthen the stability of DNA nanostructures. The physiochemical properties and stability or various DNA nanostructures may be found in Table 1.
Table 1.
Physiochemical Properties and Stability of DNA Nanostructures.
 |
 |
 |
 |
 |
 |
 |
 |
 |
||
|---|---|---|---|---|---|---|---|---|---|---|
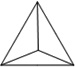
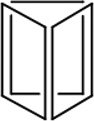
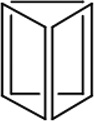
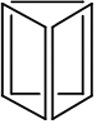
| Length of edges |
~ 7 nm | ~ 7 nm | ~ 7 nm | ~ 7 nm | ~ 7 nm | ~ 7 nm | 90 × 60 nm | 120 nm (side width ~ 30 nm) |
16 × 16 × 30 nm |
80 × 20 nm |
| Further treatment after assembly |
Hexaethylene glycol (HEG) at the end of each component strand |
T4 ligase | Decorated with CpG sequences |
|||||||
| Incubation medium |
10% FBS |
50% FBS |
10% FBS | 10% FBS | 10% FBS | 10% FBS/PBS |
Cell lysates | Cell lysates | Cell lysates | FBS containing medium |
| Mean lifetime |
42 h | 24 h | 18 h | 62 h | 200 h | 3.5 h | 12 h | 12 h | 12 h | > 9 h |
| Control | dsDNA | dsDNA | ssDNA | HEG-ssDNA | ssDNA treated with T4 ligase |
dsDNA | ssDNA & dsDNA |
ssDNA & dsDNA |
ssDNA & dsDNA |
Template DNA |
| Control’s mean lifetime |
0.8 h | < 2 h | < 1 h | ~ 28 h | ~ 39 h | 0.7 h | < 1 h | < 1 h | < 1 h | ~ 6 h |
| Reference | 65 | 66 | 66 | 66 | 66 | 67 | 68 | 70 | 70 | 64 |
In a recent study, Keum et al. first realized that for DNA nanocages with dimensions much less than 50 nm, the corresponding enzyme recognition process must be somehow different from that of linear DNA [65]. Thus, they started to follow the design of Turberfield and constructed DNA tetrahedral nanostructures with a theoretical diameter of about 7 nm. They found that, in 10% fetal bovine serum (FBS), the decay time for DNA tetrahedron was 42 h, while that of a double-stranded DNA counterpart was ~0.8 h. They demonstrated that folding of DNA strands into three-dimensional nanostructures would result in enhanced resistance to enzyme digestion when compared to their linear counterparts. The mechanism underlying this unique phenomenon was attributed to the steric hindrance introduced by each vertex shaping as three-way branch junctions, which may lower the chance of enzymatic binding with the DNA helices. Also, DNA tetrahedral nanostructures exhibited additional rigidity due to the connectivity of the 3D design, which could prevent the conformational change essential for nuclease cleavage of DNA. Moreover, if more multi-way branched junctions or shorter edges were designed into DNA nanostructures, enhanced resistance towards enzymatic digestion was achieved due to stronger steric hindrance and lesser possibility of enzyme-caused bending.
The stability of DNA tetrahedrals was examined by Li et al., who discovered that DNA tetrahedral nanostructures could effectively deliver cytosine−guanine (CpG) motifs into RAW264.7 cells while the DNA nanostructure itself triggered little immune response [66]. The stability of DNA tetrahedrals was unaffected in 50% FBS for up to 4 h when assessed by gel electrophoresis, and intact structures could still be observed after 24 h incubation. For comparison, normal double-stranded DNA (dsDNA) remained stable for <2 h in 50% FBS. Moreover, intracellular co-localization testing showed that after internalization into cells, the fluorescence resonance energy transfer (FRET) pair (Cy3 & Cy5) dual-labeled DNA tetrahedral nanostructures could be visualized for >8 h.
Conway et al. then stepped forward to construct a 3D triangular prism (TP) using chemically modified and unmodified single-stranded DNA (ssDNA) [67]. After validating the results from Keum et al., they introduced several chemical modifications (Hexaethylene glycol, HEG; Hexanediol, C6; and 5'-phosphate, P) to the end of ssDNAs and instantly observed enhanced stability when incubated in 10% FBS. However, assembly of TP with unmodified or modified ssDNAs provided more stability when comparing to all its components. This simple modification strategy suggested that significant resistance to enzyme degradation could be achieved by the simple introduction of commercially available end-substitutions of ssDNA components.
In the same group, Hamblin et al. further developed a rolling cycle amplification (RCA) method to construct a prism-like DNA nanotubes for potential intracellular drug delivery [68]. The half-life of RCA generated nanotube (RCA-NT) in 10% FBS/PBS was 2.5 h, which was five times longer than dsDNA (0.5 h), and three times longer than that of DNA nanotubes (~0.8 h) composed via a modular construction method [69]. Besides the enhanced stability against nuclease degradation in FBS, the RCA-NT also displayed a higher tendency of entering HeLa cells without the help of transfection reagents, comparing to its dsDNA counterparts. The large compact design of negatively-charged DNA and the high repeated ratio of prism-like DNA units may contribute to the impaired nuclease binding capacity.
DNA origami is another type of DNA nanostructure that has been under extensive investigation regarding their stability. For example, Mei et al. demonstrated that DNA origami with different sizes and shapes could easily tolerate cell lysates from normal cells or cancerous cells, and maintain intact for >12 h [70]. They used AGE to extract DNA origami from the incubation solution and characterized the structures with atomic force microscope (AFM). The stability was assessed by band intensity from AGE and the structure integrity was examined by AFM imaging. They also confirmed that some components of DNA origami, ssDNA or dsDNA, failed to survive in the harsh environment of cell lysates. It is the highly negative charges and limited steric accessibility provided by the compact arrangement of DNA helices that contribute to the extraordinary integrity of DNA origami, which not only enables them to remain intact, but also allows for further functionalization of DNA origami. Later in 2011, Schuller et al. also showed that DNA origami displayed enhanced stability when compared to DNA strands and DNA helices in FBS-containing medium at 37°C [64].
The incompatibility of cell culture conditions and DNA origami maintenance conditions was previously investigated by Hahn et al., who aimed to study the stability of DNA origami in normal cell culture medium and revealed the sensitivity of DNA origami to various cell culture conditions [71]. After preparing three different DNA origami including DNA nano-octahedron (DNO), 6-helix bundle nanotube (6H-NT) and 24-helix nanorods (24H-NR), the authors first examined the deformation of DNA origami due to the low concentration of cation (mainly Mg2+) in the physiological environment of cell culture medium. They found that denaturation of DNA origami due to cation shortage was shape and time-dependent. Out of all three DNA origami designs, 6H-NT showed the highest stability against Mg2+ depletion conditions. Moreover, the addition of Mg2+ into cell culture medium could effectively prevent deformation of DNA origami. Next, the authors investigated the digestion of DNA origami by nucleases from FBS in Mg2+ adjusted cell culture medium and showed that severe degradation occurred when FBS exceeded 5% or incubation time reached 24 h. The addition of actin or heat inactivation (75°C) could prevent the nuclease digestion. Also, the addition of Mg2+ allowed for enhanced DNA stability and elicited no cellular toxicity, yet these concentrations of Mg2+ are not available in vivo, further challenging the possible translation of DNA nanostructures into in vivo molecular imaging and drug delivery.
3. DNA Nanostructures for Molecular Imaging and Drug Delivery
In response to the high programmability and spatially addressable properties, researchers have gained precise control over the functionalization of DNA nanostructures. Moreover, DNA nanomaterials exhibit several valuable properties required for in vivo applications that are commonly not displayed by inorganic-organic nanostructures, including their high purity, batch-to-batch reproducibility, biocompatibility, and biodegradability [72]. DNA nanostructures may be covalently or non-covalently functionalized for in vivo imaging and drug delivery purposes through the addition of contrasts agents for visualization of diseased tissues, targeting groups for actively targeting tissues of interest, and small molecules and drugs for therapeutic delivery. Successful employment of DNA nanostructures for molecular imaging and drug delivery requires that (1) the nanomaterial be highly reproducible and substantially uniformed, (2) the nanostructure should remain intact or undergo controlled degradation within biological environments, especially within the plasma once injected into the bloodstream [73], and (3) the nanomaterial should not elicit an immune response when introduced into living organisms [64].
3.1. DNA Nanocages for Molecular Imaging and Drug Delivery
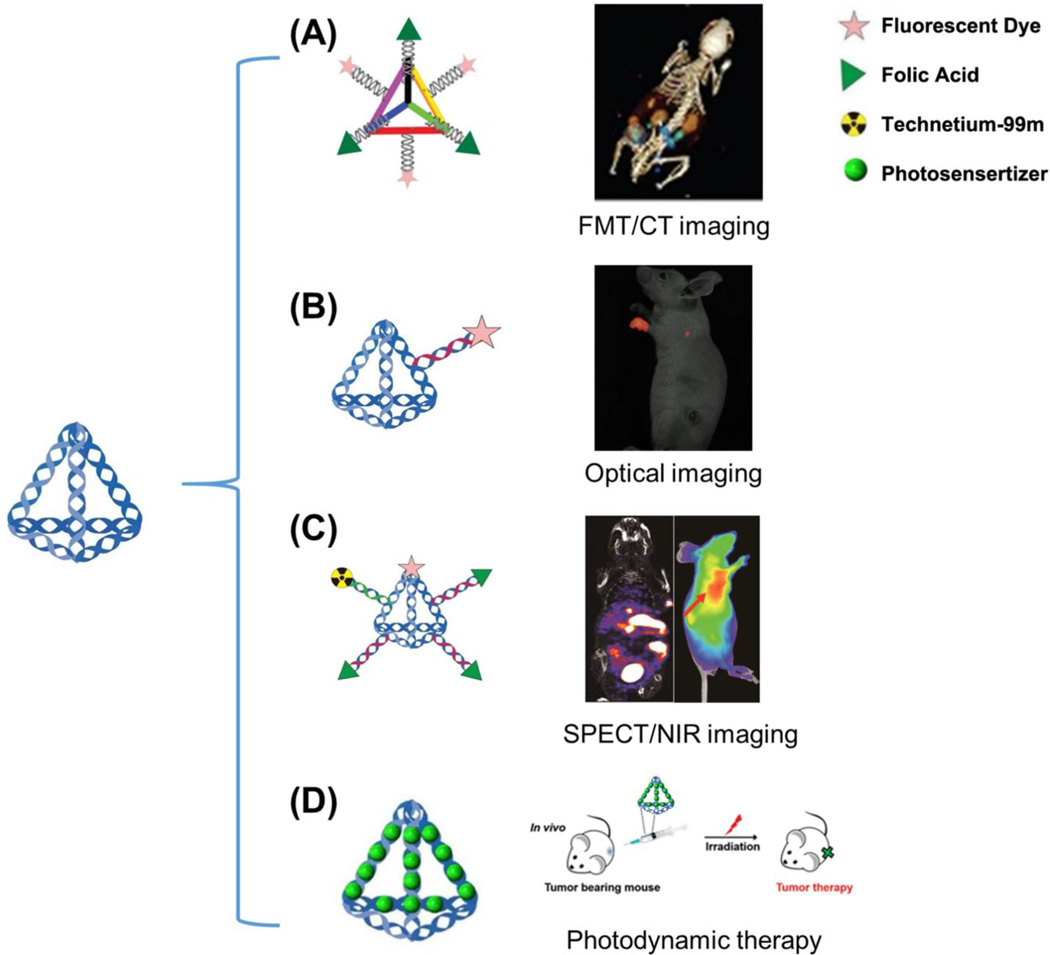
In 2012, Lee et al. took advantage of the simple design and high rigidity of oligonucleotide nanoparticles (ONPs) for targeted imaging and delivery of small interfering RNAs (siRNA) in mice [74]. ONPs showed a monodispersed size (30 base pairs per edge, about 10 nm) through the self-assembly of six complementary oligonucleotides. After the screening of four cationic peptides, one amphipathic peptide, 22 zwitterionic peptides, and one small molecule (folic acid; FA), the authors found that the FA-conjugated ONPs showed the highest gene silencing capacity. Six overhangs were designed on each edge of the ONP, three overhangs were used for hybridization of FAs, and the other three overhangs were used for ligation of a fluorescent dye (Cy5). The high controllability of the spatial orientation and density of the ligands on the nanoplatform allowed for enhanced cell internalization and gene silencing. Also, Cy5-labeled FA-ONPs were administrated to mice bearing KB tumors overexpressing the folate receptor (FR) to verify the in vivo efficacy of this nanoplatform. FA-ONPs showed primary accumulation in the tumor and kidneys, with weaker signals in other organs such as the liver, lung, heart and spleen (Fig. 5A). A detailed pharmacokinetic analysis revealed that siRNA delivered by ONPs had a longer blood circulation time (t1/2) of 24.2 min, while siRNA alone displayed a t1/2 of only 6 min. The easy self-assembly, specific designable overhangs, as well as the full control over the spatial orientation, position, and density of ligands, allowed for the DNA nanostructures to achieve enhanced in vivo tumor location for molecular imaging and siRNA delivery.
Fig. 5.
Molecular imaging and drug delivery with DNA nanocages. (A) DNA tetrahedron nanocages are loaded with three folic acid (FA) molecules and three fluorescent dyes (Cy5) utilized for tumor targeting in KB tumor-bearing mice overexpressing the folate receptor. (B) Fluorescent dye (Cy5)-conjugated DNA nanocages displayed enhanced translocation in sentinel lymph nodes and lower background in vivo. (C) With the help of both near infrared (NIR) imaging and single-photon emission computed tomography (SPECT) imaging, the detailed biodistribution and pharmacokinetic profile of DNA tetrahedron nanocages were investigated. Both imaging modalities showed excellent tumor contrast with this multifunctional probe. (D) DNA nanocages bearing photosensitizer agents could also be utilized for photodynamic therapy in vivo. Adapted with permission from [74–77].
Shortly after this discovery, Kim et al. developed a fluorescently labeled (Cy5) tetrahedral DNA nanoparticle (Td) as a probe for sentinel lymph nodes (SLN) imaging, often the first site of metastatic lesions as cancer cells detach from the primary tumor and travel through the lymphatic vessels to healthy tissues [75]. Due to the excellent biocompatibility and chemical flexibility for fluorescent modification, Cy5-Td is a promising nanomaterial for in vivo imaging. In particular, Td was shown to be an excellent SLN imaging agent due to its small size of 6–10 nm, which is within the hydrodynamic diameter required for lymphatic drainage and lymph node retention (Fig. 5B). Visualization of Cy5-Td in vivo showed enhanced translocation in SLNs and lower background signals than the double-stranded DNA counterpart, mainly due to the effective cellular uptake and improved structural integrity of DNA nanostructures.
Encouraged by this discovery, Jiang et al. developed multiple-armed tetrahedral DNA nanostructures (ma-TDNs) for dual-modality in vivo imaging, combining near-infrared (NIR) fluorescence and single-photon emission computed tomography (SPECT) together for improved tumor imaging [76]. The authors found that the existence of multiple arm-strands could significantly enhance the in vitro stability of TDNs in 80% mouse serum, allowing the structures to stay intact for >12 h, while double-stranded DNA could only survive for <2 h. Using both NIR and SPECT imaging, the plasma half-life of TDNs was measured to be 6 min, approximately two-times longer than that of single-stranded DNA (3 min). This study also demonstrated the detailed biodistribution pattern of ma-TDN in vivo, showing that the nanoplatform displayed excellent tumor contrast for imaging of oral cancer (Fig. 5C).
Recently, tetrahedral DNA nanostructures (TDNs) have been applied as carriers of photosensitizers (Methylene blue, MB) for photodynamic therapy (PDT) [77]. Based on the binding capacity of DNA helices for MB molecules, the dye could be complexed into TDNs with a 16:1 ratio. The DNA nanocarrier could deliver active MB into various cells without affecting the PDT properties of the MB molecules, proving TDNs to be a suitable carrier for delivering photosensitizers at the cellular level. Furthermore, TDN-delivered MB molecules effectively suppressed tumor growth when injected intratumorally in 4T1 tumor-bearing mice, while the free MB dye and control group showed limited effectiveness (Fig. 5D). Therefore, TDNs may be an ideal vector for in vivo delivery of PDT molecules due to their inherent biocompatibility, simple preparation, and effective intracellular delivery.
3.2. DNA Origami for Molecular Imaging and Drug Delivery
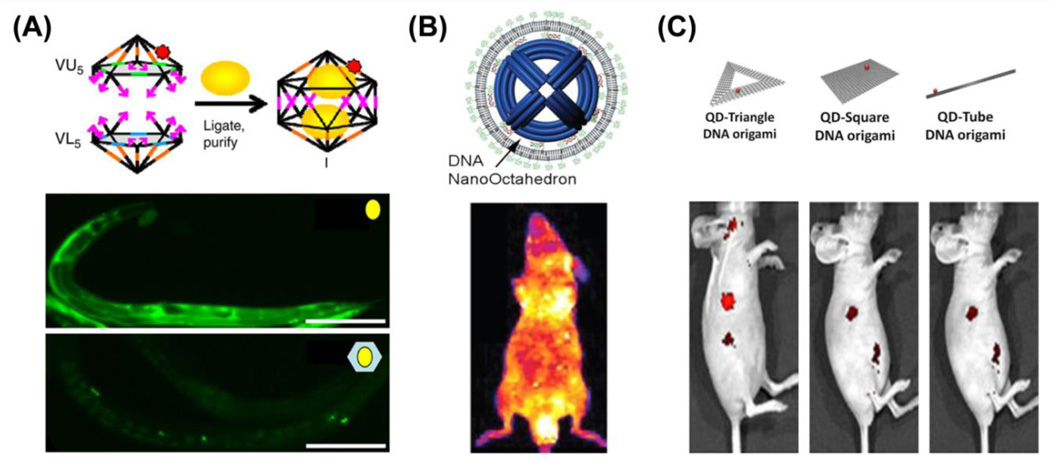
While DNA nanocages <10 nm display the diversity to accommodate different functional groups for molecular imaging and drug delivery in vivo, many efforts have also examined the utilization of DNA origami nanomaterials with various shapes and sizes to determine their pharmaceutical profiles and potential application. Bhatia et al. first utilized the synthetic icosahedral DNA nanostructures for loading of fluorescent dyes and a targeting platform for imaging of the anionic ligand-binding receptor (ALBR) pathway and mapping of pH changes in C. elegans [78]. The authors encapsulated a fluorescent biopolymer, 10-kDa fluorescein isothiocyanate (FITC)-Dextran (FD10), within a synthetic icosahedral DNA nanostructures to form a host-cargo complex without requiring any type of molecular interaction; thus, demonstrating a general strategy that may be useful for loading various types of functional molecules. The FD10-bearing DNA host effectively targeted endosomes of the ALBR pathway in coelomocytes of C. elegans, while non-functionalized FD10 showed limited localization in endosomes and non-specific cellular distribution (Fig. 6A). After internalization, the FD10 nanomaterials would break and release the fluorescent dye; thus, a correlation would provide insight into the maturation process of endosomes: a process that is characteristic with progressively narrowing of pH distributions.
Fig. 6.
Molecular imaging and drug delivery with DNA origami nanocomposites. (A) DNA icosahedron nanostructures are loaded with pH-sensitive cargo (10-kDa fluorescein isothiocyanate – Dextran, yellow spheres) for pH mapping in C. elegans. Adapted with permission from [78]. (B) Lipid-bilayer encapsulated DNA nano-octahedron (DNO) displayed significantly enhanced blood circulation and stability. Adapted with permission from [79]. (C) DNA origami loaded with quantum dots (QDs) could be used for passive tumor targeting in vivo. Among three different structures, doxorubicin-loaded QD-Triangle DNA origami showed optimal tumor retention and remarkable drug delivery efficacy. Adapted with permission from [80].
It was only two years later when Shih et al. introduced a lipid-bilayer encapsulated DNA nano-octahedron (DNO) to achieve long-term circulation and in vivo stability, which was inspired by the unique structure of common virus particles [79]. DNO (~50 nm) with 48 outer handles was self-assembled by bacteriophage-derived scaffold DNA (p7308) as the template strand and 144 short oligonucleotides as staple strands. Next, 48 DNA strands with pre-conjugated lipid molecules were arranged around DNO to form the encapsulated DNO (E-DNO) (Fig. 6B). The lipid treatment of DNO not only presented a virus-like morphology under transmission electron microscopy (TEM), but also exhibited strong protection against DNase I digestion. The pharmacokinetics of E-DNO in vivo showed an elimination half-life of about 370 ± 38 min, while the half-life of only DNO and normal oligonucleotides were only 49.5 ± 1.0 min and 38.0 ± 0.8 min, respectively. Based on the favorable in vitro stability and in vivo biodistribution pattern, E-DNO could easily become a platform for in vivo imaging and drug delivery when functionalized with active targeting agents via the outer handles or loading with contrast agents inside of the particle through DNA hybridization.
Although lipid-bilayers provided extra stability to DNA nanostructures, they can inhibit some interactions between DNA nanostructures the cell surface, potentially decreasing cell penetrating ability and drug delivery efficacy. To fully explore the biological properties of DNA origami, Zhang et al. assembled three DNA origami nanostructures and discovered that QD-conjugated DNA origami displayed higher tumor accumulation in MDA-MB-231 tumor-bearing mice, primarily through the enhanced permeability and retention (EPR) effect (Fig. 6C) [80]. Among them, triangular DNA origami possessed the most desired passive tumor targeting ability and exhibited long-term tumor uptake in vivo. After conducting fluorescent tumor imaging with DNA origami, they loaded it with the anti-cancer drug doxorubicin and observed remarkable antitumor efficacy without any noticeable toxicity in nude mice bearing orthotopic breast tumors. Compared with traditional drug nanocarriers, DNA origami presented highly sophisticated design rationality, enhanced safety, and improved biocompatibility. Also, relatively large sizes of different DNA origami, ranging from 10 to several hundred nanometers, significantly contributed to the passive targeting efficiency due to the EPR effect [81]. Moreover, compact DNA origami offered more binding sites for imaging groups and more loading capacities for drug intercalation, when compared to small DNA nanocages with sizes ranging from 7 to 20 nm.
Based on their findings, they further assembled DNA origami (DO) with gold nanorods (GNR) for in vitro and in vivo cancer theranostics [82]. The authors prepared two DNA origami, including triangular origami and tubular origami, and conjugated GNR on their surfaces, integrating the passive tumor targeting of DNA origami as well as the optical photothermal effects of GNR for cancer therapy. DNA origami, as a biocompatible platform for in vivo drug delivery, provided significant enhancement of cell uptake with MCF-7 cancer cells when compared to bare GNRs. Triangular DO-GNR further exhibited better tumor accumulation than tubular origami, and GNR accomplished photothermolysis against tumor cells. The organization of the GNR at predesignated locations on the surface of DNA origami allowed for enhanced photothermal efficacy.
Overall, the extraordinary programmability and addressability of DNA nanostructures provide a practical biodegradable platform for in vivo investigation of biological processes, including both noninvasive imaging as well as delivery of functional groups to sites of interest in living animals. DNA nanostructures could be easily functionalized with NIR dyes, radioactive isotopes, or inorganic nanoparticles. Most importantly, the rational design of DNA nanostructures allowed for site-specific modification with various targeting entities.
4. Potential Toxicities of DNA Nanostructures
Living organisms display unique surveillance systems that can recognize and destroy exogenous invaders, including foreign DNA and as-formed DNA nanomaterials. The interactions between external DNA and cells are highly complex. DNA nanomaterials can enhance cellular uptake, which may result in activation of the immune system in some cases. Several groups have investigated the potential immune response elicited by incubation of DNA nanomaterials in vitro and injection of DNA nanostructures in vivo. The toxicity of DNA nanostructures must be correlated with its biological application, as naked DNA nanostructures should elicit minimal immune activation. However, in some instances, DNA nanostructures are employed to delivery drugs or other agents that can elicit an immune response. For example, Liu et al. loaded CpG and a model antigen into DNA tetrahedral nanostructures to show that the nanostructure induced strong and persistent response against the antigen, while naked DNA nanostructures triggering no immune response [83].
In another study, Li et al. demonstrated that DNA tetrahedrons did not stimulate cytokine release from RAW264.7 cells [66]. However, when functionalized with CpG oligonucleotides, a type of nucleic acids with therapeutic effects via its strong immune-stimulatory activities, DNA tetrahedron dramatically induced the secretion of tumor necrosis factor (TNF), interleukin-6 (IL-6), and interlukin-12 (IL-12), excelling those of free CpG or free DNA tetrahedron by 9–18 times higher. The authors then exploited the controlled self-assembly of CpG-modified DNA nanostructures for intracellular delivery of CpG oligonucleotides to achieve greater immune stimulation.
Similarly, Mohri et al. constructed polypod-like DNA nanostructures as the delivery platform of CpG motifs to immune cells [84]. They demonstrated that polypod-like DNA nanomaterials with 3–8 pods display different properties regarding thermal stability, cell uptake, and immune response on toll-like receptor 9 (TLR9)-positive cells. A positive correlation was found between the number of pods and decreased cellular uptake of the DNA nanostructures and increased cytokine production and secretion. Later, Schuller et al. set forth to use DNA origami as the delivery platform of CpG [64]. When incubated with CpG-modified DNA origami, immune cells were stimulated via TLR9 to produce and secrete cytokines, including IL-6 and IL-12, which are cytokines commonly upregulated during initial immune stimulation. Pure DNA origami built with the M13mp18 template and 227 staple DNA strands triggered minimal immune reactivity; however, the coupling of 62 CpG sequences on DNA origami resulted in a significant enhancement of cytokine secretion, equal to that of the commonly employed Lipofectamine® carrier system.
Although many studies reported that free DNA nanostructures elicited minimal immune response from cells, Perrault et al. further examined the potential toxicity by developing a method for applying a biocompatible PEGylated lipid bilayer to encapsulate DNO [79]. This was expected to help maintain the PEG-DNO structural integrity, while also minimizing potential immune recognition. Similarly, this virus-inspired membrane enveloped DNA nanostructure triggered minimal production and secretion of IL-6 and IL-12, when compared with immune response elicited by non-encapsulated DNA nanostructures. Cells have the ability to detect and remove foreign DNA through various mechanisms, including lysosomal degradation, DNase digestion, and cytosolic receptor internalization [85]. Oftentimes, the cells respond to foreign DNA by activating an immune response, including upregulation of interferon genes and stimulation of caspace-1 activity [86, 87]. For these reasons, investigations into potential toxicities associated with DNA nanostructures remains critical for potential translation of these structures to the clinic. Additional data are needed to verify that naked DNA nanostructures are intrinsically non-toxic in vivo. As researchers continue to the examine DNA nanostructures for biological purposes, the use of accurate control structures will assist in this endeavor.
5. Conclusions and Future Perspectives
Molecular imaging has harnessed the ability to monitor biological processes non-invasively; thus, it has been widely applied for detection of disease occurrence and progression in vivo. Better imaging abilities require powerful contrast agents to achieve higher imaging specificity and lower possible toxicity. DNA nanoparticles have emerged as a new horizon for in vivo imaging applications based on their unique structure and high designability, simple functionalization, and low toxicity concerns. It is widely believed that the position and density of surface ligands can significantly influence the interactions between nanoparticles and cell membranes. However, it's hard to control the specific spatial location and orientation of functional groups during the synthesis of DNA nanomaterials. This review summarized recent advancements in molecular imaging and drug delivery with the assistance of DNA nanotechnology.
DNA nanoparticles display several unique properties that make them excellent imaging and therapy candidates. First, the Watson-Crick base-pairing principle enables the precise three-dimensional design of DNA nanostructures and the rapid development of DNA modification methods, including but not limited to fluorescent dyes, small molecules, peptides, proteins, and drug molecules. This makes it possible to obtain multifunctional DNA nanostructures with high accuracy. Next, the self-assembly process of multifunctional DNA constructions can be completed by simply mixing pre-designed DNA components with a single annealing step, which is much easier than some other nanoplatforms like dendrimers. Also, DNA nanostructures presented few toxicity concerns for in vitro and in vivo application. Moreover, negatively charged DNA nanostructures can enter cells without the help of transfection agents via the anionic ligand-binding receptor (ALBR) pathway, which provides more possibilities for in vivo applications. While the design of DNA nanoparticles can be better controlled during synthesis than other nanoparticles, the stability of DNA nanostructures remains an unavoidable issue. However, several studies have suggested that folding of DNA strands into polyhedron scaffolds can effectively prevent the enzymatic digestion of DNA nanomaterials. Also, stacking of DNA helix layers into DNA origami not only inhibits the enzyme digestion but offers more binding sites for functional groups and more intercalation space for drug molecules.
A very different and complex environment must be examined when moving from in vitro to in vivo studies with DNA nanomaterials. First, stability testing of DNA nanostructures is often performed in 10% FBS, which does not truly represent the physiological environment found in vivo. Thus, stability testing will hold more benefit when performed in vivo. Secondly, a comprehensive understanding of the biodistribution profiles and pharmacokinetic patterns of different DNA nanostructures in vivo should be examined. Then, improved strategies for enhancing the delivery of DNA nanostructures should be considered. Lastly, the influence of shape, size, surface charge, functional group, coating materials, and base sequence on the overall delivery and efficacy of DNA nanomaterials should be thoroughly studied, as it will provide vital insight into developing novel DNA nanomaterials in the future. The emergence of DNA nanostructures provides us a potential alternative to design and construct nanostructures with unprecedented precision, which is an opportunity that many envision with new horizons to investigate unsolved biomedical issues regarding disease diagnosis as well as treatment. Currently, toxicity concerns and in vivo integrity remain two limiting factors to the successful translation of DNA nanostructures to the clinic; yet, extensive investigations into the two topics are currently underway. To date, several DNA-based antisense therapies have gained federal drug administration (FDA)-approval, including fomivirsen (Vitravene®) for the treatment of cytomegalovirus retinitis and mipomersen (Kynamro®) for treating familial hypercholesterolemia. Thus, with the current progress and further studies, we expect that DNA nanotechnology will play a significant role in the development of molecular imaging, imaging-guided drug delivery, and therapy.
Acknowledgments
This work was supported, in part, by the University of Wisconsin - Madison, the National Institutes of Health (NIBIB/NCI 1R01CA169365, 1R01EB021336, P30CA014520, T32CA009206), and the American Cancer Society (125246-RSG-13-099-01-CCE).
References
- 1.Youn H, Hong KJ. In vivo noninvasive small animal molecular imaging. Osong. Public Health Res. Perspect. 2012;3:48–59. doi: 10.1016/j.phrp.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoelder S, Clarke PA, Workman P. Discovery of small molecule cancer drugs: successes, challenges and opportunities. Mol. Oncol. 2012;6:155–176. doi: 10.1016/j.molonc.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams JL, Smothers J, Srinivasan R, Hoos A. Big opportunities for small molecules in immuno-oncology. Nat. Rev. Drug Discov. 2015;14:603–622. doi: 10.1038/nrd4596. [DOI] [PubMed] [Google Scholar]
- 4.Kaspar AA, Reichert JM. Future directions for peptide therapeutics development. Drug Discov. Today. 2013;18:807–817. doi: 10.1016/j.drudis.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Gongora-Benitez M, Tulla-Puche J, Albericio F. Multifaceted roles of disulfide bonds. Peptides as therapeutics. Chem. Rev. 2014;114:901–926. doi: 10.1021/cr400031z. [DOI] [PubMed] [Google Scholar]
- 6.Boohaker RJ, Lee MW, Vishnubhotla P, Perez JM, Khaled AR. The use of therapeutic peptides to target and to kill cancer cells. Curr. Med. Chem. 2012;19:3794–3804. doi: 10.2174/092986712801661004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nat. Rev. Cancer. 2002;2:683–693. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 8.Mammatas LH, Verheul HM, Hendrikse NH, Yaqub M, Lammertsma AA, Menke-van der Houven van Oordt CW. Molecular imaging of targeted therapies with positron emission tomography: the visualization of personalized cancer care. Cell Oncol. (Dordr.) 2015;38:49–64. doi: 10.1007/s13402-014-0194-4. [DOI] [PubMed] [Google Scholar]
- 9.England CG, Hernandez R, Eddine SB, Cai W. Molecular imaging of pancreatic cancer with antibodies. Mol. Pharm. 2016;13:8–24. doi: 10.1021/acs.molpharmaceut.5b00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 11.Farokhzad OC, Langer R. Impact of nanotechnology on drug delivery. ACS Nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 12.LaVan DA, McGuire T, Langer R. Small-scale systems for in vivo drug delivery. Nat. Biotechnol. 2003;21:1184–1191. doi: 10.1038/nbt876. [DOI] [PubMed] [Google Scholar]
- 13.Anselmo AC, Mitragotri S. An overview of clinical and commercial impact of drug delivery systems. J. Control. Release. 2014;190:15–28. doi: 10.1016/j.jconrel.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrne JD, Betancourt T, Brannon-Peppas L. Active targeting schemes for nanoparticle systems in cancer therapeutics. Adv. Drug Deliv. Rev. 2008;60:1615–1626. doi: 10.1016/j.addr.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Lukyanov AN, Torchilin VP. Micelles from lipid derivatives of water-soluble polymers as delivery systems for poorly soluble drugs. Adv. Drug Deliv. Rev. 2004;56:1273–1289. doi: 10.1016/j.addr.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Kabanov AV, Batrakova EV, Alakhov VY. Pluronic (R) block copolymers as novel polymer therapeutics for drug and gene delivery. J. of Control. Release. 2002;82:189–212. doi: 10.1016/s0168-3659(02)00009-3. [DOI] [PubMed] [Google Scholar]
- 17.Schmaljohann D. Thermo- and pH-responsive polymers in drug delivery. Adv. Drug Deliv. Rev. 2006;58:1655–1670. doi: 10.1016/j.addr.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 18.Bianco A, Kostarelos K, Prato M. Applications of carbon nanotubes in drug delivery. Curr. Opin. Chem. Biol. 2005;9:674–679. doi: 10.1016/j.cbpa.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Prato M, Kostarelos K, Bianco A. Functionalized carbon nanotubes in drug design and discovery. Acc. Chem. Res. 2008;41:60–68. doi: 10.1021/ar700089b. [DOI] [PubMed] [Google Scholar]
- 20.Feng L, Liu Z. Graphene in biomedicine: opportunities and challenges. Nanomedicine (Lond.) 2011;6:317–324. doi: 10.2217/nnm.10.158. [DOI] [PubMed] [Google Scholar]
- 21.Smith AM, Duan H, Mohs AM, Nie S. Bioconjugated quantum dots for in vivo molecular and cellular imaging. Adv. Drug Deliv. Rev. 2008;60:1226–1240. doi: 10.1016/j.addr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zrazhevskiy P, Sena M, Gao X. Designing multifunctional quantum dots for bioimaging, detection, and drug delivery. Chem. Soc. Rev. 2010;39:4326–4354. doi: 10.1039/b915139g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamkaew A, Sun H, England CG, Cheng L, Liu Z, Cai W. Quantum dot-NanoLuc bioluminescence resonance energy transfer enables tumor imaging and lymph node mapping in vivo. Chem. Commun. (Camb.) 2016;52:6997–7000. doi: 10.1039/c6cc02764d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad MZ, Akhter S, Jain GK, Rahman M, Pathan SA, Ahmad FJ, Khar RK. Metallic nanoparticles: technology overview & drug delivery applications in oncology. Expert Opin. Drug Deliv. 2010;7:927–942. doi: 10.1517/17425247.2010.498473. [DOI] [PubMed] [Google Scholar]
- 25.Boisselier E, Astruc D. Gold nanoparticles in nanomedicine: preparations, imaging, diagnostics, therapies and toxicity. Chem. Soc. Rev. 2009;38:1759–1782. doi: 10.1039/b806051g. [DOI] [PubMed] [Google Scholar]
- 26.Duncan B, Kim C, Rotello VM. Gold nanoparticle platforms as drug and biomacromolecule delivery systems. J. Control. Release. 2010;148:122–127. doi: 10.1016/j.jconrel.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.England CG, Miller MC, Kuttan A, Trent JO, Frieboes HB. Release kinetics of paclitaxel and cisplatin from two and three layered gold nanoparticles. Eur. J. Pharm. Biopharm. 2015;92:120–129. doi: 10.1016/j.ejpb.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slowing, Vivero-Escoto JL, Wu CW, Lin VS. Mesoporous silica nanoparticles as controlled release drug delivery and gene transfection carriers. Adv. Drug Deliv. Rev. 2008;60:1278–1288. doi: 10.1016/j.addr.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 29.Cheng Y, A CS, Meyers JD, Panagopoulos I, Fei B, Burda C. Highly efficient drug delivery with gold nanoparticle vectors for in vivo photodynamic therapy of cancer. J. Am. Chem. Soc. 2008;130:10643–10647. doi: 10.1021/ja801631c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi KY, Liu G, Lee S, Chen X. Theranostic nanoplatforms for simultaneous cancer imaging and therapy: current approaches and future perspectives. Nanoscale. 2012;4:330–342. doi: 10.1039/c1nr11277e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niemeyer CM. Self-assembled nanostructures based on DNA: towards the development of nanobiotechnology. Curr. Opin. Chem. Biol. 2000;4:609–618. doi: 10.1016/s1367-5931(00)00140-x. [DOI] [PubMed] [Google Scholar]
- 32.Pinheiro AV, Han D, Shih WM, Yan H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sacca B, Niemeyer CM. Functionalization of DNA nanostructures with proteins. Chem. Soc. Rev. 2011;40:5910–5921. doi: 10.1039/c1cs15212b. [DOI] [PubMed] [Google Scholar]
- 34.Chen YJ, Groves B, Muscat RA, Seelig G. DNA nanotechnology from the test tube to the cell. Nat. Nanotechnol. 2015;10:748–760. doi: 10.1038/nnano.2015.195. [DOI] [PubMed] [Google Scholar]
- 35.Zhang F, Nangreave J, Liu Y, Yan H. Structural DNA nanotechnology: state of the art and future perspective. J. Am. Chem. Soc. 2014;136:11198–11211. doi: 10.1021/ja505101a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Fan C, Pei H, Shi J, Huang Q. Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Adv. Mater. 2013;25:4386–4396. doi: 10.1002/adma.201300875. [DOI] [PubMed] [Google Scholar]
- 37.Chao J, Liu H, Su S, Wang L, Huang W, Fan C. Structural DNA nanotechnology for intelligent drug delivery. Small. 2014;10:4626–4635. doi: 10.1002/smll.201401309. [DOI] [PubMed] [Google Scholar]
- 38.Yang D, Hartman MR, Derrien TL, Hamada S, An D, Yancey KG, Cheng R, Ma M, Luo D. DNA materials: bridging nanotechnology and biotechnology. Acc. Chem. Res. 2014;47:1902–1911. doi: 10.1021/ar5001082. [DOI] [PubMed] [Google Scholar]
- 39.Duckett DR, Lilley DMJ. The 3-way DNA junction is a Y-Shaped molecule in which there is no helix helix stacking. Embo J. 1990;9:1659–1664. doi: 10.1002/j.1460-2075.1990.tb08286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seeman NC. Nanotechnology and the double helix. Sci. Am. 2004;290:64–69. doi: 10.1038/scientificamerican0604-64. 72-65. [DOI] [PubMed] [Google Scholar]
- 41.Seeman NC. Nucleic acid junctions and lattices. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 42.Kallenbach NR, Ma RI, Seeman NC. An immobile nucleic-acid junction constructed from oligonucleotides. Nature. 1983;305:829–831. [Google Scholar]
- 43.Ma RI, Kallenbach NR, Sheardy RD, Petrillo ML, Seeman NC. Three-arm nucleic acid junctions are flexible. Nucleic Acids Res. 1986;14:9745–9753. doi: 10.1093/nar/14.24.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu TJ, Seeman NC. DNA double-crossover molecules. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 45.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 46.Liu D, Wang M, Deng Z, Walulu R, Mao C. Tensegrity: construction of rigid DNA triangles with flexible four-arm DNA junctions. J. Am. Chem. Soc. 2004;126:2324–2325. doi: 10.1021/ja031754r. [DOI] [PubMed] [Google Scholar]
- 47.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 48.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 49.Matthies M, Agarwal NP, Schmidt TL. Design and synthesis of triangulated DNA origami trusses. Nano. Lett. 2016;16:2108–2113. doi: 10.1021/acs.nanolett.6b00381. [DOI] [PubMed] [Google Scholar]
- 50.Selnihhin D, Andersen ES. Computer-aided design of DNA origami structures. Methods Mol. Biol. 2015;1244:23–44. doi: 10.1007/978-1-4939-1878-2_2. [DOI] [PubMed] [Google Scholar]
- 51.Kuzuya A, Komiyama M. Design and construction of a box-shaped 3D-DNA origami. Chem. Commun. (Camb.) 2009:4182–4184. doi: 10.1039/b907800b. [DOI] [PubMed] [Google Scholar]
- 52.Goodman RP, Schaap IA, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 53.Goodman RP, Berry RM, Turberfield AJ. The single-step synthesis of a DNA tetrahedron. Chem. Commun. (Camb.) 2004:1372–1373. doi: 10.1039/b402293a. [DOI] [PubMed] [Google Scholar]
- 54.Erben CM, Goodman RP, Turberfield AJ. A self-assembled DNA bipyramid. J. Am. Chem. Soc. 2007;129:6992–6993. doi: 10.1021/ja071493b. [DOI] [PubMed] [Google Scholar]
- 55.Ke Y, Ong LL, Shih WM, Yin P. Three-dimensional structures self-assembled from DNA bricks. Science. 2012;338:1177–1183. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ke Y, Ong LL, Sun W, Song J, Dong M, Shih WM, Yin P. DNA brick crystals with prescribed depths. Nat. Chem. 2014;6:994–1002. doi: 10.1038/nchem.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goodman RP, Heilemann M, Doose S, Erben CM, Kapanidis AN, Turberfield AJ. Reconfigurable, braced, three-dimensional DNA nanostructures. Nat. Nanotechnol. 2008;3:93–96. doi: 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- 58.Stellwagen NC. Electrophoresis of DNA in agarose gels, polyacrylamide gels and in free solution. Electrophoresis. 2009;30(Suppl 1):S188–S195. doi: 10.1002/elps.200900052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bellot G, McClintock MA, Lin C, Shih WM. Recovery of intact DNA nanostructures after agarose gel-based separation. Nat. Methods. 2011;8:192–194. doi: 10.1038/nmeth0311-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xing S, Jiang D, Li F, Li J, Li Q, Huang Q, Guo L, Xia J, Shi J, Fan C, Zhang L, Wang L. Constructing higher-order DNA nanoarchitectures with highly purified DNA nanocages. ACS Appl. Mater. Interfaces. 2015;7:13174–13179. doi: 10.1021/am505592e. [DOI] [PubMed] [Google Scholar]
- 61.Lin C, Perrault SD, Kwak M, Graf F, Shih WM. Purification of DNA-origami nanostructures by rate-zonal centrifugation. Nucleic Acids Res. 2013;41:e40. doi: 10.1093/nar/gks1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei X, Nangreave J, Liu Y. Uncovering the self-assembly of DNA nanostructures by thermodynamics and kinetics. Acc. Chem. Res. 2014;47:1861–1870. doi: 10.1021/ar5000665. [DOI] [PubMed] [Google Scholar]
- 63.Shaw A, Benson E, Hogberg B. Purification of functionalized DNA origami nanostructures. ACS Nano. 2015;9:4968–4975. doi: 10.1021/nn507035g. [DOI] [PubMed] [Google Scholar]
- 64.Schuller VJ, Heidegger S, Sandholzer N, Nickels PC, Suhartha NA, Endres S, Bourquin C, Liedl T. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano. 2011;5:9696–9702. doi: 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- 65.Keum JW, Bermudez H. Enhanced resistance of DNA nanostructures to enzymatic digestion. Chem. Commun. (Camb.) 2009:7036–7038. doi: 10.1039/b917661f. [DOI] [PubMed] [Google Scholar]
- 66.Li J, Pei H, Zhu B, Liang L, Wei M, He Y, Chen N, Li D, Huang Q, Fan C. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano. 2011;5:8783–8789. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 67.Conway JW, McLaughlin CK, Castor KJ, Sleiman H. DNA nanostructure serum stability: greater than the sum of its parts. Chem. Commun. (Camb.) 2013;49:1172–1174. doi: 10.1039/c2cc37556g. [DOI] [PubMed] [Google Scholar]
- 68.Hamblin GD, Carneiro KM, Fakhoury JF, Bujold KE, Sleiman HF. Rolling circle amplification-templated DNA nanotubes show increased stability and cell penetration ability. J. Am. Chem. Soc. 2012;134:2888–2891. doi: 10.1021/ja2107492. [DOI] [PubMed] [Google Scholar]
- 69.Aldaye FA, Lo PK, Karam P, McLaughlin CK, Cosa G, Sleiman HF. Modular construction of DNA nanotubes of tunable geometry and single- or double-stranded character. Nat. Nanotechnol. 2009;4:349–352. doi: 10.1038/nnano.2009.72. [DOI] [PubMed] [Google Scholar]
- 70.Mei Q, Wei X, Su F, Liu Y, Youngbull C, Johnson R, Lindsay S, Yan H, Meldrum D. Stability of DNA origami nanoarrays in cell lysate. Nano Lett. 2011;11:1477–1482. doi: 10.1021/nl1040836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hahn J, Wickham SF, Shih WM, Perrault SD. Addressing the instability of DNA nanostructures in tissue culture. ACS Nano. 2014;8:8765–8775. doi: 10.1021/nn503513p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wen YL, Li LY, Wang LL, Xu L, Liang W, Ren SZ, Liu G. Biomedical applications of DNA-nanomaterials based on metallic nanoparticles and DNA self-assembled nanostructures. Chin. J. Chem. 2016;34:283–290. [Google Scholar]
- 73.Yang F, Zhang X, Song L, Cui H, Myers JN, Bai T, Zhou Y, Chen Z, Gu N. Controlled drug release and hydrolysis mechanism of polymer-magnetic nanoparticle composite. ACS Appl. Mater. Interfaces. 2015;7:9410–9419. doi: 10.1021/acsami.5b02210. [DOI] [PubMed] [Google Scholar]
- 74.Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim KR, Lee YD, Lee T, Kim BS, Kim S, Ahn DR. Sentinel lymph node imaging by a fluorescently labeled DNA tetrahedron. Biomaterials. 2013;34:5226–5235. doi: 10.1016/j.biomaterials.2013.03.074. [DOI] [PubMed] [Google Scholar]
- 76.Jiang D, Sun Y, Li J, Li Q, Lv M, Zhu B, Tian T, Cheng D, Xia J, Zhang L, Wang L, Huang Q, Shi J, Fan C. Multiple-armed tetrahedral DNA nanostructures for tumor-targeting, dual-modality in vivo imaging. ACS Appl. Mater. Interfaces. 2016;8:4378–4384. doi: 10.1021/acsami.5b10792. [DOI] [PubMed] [Google Scholar]
- 77.Kim KR, Bang D, Ahn DR. Nano-formulation of a photosensitizer using a DNA tetrahedron and its potential for in vivo photodynamic therapy. Biomater. Sci. 2016;4:605–609. doi: 10.1039/c5bm00467e. [DOI] [PubMed] [Google Scholar]
- 78.Bhatia D, Surana S, Chakraborty S, Koushika SP, Krishnan Y. A synthetic icosahedral DNA-based host-cargo complex for functional in vivo imaging. Nat. Commun. 2011;2:339. doi: 10.1038/ncomms1337. [DOI] [PubMed] [Google Scholar]
- 79.Perrault SD, Shih WM. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano. 2014;8:5132–5140. doi: 10.1021/nn5011914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Q, Jiang Q, Li N, Dai L, Liu Q, Song L, Wang J, Li Y, Tian J, Ding B, Du Y. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano. 2014;8:6633–6643. doi: 10.1021/nn502058j. [DOI] [PubMed] [Google Scholar]
- 81.Kobayashi H, Watanabe R, Choyke PL. Improving conventional enhanced permeability and retention (EPR) effects; what is the appropriate target? Theranostics. 2013;4:81–89. doi: 10.7150/thno.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang Q, Shi Y, Zhang Q, Li N, Zhan P, Song L, Dai L, Tian J, Du Y, Cheng Z, Ding B. A Self-assembled DNA origami-gold nanorod complex for cancer theranostics. Small. 2015;11:5134–5141. doi: 10.1002/smll.201501266. [DOI] [PubMed] [Google Scholar]
- 83.Liu X, Xu Y, Yu T, Clifford C, Liu Y, Yan H, Chang Y. A DNA nanostructure platform for directed assembly of synthetic vaccines. Nano Lett. 2012;12:4254–4259. doi: 10.1021/nl301877k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mohri K, Nishikawa M, Takahashi N, Shiomi T, Matsuoka N, Ogawa K, Endo M, Hidaka K, Sugiyama H, Takahashi Y, Takakura Y. Design and development of nanosized DNA assemblies in polypod-like structures as efficient vehicles for immunostimulatory CpG motifs to immune cells. ACS Nano. 2012;6:5931–5940. doi: 10.1021/nn300727j. [DOI] [PubMed] [Google Scholar]
- 85.Surana S, Shenoy AR, Krishnan Y. Designing DNA nanodevices for compatibility with the immune system of higher organisms. Nat. Nanotechnol. 2015;10:741–747. doi: 10.1038/nnano.2015.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]