Abstract
Virtual screening has become an indispensable procedure in the drug discovery. Virtual screening methods can be classified into two categories, ligand-based and structure-based methods. While the former have advantages, including being quick to compute, in general they are relatively weak at discovering novel active compounds because they use known actives as references. On the other hand, structure-based methods have higher potential to find novel compounds because they directly predict binding affinity of a ligand in a target binding pocket, albeit with substantially slower speed than ligand-based methods. Here, we report a novel structure-based virtual screening method, PL-PatchSurfer2. In PL-PatchSurfer2, protein and ligand surfaces are represented by a set of overlapping local patches, each of which are represented by 3D Zernike descriptors (3DZDs). By using 3DZDs, shape and physicochemical complementarity of local surface regions of a pocket surface and a ligand molecule can be concisely and effectively computed. Compared to the previous version of the program, the performance of PL-PatchSurfer2 is substantially improved by adding two more features, atom-based hydrophobicity and hydrogen bond acceptors and donors. Benchmark studies showed that PL-PatchSurfer2 performed better than or comparable to popular existing methods. Particularly, PL-PatchSurfer2 significantly outperformed existing methods when apo form or template-based protein models were used for queries. The computational time of PL-PatchSurfer2 is about 20 times faster than conventional structure-based methods. The PL-PatchSurfer2 program are available at kiharalab.org/plps2/.
Graphical abstract

INTRODUCTION
Virtual screening is a computational technique that searches a molecule library to find candidate active compounds for a target protein1. This technique has been widely used in drug discovery2–4. It can be classified into two categories based on the strategy it adopts: ligand-based virtual screening (LBVS) or structure-based virtual screening (SBVS). LBVS methods rank ligands in a library by calculating similarity between a ligand and a template active compound. Depending on how compounds are represented, LBVS methods can be further classified into those which use 1D5,6, 2D7–9, or 3D representation10–12. LBVS methods are generally fast, and only require template compounds. That these methods to not require receptor structures can be considered among their advantages; however, this disregarding of receptor structure weakens their ability to discover novel scaffold compounds13. On the other hand, SBVS methods predict a binding pose of a ligand in a target receptor pocket and calculate binding affinity from that pose. Thus, protein-ligand docking14–17 and receptor pharmacophore search18,19 are common strategies for SBVS. Since it uses a receptor structure and does not require a priori knowledge of known drugs, hits found from a ligand library could be more diverse than those from LBVS. However, SBVS methods usually takes more time than LBVS methods because they search docking conformational space and calculate binding energy. Thus, an ideal VS method would have advantages from both types of methods: it would find diverse active compounds quickly, and would be applicable to target receptors that do not have experimentally solved structures.
Here, we report a novel SBVS program, PL-PatchSurfer2, which is a substantial upgrade from the prototype of the program we developed earlier20. Unlike conventional SBVS methods that use atomic-detailed descriptions of molecules, PL-PatchSurfer2 uses a molecular surface representation, which has several notable advantages, as we will show in the benchmark results later in this report. Surfaces of a target binding pocket and a ligand are segmented into overlapping local surface patches to enable evaluation of the local compatibility of the pocket and the ligand. In addition to the surface shape, physicochemical properties are mapped onto the surface, and represented with three-dimensional Zernike descriptors (3DZDs). A 3DZD is a rotation-invariant vector representation of a 3D scalar-valued function in Euclidean space (e.g. shape, properties mapped in the space), derived from a series expansion of the 3D function based on the 3D Zernike moments21,22. The 3DZD representation of molecular surface has been applied to solve many structural biology problems, including ligand-ligand comparison12, pocket-pocket comparison23–26, electron density maps from electron microscopy27, and protein structure similarity search28. It has been shown that representing local surface patches with multiple 3DZDs is effective to capture conformational changes of pockets and ligands in pocket-pocket comparison and ligand-ligand comparison12,25.
The largest difference between PL-PatchSurfer2 and its earlier version is that atom-based hydrophobicity and hydrogen-bond donor/acceptor positions, which are important chemical characteristics in protein-ligand interactions, were newly implemented to describe the molecular surface in addition to the shape and surface electrostatics used from the earlier version. Moreover, weights that combine scoring terms were now trained using the BEDROC score29 as the target function rather than the area-under receiver operating characteristic curve (AUC) used in the previous version, because AUC is known to fail to promote early recognition of active compounds29,30. The performance of PL-PatchSurfer2 was tested extensively against four benchmark datasets in comparison with three popular virtual screening methods, AutoDock Vina16, DOCK617, and ROCS11. In the first test on the DUD31 dataset, PL-PatchSurfer2 performed better than or comparably to the three other methods compared. Remarkably, when tested on a dataset with apo (ligand free) binding pocket structures and another dataset with template-based computational models of receptor proteins, PL-PatchSurfer2 significantly outperformed the existing methods with almost twice the enrichment factor (EF) at 1% on the former set and over five times the EF1% for the latter dataset than AutdockVina. PL-PatchSurfer2 also showed better or comparable results than the existing methods on the WOMBAT dataset32, which contains more diverse active compounds than the DUD set. PL-PatchSurfer2 runs about 20 times faster than AutoDock Vina and DOCK6. Based on the benchmark results, unique characteristics of PL-PatchSurfer2 are discussed with a focus on the possibility of expanding target proteins that can be handled by applying computational structure modeling.
METHODS
Overview of PL-PatchSurfer2
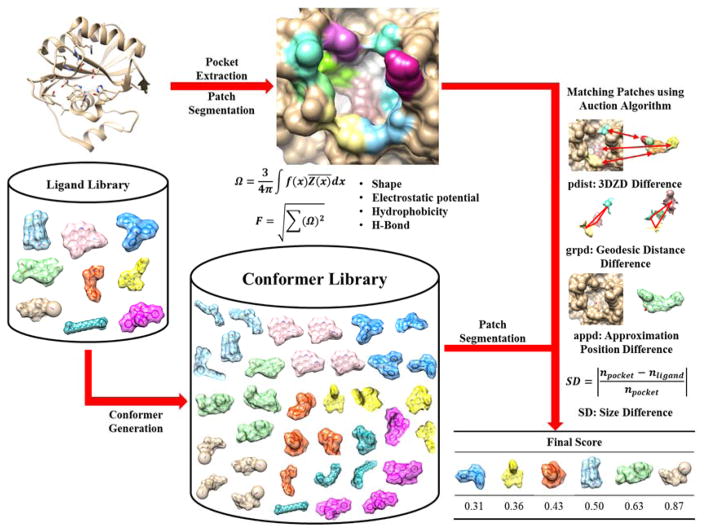
PL-PatchSurfer2 is a virtual screening program that ranks ligands in a library for a target pocket by identifying complementarity between pocket and ligand local surface regions. The overall workflow of PL-PatchSurfer2 is shown in Figure 1. The first step of PL-PatchSurfer2 is to generate multiple conformations of each ligand using OMEGA33 to consider conformational flexibility of the ligand. Then, molecular surface of a target protein and the ligands in different conformations are generated by APBS34. Four features are assigned onto the surface of molecules: shape, electrostatic potential, hydrophobicity, and hydrogen bond acceptor/donor positions. The surface of the protein pocket and ligand are segmented into a number of overlapping patches, and the four physicochemical features of the patches are represented with 3DZDs. For a target pocket and a ligand molecule, surface patches are compared to identify compatible patch pairs, and evaluated by a score. Then, ligands in the library are ranked according to the score. Comparing with the previous version of PL-PatchSurfer (PL-PatchSurfer1), PL-PatchSurfer2 was improved in five aspects: (1) Atom-based hydrophobicity and hydrogen bonding acceptors/donor information are added for features of molecules. (2) The maximum number of conformations to be generated for ligands is increased from 20 to 50. (3) Weighting factors for scoring terms in the scoring function named the Conformer Score, which is important for summarizing individual scoring terms for assessing compatibility for a ligand to a target pocket, were more thoroughly optimized. (4) The target function for optimizing Conformer Score is BEDROC instead of AUC to put more emphasis on early-recognition. (5) We devised a Boltzmann averaging in computing a score for a ligand, instead of taking average of top ten docking conformations in the previous version. The PL-PatchSurfer2 program are available at kiharalab.org/plps2/. We explain each step in greater details in the following sections.
Figure 1.
Workflow of PL-PatchSurfer2.
Assigning physicochemical properties on molecule surface
Four features - geometric shape, the electrostatic potential, hydrophobicity, and hydrogen bond acceptor/donor positions, are used to characterize molecular surfaces. The molecular surface of a protein or a ligand is computed with APBS34. It also computes electrostatic potential on the molecular surface by solving the Poisson-Boltzmann equation. The computed shape and electrostatic potential information are mapped to 3D voxels. A protein structure is mapped onto a 3D grid, such that a voxel point is marked with 1 if it overlaps with the protein surface, and 0 otherwise. Likewise, the computed electrostatic potential, hydrophobicity, and hydrogen-bond acceptors/donors at each position on molecular surface are mapped at corresponding voxels.
As for the newly introduced hydrophobicity in PL-PatchSurfer2, a hydrophobic field was calculated and assigned on the surface in the similar way as the electrostatic potential. Similar to a work by Kellogg et al., who calculated a molecular hydrophobic field as a distance-weighted atomic logP value35, we calculated the hydrophobicity of a surface grid point using a Fermi-like distance-based weight as follows36:
| (1) |
where i, j, and N are a voxel point, an atom, and the number of atoms in the molecule, respectively. fj is the atomic hydrophobicity and rij is the distance between the voxel point i and the atom j. α was set to 1.0 Å−1, so the effective distance between a voxel point and a ligand atom is 4.0 Å. Atomic hydrophobicity, fj, is computed by the XlogP337 program.
The hydrogen bond property, another new feature in PL-PatchSurfer2, is assigned to a voxel point based on the character of the closest atom. If the closest atom is a hydrogen bond donor (or acceptor), 1 (or −1) is assigned to the voxel point. If the closest atom is neither a donor nor an acceptor, 0 is assigned.
Generating multiple conformations of ligands, protein pocket extraction, and surface segmentation
Conformations of a ligand were generated with OMEGA33. The maximum number of conformations was set to 50, the maximum energy difference between the highest energy conformation and the lowest energy conformation was set to 15.0 kcal/mol, and root-mean square deviation (RMSD) cutoffs between generated conformers was set to 0.5 Å, 0.8 Å, and 1.0 Å for a ligand with less than six, six to ten, and more than 10 rotatable bonds, respectively.
A protein pocket surface was extracted after all four features were assigned to the protein surface. The pocket region was determined by casting rays from the center of the pocket to the pocket wall24. The surfaces of the pocket and ligands are segmented into a number of overlapping patches as follows: First, a set of centers of patches, called seed points, were spread on the surface. Seed points were selected iteratively so that points were separated by at least 3.0 Å. Then, spheres of 5.0 Å radius centered at each seed point were placed and the surface regions within the spheres were segmented. These segmented local surface regions, called patches, were represented with 3DZDs as described in the next section.
3D Zernike descriptors
A 3DZD represents a 3D function using a series expansion based on Zernike-Canterakis basis functions21,22. Here, a brief introduction of 3DZD is given. More detailed explanations of the descriptor are found in previous works21,22. The Zernike-Canterakis basis function, , is shown in Eq. 2:
| (2) |
Rnl(r) is a radial function, where r is the distance from the origin, while Ylm(θ,φ) is a spherical harmonic. The integers, n, l, m, are referred to as order, degree, and repetition, respectively. Conditions for these numbers are −l < m < l, 0 ≤ l ≤ n, and (n − l) is an even number. Thus, the ranges of l and m are determined by the order n.
To represent 3D molecular surface shape or a physicochemical property on the surface with 3DZD, 3D voxels with mapped property values are considered as a 3D function, f(x,y,z). 3D Zernike moments of this function, , is obtained as follows (Eq. 3):
| (3) |
where x = (x, y, z). Taking the norm of moments, with the same n and l yields a rotation-invariant (2l + 1)-dimensional vector (Eq. 4):
| (4) |
The dimension of Fnl depends on the order n that determines the “resolution” of the 3DZD representation. In PL-PatchSurfer, n is set to 15, which makes 3DZD a 72-dimensional vector. Thus, 3DZD provides a compact and rotationally invariant description of 3D shapes and numerical features mapped on a 3D shape.
Matching patches between a target pocket and a ligand
To search optimal complementary patch pairs between a receptor pocket and a ligand in a certain conformation, a modified auction algorithm is used25. The algorithm minimizes Patch Score (Eq. 5) by selecting proper patch pairs in an iterative fashion. Patch Score between a protein patch p and a ligand patch l is computed as:
| (5) |
pdist is a weighted sum of the Euclidean distance between 3DZDs for p and l (Eq. 6):
| (6) |
where i represents a physicochemical feature, shape, electrostatic potential, hydrophobicity, or hydrogen bond acceptor/donor and ν is a relative weight assigned to the corresponding feature. The weights ν were trained so that the score pdist can correctly identify the interacting protein patch for each query ligand patch in the benchmark dataset of 195 protein-ligand complex structures from the PDBBind set38. A protein-ligand patch pairs are defined as physically interacting if their patch center distance is less than 3.0 Å in the crystal structures. The optimization yielded weight values of 1.0, 0.4, 0.1, and 0.2 for shape, electrostatic potential, hydrophobicity, and hydrogen bond acceptor/donor, respectively.
The second term, grpd, geodesic relative position difference, is defined as the summation of geodesic distance difference of matched pairs to the other patch pairs (Eq. 7):
| (7) |
where M is a set of matched patch pairs in the previous iteration of the modified auction algorithm, and n is the number of pairs. G(a, a′) is a geodesic distance (i.e. distance along the molecular surface) between patch centers of a and a′. In the initial round of the modified auction algorithm, this term is set to zero, because there are no matched pairs from a previous round.
The last term of Eq. 5 considers Approximate Patch Position Difference, appd (Eq. 8):
| (8) |
APP is a vectorized histogram of the geodesic distance from a patch center to other patch centers in the pocket or the ligand. The bin size was set to 1.0 Å. An approximate position of a patch in a pocket or a ligand, e.g. whether a patch locates in the middle or at the edge of the molecule, can be represented by this vector.
The weights in Patch Score (Eq. 5) were optimized to maximize the success rate of finding correct patch pairs from a target pocket and a ligand. The weights were trained on the PDBBind set38 and tested on the ASTEX diverse set39, which consists of 85 protein-ligand complex structures. The result is shown in Table S1 in Supporting Information. We trained weights for large pockets (whose surface is segmented to 40 or more patches) and small pockets (less than 40 surface patches) separately, because that was found to improve the overall success rate than using a single weight set for the entire pockets in the datasets.
Scoring function to rank ligands for a target pocket
As described in the previous section, for a target pocket P and a conformation C of a ligand, complementary patches are matched using Eq. 5. Then, overall fitness of the ligand conformer C to the pocket P is calculated as follows (Eq. 9):
| (9) |
where avg() is the average of a term in Eq. 5 over all matched patch pairs. The last term, SD is a size difference between the pocket and the ligand conformer, which is quantified by the difference of the number of patches covering the pocket and the conformer (Eq. 10). It is to capture the similarity of an approximate surface area between the pocket and the conformer:
| (10) |
npocket and nligand are the number of patches in the pocket and the ligand conformer, respectively.
All the weight parameters in Eq. 9 were determined to maximize a virtual screening performance measured by BEDROC29. The DUD set31, which composed of 40 proteins, were used to train and evaluate the virtual screening performance. As before, a grid search was used to find the optimum weight set. The weights were changed from 0.0 to 1.0 with an interval of 0.1. For each ligand molecule, up to 50 conformations were generated and each of which obtains Conformer Score for a target pocket. Using the scores given to each of the conformers of a ligand, the final score of a ligand for a pocket was computed in two different ways. First, the lowest Conformer Score (LCS) was taken as the final score to the ligand. Second, we computed a weighted average of Conformer Score which is inspired by the Boltzmann distribution (BS)40,41:
| (11) |
Here PS(P,C) is Conformer Score of pocket P and conformer C, Nconf is the number of top-scoring conformations considered in computing the score. β is a parameter. In the Result section, we tried different values of β and Nconf.
BENCHMARK DATASETS
The performance of PL-PatchSurfer2 was evaluated in four datasets: the DUD dataset, apo structures of targets in DUD, template-based structure model of targets in DUD, and the WOMBAT dataset. We briefly describe the datasets in this section.
The DUD Benchmark Set
The DUD set31 is composed of 40 important pharmaceutical targets. The ligand library was constructed for individual protein targets, a combination of known active compounds and decoy compounds that have similar physicochemical properties to actives. 15 targets were removed from the original DUD set, which have cofactors, ions, or disordered regions and thus cannot be handled by APBS to compute molecular surface and the surface electrostatic potential. They are angiotensin-converting enzyme, adenosine deaminase, catechol O-methyltransferase, phosphodiesterase 5, dihydrofolate reductase, glycinamide ribonucleotide transformylase, aldose reductase, enoyl ACP reductase, glycogen phosphorylase β, purine nucleoside phosphorylase, and S-adenosyl-homocysteine hydrolase, hydroxymethylglutaryl-CoA reductase, trypsin, human shock protein 90, and thymidine kinase. The remaining 25 targets were divided into two sets to perform a two-fold cross-validation to train and test the weighting parameters. The full list of training and test set can be found in Table S2 (Supporting Information). To speed up the virtual screening process, the ratio between the number of actives and decoys was changed to 1: 29 from the original 1:36 by randomly selecting decoy compounds from library. If the number of molecules in whole library is higher than 3000, the library was re-constructed by selecting 60 active compounds and 1740 decoy compounds randomly from the original DUD library.
Apo structure dataset
In addition to the holo form (target bound form) of the target proteins in DUD, we also tested PL-PatchSurfer2 on apo form (ligand free form) of targets. Using the apo form of target pockets is generally difficult for virtual screening because pockets change their shape upon binding42. Out of the 25 targets, 19 of them have available crystal structures in apo form. The PDB IDs of the apo forms are listed in Table S2.
Template-based structure model dataset
We have further tested the performance of PL-PatchSurfer2 on computational models of target proteins. The applicability of SBVS can dramatically enlarge if a SBVS method can show sufficiently good performance with computational protein structure models42–44. Particularly, due to the expansion of the protein structure database, PDB45, an increasing number of protein structures can be modeled using template-based modeling (TBM) methods. For each of the 25 targets in DUD, we used PSI-BLAST46 to find homologous proteins to the targets in PDB. For 18 targets, a template with a sequence identity of over 30% and an alignment coverage of over 90% to the target was found. Moreover, among them, for six targets, three templates, one each at a sequence identity of 30–50%, 50–70%, and 70–90% with a coverage of over 90% were found. Identified templates are listed in Table S2. Using the templates, template-based models were constructed using Modeller9v1147.
The WOMBAT dataset
The last dataset used is the WOMBAT dataset. This dataset is intended to collect more diverse active compounds than the DUD dataset32. In the WOMBAT dataset, active compounds that have lead-like conditions (logP < 4.5 and molecular weight < 450) were collected mainly from literature and similar compounds were removed by clustering using 2D graph comparison32. Thus, the resulting dataset is more diverse than the DUD set and therefore more suitable for testing scaffold hopping ability of virtual screening methods.
We used nine targets out of thirteen proteins in the WOMBAT set, which overlap with the DUD (Supporting Information, Table S3). Since the WOMBAT set only provides active compounds, we supplemented the dataset with decoy compounds from the DUD. The ratio between actives and decoys was set to 1:29, the same as in the DUD dataset we used, by randomly selecting compounds, except for the cases where there are not enough decoy compounds available. In the three such cases, all decoy compounds were used.
Existing virtual screening methods compared
PL-PatchSurfer2 was compared with its original version, PL-PatchSurfer120, as well as three popular virtual screening programs, AutoDock Vina16, DOCK617, and ROCS11. The former two programs are SBVS methods while ROCS is an LBVS method. Since ROCS is for LBVS, which does not use target protein structures, its performance was tested only on the DUD and the WOMBAT dataset.
To run five SBVS programs including PL-PatchSurfer1 and 2, a binding pocket of a target was defined from the geometrical center of the co-crystallized ligand. For apo and template-based model datasets, a target structure was aligned with the holo form structure in the DUD set using TM-align48, and the cognate ligand copied to the target structure from the holo structure was used to determine the geometrical center of the ligand, from which a binding pocket was defined. As for ROCS, which needs a template (query) ligand for screening, we used the same multiple conformers of the native ligand in the holo structure generated by OMEGA for running PL-PatchSurfer2.
Protonation states and atomic charges of proteins and ligands were kept the same as original assignments in the DUD and the WOMBAT dataset. For apo and template-based structures, hydrogen atoms and Gasteiger charges were added by using the DockPrep module of Chimera50.
To run AutoDock Vina, the docking box was defined as a 25 Å3 cube centered at the geometrical center of the cognate ligand. For DOCK6, the docking box was constructed so that it has a 5 Å margin from a space occupied by the cognate ligand, which is defined as a set of spheres of a 10.0 Å radius centered at each atom of the cognate ligand. The other parameters of AutoDock Vina and DOCK6 were set to their default values.
Evaluation Metrics
The performance of virtual screening was evaluated by the enrichment factor (EF), area under receiver operating characteristic curve (AUC), and BEDROC. EF at top x% subset is calculated as follows:
| (12) |
where Activesx% is the number of actives found in the top x% score-ranked compounds while Compoundsx% is the number of all the compounds within the top x% subset. ActivesLibrary and CompoundsLibrary are the number of all active compounds and the entire compounds in the library, respectively. A random retrieval would yield EFx% = 1. We examined EFs at 1%, 5%, and 10%.
We also used Area Under the Curve (AUC) of Receiver operating Characteristic (ROC) curve, which shows a performance of a virtual screening program by plotting the true positive rate (rate of finding active compounds) relative to the false positive rate (rate of finding decoy compounds). AUC is 0.5 for random retrieval, and 1.0 for the maximum possible value when a program finds all active compounds before ranking any decoy compounds.
BEDROC assigns larger weights to actives detected earlier in a rank to reward early detection of actives29,30:
| (13) |
where N is the number of compounds, n is the number of hits, Ra is n divided by N, ri is the ranking of the ith hit, and α is a weight parameter, which is set to 20 as suggested by Truchon & Bayly29.
RESULTS AND DISCUSSION
Improvements of PL-PatchSurfer2 over PL-PatchSurfer1
First, we compare PL-PatchSurfer2 with its previous version PL-PatchSurfer1. In Table 1, we investigated the effect of each of the following three new main developments of PL-PatchSurfer2 relative to PL-PatchSurfer1: (1) adding a hydrophobicity feature; (2) adding a hydrogen bond feature; and (3) an increase of the number of maximum conformations of a ligand to generate from 20 to 50. The results were obtained for the 25 targets in the DUD set. The lowest Conformer Score (LCS) (Eq. 9) was used for PL-PatchSurfer2.
Table 1.
Comparison between PL-PatchSurfer1 and PL-PatchSurfer2. Three new features, a hydrophobicity term (HP), a hydrogen-bond term (HB) are added one by one to the original PL-PatchSurfer1. The last row, PL-PatchSurfer2 (PL-PS2) further uses up to 50 ligand conformations rather than 20, which was originally used in PL-PatchSurfer1. The p-values of difference between the methods are provided in Table S4 in Supporting Information.
| Method | EF1% | EF5% | EF10% | AUC | BEDROC |
|---|---|---|---|---|---|
| PL-PS1 | 10.85 | 3.65 | 2.39 | 0.476 | 0.225 |
| PL-PS1 + HP | 12.72 | 4.54 | 2.67 | 0.530 | 0.266 |
| PL-PS1 + HB | 13.24 | 4.65 | 2.79 | 0.535 | 0.276 |
| PL-PS1 + HB + HP | 13.41 | 4.68 | 2.80 | 0.537 | 0.280 |
| PL-PS2 (LCS) | 13.74 | 4.71 | 2.84 | 0.539 | 0.280 |
All the four new schemes showed improvements over PL-PatchSurfer1 in all five metrics. Adding the hydrogen-bond term was more effective than the hydrophobicity feature. Increasing the number of ligand conformations showed improvement on average (PL-PS2 over PL-PS1 + HP + HB) shows marginal improvement. In Table S4 in Supporting information, the p-values of pairwise comparisons are shown. Considering Bonferroni correction for ten comparisons among five methods, which makes the p < 0.1 level to p-value < 0.01 (= 0.1/10 comparisons), improvement by PL-PatchSurfer2 over the original PL-PatchSurfer1 was significant by all the other variations for EF5%, AUC, and BEDROC. In terms of EF1%, performance of PLPS1+HB+HP and PL-PatchSurfer2 was significantly different over PL-PathSurfer1.
Among the 25 targets, the largest improvement by PL-PatchSurfer2 was observed for RXRα in terms of BEDROC (PL-PS1: 0.029, PL-PS1 with HB: 0.291, PL-PS1 with HP: 0.291, PL-PS with HB and HP: 0.301, PL-PS2: 0.312). This is probably due to the new implementation of hydrophobicity and hydrogen bond terms in PL-PatchSurfer2, because these two are the main interactions between RXRα and its ligands (Supporting Information Figure S1).
Performance on the DUD dataset
Twenty-five targets of the DUD benchmark set were divided into two subsets, Set 1 and 2 for performing a two-fold cross validation (Table 2). Training of weights for Conformer Score (Eq. 9) using Set1 and Set2 for training yielded (ωcp, ωcg, ωca, ωcs) = (0.6, 0.4, 0.7, 0.6) and (0.4, 0.3, 0.5, 0.3), respectively. Table 2 summarizes the results of PL-PatchSurfer2 (PL-PS2) on Set 1, Set 2, and the whole set in comparison with Autdock Vina, DOCK6, and ROCS. For PL-PatchSurfer2, two scoring functions were used, the lowest Conformer Score (LCS) (Eq. 9) and the Boltzmann-weighted Ligand Score (BS) (Eq. 11). Results of individual targets are provided in Table S5 in Supporting Information.
Table 2.
Results on the DUD benchmark dataset.
| EF 1% | EF 5% | EF 10% | AUC | BEDROC | |
|---|---|---|---|---|---|
| Set 1 (12 targets) | |||||
|
| |||||
| PL-PS2 (LCS) | 12.18 (13.22) | 4.27 (4.33) | 2.66 (2.74) | 0.551 (0.541) | 0.254 (0.260) |
| PL-PS2 (BS) | 12.51 (13.93) | 4.51 (4.67) | 2.76 (2.81) | 0.573 (0.564) | 0.266 (0.277) |
| AutoDock Vina | 9.54 | 5.78 | 3.84 | 0.659 | 0.317 |
| DOCK6 | 13.10 | 4.15 | 2.50 | 0.518 | 0.253 |
| ROCS | 14.74 | 6.86 | 4.37 | 0.690 | 0.393 |
|
| |||||
| Set 2 (13 targets) | |||||
|
| |||||
| PL-PS2 (LCS) | 15.18 (15.33) | 5.12 (5.30) | 3.00 (3.04) | 0.527 (0.546) | 0.303 (0.307) |
| PL-PS2 (BS) | 16.90 (16.58) | 5.80 (5.73) | 3.39 (3.54) | 0.581 (0.599) | 0.346 (0.345) |
| AutoDock Vina | 6.42 | 4.38 | 2.95 | 0.606 | 0.238 |
| DOCK6 | 9.96 | 3.90 | 2.45 | 0.486 | 0.227 |
| ROCS | 9.01 | 4.33 | 2.75 | 0.529 | 0.246 |
|
| |||||
| All targets (25 Targets) | |||||
|
| |||||
| PL-PS2 (LCS) | 13.74 | 4.71 | 2.84 | 0.539 | 0.280 |
| PL-PS2 (BS) | 14.79 | 5.18 | 3.09 | 0.577 | 0.308 |
| AutoDock Vina | 7.92 | 5.05 | 3.37 | 0.632 | 0.276 |
| DOCK6 | 11.47 | 4.02 | 2.47 | 0.502 | 0.239 |
| ROCS | 11.76 | 5.54 | 3.52 | 0.606 | 0.316 |
For Set 1 and Set 2 results of PL-PatchSurfer2, values that were obtained when the dataset was used as the testing set are primarily shown while values obtained when the dataset was used for training are reported in parentheses. The overall set results reported for PL-PatchSurfer2 are the average of the 25 targets when they were handled as test data.
Comparing the two scoring schemes of PL-PatchSurfer2, BS showed better performance than LCS in all the metrics used. This improvement is consistent with a previous study, which used the Boltzmann weighting scheme for scoring40. The performance of BS with different parameter values, β and Nconf, is provided in Supporting Information (Table S6). We used β =1 Nconf = 50, because it gave the largest BEDROC and - EF1% among parameter combinations tested. For a fixed value of β, performance improved as Nconf increased. For a fixed value of Nconf, using the largest β value, β =100, gave the worst results except for one case (BEDROC, Nconf = 5).
Subsequently, we compared the performance of PL-PatchSurfer2 with two SBVS methods, AutoDock Vina and DOCK6 as well as a LBVS method, ROCS (The last Overall 25 target results in Table 1). In Supporting Information Table S7, statistical significance of performance difference of the methods observed in Table 1, i.e. p-values of student’s pairwise t-test for EF1%, AUC, and BEDROC, are provided. PL-PatchSurfer2 showed the best performance at EF1%, while ROCS performed best in three metrics, EF5%, EF10%, and BEDROC. Statistically speaking, PL-PatchSurfer2’s performance at EF1% was significantly better that AutoDock Vina at p < 0.1 level (which corresponds to p < 1.67E-02 when Bonferroni correction is applied, 1.67E-02 = 0.1/6 [the number of pairwise comparisons between four methods]). The distinction of PL-PatchSurfer2 is large at EF1%, and the values of all the programs became similar as the cutoff of the enrichment is increased. In terms of AUC, AutoDock Vina showed the best performance. These results indicate that PL-PatchSurfer2 has an advantage in the early detection of actives.
We further benchmarked the methods by dividing the DUD dataset based on the average dissimilarity between the active compounds in the library and the cognate ligand in the crystal structure of targets. The dissimilarity was calculated by SIMCOMP9, which compares 2D structures of compounds as graphs. With a dissimilarity threshold of 0.75, the DUD dataset was split into 12 and 13 targets, which have average dissimilarity of less than 0.75 or else, respectively. Furthermore, five targets were selected that have a dissimilarity value higher than 0.8. The box plots of EF1%, AUC, and BEDROC on the three subsets are reported in Figure 2.
Figure 2.
Performance depending on the dissimilarity between active compounds and the co-crystalized ligand of a target. Three subsets from the DUD targets were used, those which have dissimilarity less than 0.75, 0.75 or higher, or higher than 0.8. (A) EF1%, (B) AUC, and (C) BEDROC. Average is the values taken from Table 1, results of the 25 targets. Average values are marked as white stars on boxes.
Comparing the results on the three subsets (Figure 2), performance of AutoDock Vina, DOCK6, and ROCS declined sharply as the dissimilarity of active compounds increased to 0.8 to 1.0 in all three metrics. Drop of performance of ROCS was expected, because LBVS programs identify compounds similar to the template (query) active compound50. In contrast, notably the performance of PL-PatchSurfer2 did not deteriorate; rather, showed even better results as the dissimilarity increases on these datasets. The stable performance of PL-PatchSurfer2 would be due to its local surface matching between a target pocket and a ligand, which is more tolerant to global shape differences than other virtual screening programs.
An example where PL-PatchSurfer2 showed better performance than the other three methods is shown in Figure 3. PL-PatchSurfer2 demonstrated higher earlier active compound detection rate than the other three methods for this target, HIVPR (Figure 3A), which also reflected to larger BEDROC scores: BEDROC values of PL-PatchSurfer2 (LCS), PL-PatchSurfer2 (BS), AutoDock Vina, DOCK6, and ROCS are 0.503, 0.559, 0.286, 0.121, and 0.064, respectively. The three active compounds shown in Figure 3C are highly dissimilar to the target’s template compounds (Figure 3B), and thus AutoDock Vina, DOCK6, and ROCS could not retrieve them at an earlier rank. ZINC03815630 was ranked by AutoDock Vina, DOCK6, and ROCS at very low ranks, 152nd, 1535th, and 614th, respectively. Similarly, ZINC03833855 was ranked at 1587th, 1549th, 1547th and ZINC03815642 were identified at 405th, 290th, 525th by AutoDock Vina, DOCK6, and ROCS, respectively. In contrast, unlike the three methods, PL-PatchSurfer2 ranked them very high: ZINC03815630: 3rd by LCS, 29th by BS; ZINC03833855: 8th by LCS, 4th by BS, ZINC03815642: 54th by LCS, 28th by BS.
Figure 3.
Screening results for HIVPR. (A) ROC curves of the five methods. (B) The structure of the cognate ligand of the target (PDB ID: 1HPX). (C) Structures of three active compounds, ZINC03815630, ZINC03833855, and ZINC03815642. Dissimilarities of these three compounds from the template ligand are 0.805, 0.872, and 0.852, respectively.
Benchmark results on the apo structure dataset
Considering protein structure flexibility is important for protein-ligand docking, not only for predicting binding poses51, but also for virtual screening52. Since a binding pocket changes its conformation upon docking with a ligand, it is more difficult in general to identify active compounds if a target is in its apo form. To examine how receptor structure change affects the virtual screening performance, in this section we benchmarked using 19 apo target structures. The result is summarized in Table 3 and the detailed results are shown in Supporting Information (Table S8). The p-values of pairwise method comparison results are shown in Supporting Information Table S9.
Table 3.
Virtual screening benchmark results on the apo structure dataset (19 targets). The results of the same targets in their holo form are also shown for comparison. The numbers in parentheses are averages of three apo form targets whose binding site has a larger than 2.0 Å Cα-RMSD from its holo form.
| EF 1% | EF 5% | EF 10% | AUC | BEDROC | |
|---|---|---|---|---|---|
| Apo Structures | |||||
|
| |||||
| PL-PS2 (LCS) | 10.51 (14.11) | 3.98 (5.26) | 2.58 (3.60) | 0.532 (0.596) | 0.245 (0.340) |
| PL-PS2 (BS) | 11.67 (17.78) | 4.42 (6.55) | 2.84 (4.18) | 0.559 (0.654) | 0.267 (0.399) |
| AutoDock Vina | 3.86 (1.33) | 2.55 (3.41) | 2.00 (2.67) | 0.549 (0.604) | 0.155 (0.180) |
| DOCK6 | 7.69 (1.67) | 3.58 (1.60) | 2.48 (1.32) | 0.511 (0.402) | 0.212 (0.103) |
|
| |||||
| Holo Structures | |||||
|
| |||||
| PL-PS2 (LCS) | 12.90 (19.67) | 4.74 (7.13) | 2.88 (4.20) | 0.557 (0.642) | 0.278 (0.419) |
| PL-PS2 (BS) | 14.00 (19.22) | 5.07 (7.74) | 3.02 (4.89) | 0.577 (0.692) | 0.299 (0.451) |
| AutoDock Vina | 7.35 (10.78) | 4.95 (8.22) | 3.39 (5.11) | 0.653 (0.780) | 0.272 (0.417) |
| DOCK6 | 12.49 (18.44) | 4.60 (6.19) | 2.86 (3.84) | 0.548 (0.516) | 0.270 (0.367) |
As expected, the performance of all the methods deteriorated for the apo structures from the holo structure results. However, remarkably, PL-PatchSurfer2 did not deteriorate largely when compared with the other two conventional SBVS programs. For example, the EF1% values of AutoDock Vina and DOCK6 decreased over 40% (AutoDock Vina: from 7.35 to 3.86, DOCK6: from 12.49 to 7.69), while PL-PatchSurfer2 with the two scoring schemes dropped only about 20% (LCS: from 12.90 to 10.51, BS: from 14.00 to 11.67). In terms of EF1%, PL-PatchSurfer2 is significantly better than the other two programs at p < 0.1 level, even Bonferroni correction is applied (p-value becomes 3.33E-02 = 0.1/3 when the correction was applied for comparing three comparisons). For AUC, the three programs are distinguishable, but for BEDROC, PL-PatchSurfer2 is significantly better than AutoDock Vina (Table S9). The difference of EF1%, BEDROC, and AUC observed for holo and apo form of the individual targets are shown in Figure S2 in Supporting Information. Overall, performance difference between apo and holo form tends to increase as the binding site RMSD between the two forms becomes larger for AutoDock Vina and DOCK6. In contrast, PL-PatchSurfer2 is more tolerant to structural change of the binding pockets. When the three methods were compared, PL-PatchSurfer2 clearly shows highest performance for all the metrics.
In Table 3, we also reported results on three targets that underwent a large conformation change at the binding sites from holo to apo forms by more than 2 Å Cα-RMSD (RXRα, HIVPR, and SRC). The average values of the three proteins are shown in the parentheses in Table 3. The EF1% of AutoDock Vina and DOCK6 decreased by more than 90% (AutoDock Vina: from 10.78 to 1.33; DOCK6: from 18.44 to 1.67) and by over 50% for BEDROC (AutoDock Vina: from 0.417 to 0.180, DOCK6: from 0.367 to 0.103). In contrast, PL-PatchSurfer2 with the LCS and BS scoring schemes retained over 70% and 90% of the performance, respectively.
Figure 4 illustrates a typical situation that highlights the performance difference between PL-PatchSurfer2 and the other two methods on the apo form of a target binding site. It shows SRC kinase, which exhibits a large conformational change of 2.14 Å Cα-RMSD between the two forms. This large conformational change takes place in two loops, the glycine-rich flap and the activation loop, in the same way as other kinases52,53. When the two structures is superimposed, the cognate ligand in the holo structure causes steric crashes with the glycine rich flap of the apo structure (Figure 4A). This makes it difficult for AutoDock Vina and DOCK6 to find active compounds with the target’s apo form. A substantial drop of EF1% and BEDROC were observed for AutoDock Vina and DOCK6 on this target between the holo and the apo form: EF1% dropped from 3.33 to 0.0 (−3.33) and 23.33 to 5.00 (−18.33) for AutoDock Vina and DOCK6, respectively, and BEDROC dropped from 0.217 to 0.069 (−0.149) and 0.432 to 0.196 (−0.236) for AutoDock Vina and DOCK6, respectively. In contrast, PL-PatchSurfer2 did not exhibit much performance deterioration, and in fact often somehow exhibited a better performance for the apo form. With the LCS/BS scoring scheme, EF1% of the holo and the apo form were 25.00/21.67 and 23.33/23.33, respectively. BEDROC values were 0.441/0.437 and 0.447/0.440 (LCS/BS) for the holo and the apo form, respectively.
Figure 4.
(A) Structure superimposition of SRC holo (gold) and apo (cyan) structures. The cognate ligand from the holo structure is colored in pink. (B) The top-scored conformation of an active compound, ZINC03815530, by PL-PatchSurfer2 with the LCS scoring scheme. The two upper panels are from the holo results, while the two lower panels are from the apo results. Matching local regions between the compound and the binding pocket in the receptor are indicated in the same color. (C) The predicted binding pose by AutoDock Vina on the holo (left panel, the predicted pose is in green) and the apo (right panel, the predicted pose is in magenta) structures. The compound in pink is the cognate ligand of the holo structure. (D) The predicted binding pose by DOCK6 on the holo (left) and the apo (right) structures. The color codes are the same as in C.
Figure 4B, C, D show the docking poses of an active compound, ZINC03815530, predicted by the three methods for the holo and apo forms. This compound was ranked within top 5% by the three methods when the holo structure was used: it was ranked at 70th, 92nd, 83rd, and 40th by PL-PatchSurfer2 (LCS), PL-PatchSurfer2 (BS), AutoDock Vina, and DOCK6, respectively. However, when the apo structure was used, the rank changed to 98th, 48th, 1692nd, and 1040th, for each method in the same order, showing deterioration with AutoDock Vina, and DOCK6 in contrast to PL-PatchSurfer2. PL-PatchSurfer2 found similar compatible surface patch pairs between the compound and the binding site in the holo (the upper panels) and the apo form (the lower panels) despite a large conformational change between them (Figure 4B). In the cases of AutoDock Vina and DOCK6, although the predicted docking poses for the holo form binding site were reasonably good (the left panels in Figure 4C and 4D), predicted poses for the apo form were toward the outside of the binding pocket due to the conformation change in the glycine rich flap, which blocked the compound from being placed inside of the binding pocket (the right panels in Figure 4C and 4D).
Results on the template-based structure model dataset
Next, we examined performance of the SBVS methods on template-based models of target proteins. Using computational models for target proteins can substantially enlarge application of SBVS methods, particularly for receptors whose structures are difficult to solve by experimental means42–44, such as G-protein-coupled receptors54. However, using models is a challenge for SBVS methods because models are not perfectly accurate43. As shown in Table S2 in Supporting Information, 18 DUD targets that have appropriate template structures available were modelled using Modeller9v1147. The VS results are shown in Table 4 and Supplemental Information Table S10 gives results for individual targets. Statistical significance of performance difference of the three methods is provided in Table S11.
Table 4.
Virtual screening benchmark on the template-based models of 18 DUD targets. The results for the holo form of the same targets are shown in the bottom half of the table. When a target has multiple templates in Table S2, the template with the lowest sequence identity to the target was used for modeling. The numbers in parentheses are the averages for models of the nine targets, which were built using a template with 30% sequence identity.
| EF 1% | EF 5% | EF 10% | AUC | BEDROC | |
|---|---|---|---|---|---|
| Template-based models | |||||
|
| |||||
| PL-PS2 (LCS) | 11.48 (9.16) | 4.40 (4.56) | 2.70 (2.95) | 0.540 (0.610) | 0.255 (0.255) |
| PL-PS2 (BS) | 12.02 (10.69) | 4.72 (5.11) | 2.90 (3.28) | 0.563 (0.642) | 0.280 (0.297) |
| AutoDock Vina | 1.86 (1.68) | 1.39 (1.30) | 1.21 (1.30) | 0.478 (0.556) | 0.089 (0.092) |
| DOCK6 | 6.91 (2.58) | 3.07 (1.88) | 2.06 (1.58) | 0.484 (0.494) | 0.180 (0.099) |
|
| |||||
| Holo structures | |||||
|
| |||||
| PL-PS2 (LCS) | 12.51 (10.91) | 4.70 (4.78) | 2.85 (3.10) | 0.554 (0.606) | 0.274 (0.278) |
| PL-PS2 (BS) | 13.67 (11.97) | 4.96 (5.24) | 2.97 (3.29) | 0.573 (0.634) | 0.295 (0.306) |
| AutoDock Vina | 7.34 (8.63) | 5.00 (6.14) | 3.42 (4.09) | 0.654 (0.730) | 0.273 (0.331) |
| DOCK6 | 12.90 (11.70) | 4.69 (4.40) | 2.92 (2.98) | 0.554 (0.569) | 0.277 (0.266) |
As observed for the previous datasets, the BS scheme showed higher values than the LCS scheme in all the metrics, and both scoring methods of PL-PatchSurfer2 outperformed AutoDock Vina and DOCK6. Comparing with the performance on the holo structures, PL-PatchSurfer2 retained about 90% of its performance on computational models in all the metrics, while the other two programs decreased their performance by more than 40%. The better performance of PL-PatchSurfer2 against AutoDock Vina is statistically significant (Supporting Information Table S11) at p < 0.05 level (p < 0.05 becomes 1.67E-02 when Bonferroni correction is applied) for EF1%, AUC, and BEDROC, and significant for EF1% and BEDROC against DOCK6.
To further understand how the quality of structure models influences the virtual screening performance of each program, we constructed three models of different qualities for each of the six target proteins using templates with a sequence identity to the target in the range of 70–90%, 50–70%, and 30–50%. The result is presented in Figure 5 and Table S12.
Figure 5.
Box plots of virtual screening results for template-based models constructed with templates with varying sequence identity to the targets. Average over six targets are shown. (A) EF1%, (B) EF5%, (C) EF10%, (D) AUC, and (E) BEDROC. For comparison, results on the holo structure of the targets are also shown (X-ray, on the left). On each box, the mean values is shown with a white star.
Overall, the performance of all the methods deteriorates as more distant templates were used for target structure modeling. The methods showed relatively stable performance in AUC (Figure 5D). The severity of the performance decline differs between the methods. Compared to AutoDock Vina and Dock6, which exhibited performance drop, results from PL-PatchSurfer2 did not decline much, demonstrating more tolerance to inaccuracy of structure models of targets.
In Figure 6, we further examined the performance of PL-PatchSurfer2 on template-based models from a different angle, relative to the RMSD of the models. 30 template-based models built with template structures in Table S2 were used. RMSD of models (x-axis) and the drop of the performance (y-axis) do not show clear correlation, particularly in terms of EF1% (Figure 6A). PL-PatchSurfer2 did not deteriorate its performance in terms of EF1% for substantial number (16) of models, which include one with an RMSD of 3.73 Å for which PL-PatchSurfer2 even somehow worked better than the holo form of the target. If we consider that a 10% decrease in EF1%, AUC, or BEDROC is acceptable by using template-based models, 75% of models (six out of eight) up to an RMSD of 1.5 Å PL-PatchSurfer2’s performance was acceptable. If up to 20% decrease of the performance is acceptable, actually, overall 76.7%, 90.0%, and 90.0% (23, 27, and 27 out of 30) of template models were within the decrease range in terms of EF1%, AUC, and BEDROC, respectively.
Figure 6.
Performance difference between holo and template-based target models. (A) EF1%, (B) AUC, and (C) BEDROC difference between holo and template-based models (model - holo)/holo. The x-axis is backbone RMSD between models and the target in its holo form. PL-PatchSurfer2 (BS) was used. The x-axis is binding site Cα RMSD.
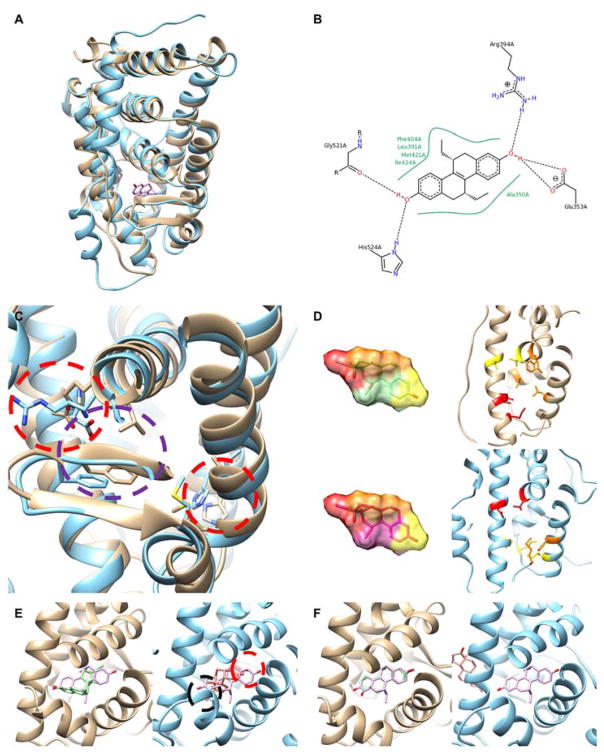
Figure 7 is an example of docking to template-based models, a model of ERα with its agonists. The template-based model shown was built based on a template (PDB ID: 1Z5X) that has a sequence identity of 30% to the target (PDB ID: 1L2I) and has a backbone RMSD of 2.69 Å. Superimposition shows that a long loop (at the left bottom) which is a part of the ligand binding site55 is misplaced in the model (Figure 7A). The cognate ligand is structurally symmetrical, and its interaction with the binding pocket is mainly governed by hydrophobic interaction at the middle and hydrogen bonds at the both termini of the molecule (Figure 7B).
Figure 7.
Example of ligand docking to a template-based model. (A) Structure superimposition of ER alpha holo (gold) and template-based model (cyan). The cognate ligand from the holo structure is shown in pink. (B) The cognate ligand, (R,R)-5,11-cis-Diethyl-5,6,11,12-tetrahydrochrysene-2,8-diol (PDB ID: ETC) in the holo structure. This diagram was drawn with PoseView56. (C) The side-chains of the binding site of ER alpha. Two regions that form hydrogen bonds with the cognate ligand are marked by red circles and a region for hydrophobic interaction is marked by a purple circle. (D) The top-scoring conformations of ZINC01999257 by the PL-PatchSurfer2 LCS scoring scheme. The two upper panels are results using the holo structure, while the lower panel are results using a template-based model of the target. Local regions that matched between the compound and the binding pocket are indicated in the same colors. (E) The predicted binding pose of ZINC01686128 by AutoDock Vina in the holo (left, the compound in the predicted pose is in green) and a template-based model (right, the predicted pose is magenta). The compound forms hydrogen bonds at one end (black circle) but no bonds were formed on the other end (red circle). (F) The predicted binding pose of ZINC03815416 by DOCK6 on the holo structure (left) and a model (right). The colors of the compounds are the same as in E.
The side-chain conformations of the residues at the binding site differs in the model relative to the holo form, particularly the residues that participate in forming hydrogen bonds (red circles, Figure 7C). Using the model inevitably decreased the VS methods’ performance due to its structural difference to the holo form. With PL-PatchSurfer2, EF1% decreased from 9.0/15.0 to 7.5/13.5 (using LCS/BS scheme) by using the model instead of the holo form target, AutoDock Vina from 13.5 to 4.5, and DOCK6 reduced its EF1% from10.5 to 3.0.
The latter three panels (Figure 7D, E, F) explain what happened to predicted binding modes of top-ranked active compounds when the template-based model was used. ZINC01999257 was ranked 2nd (out of 2010 compounds) in both scoring schemes of PL-PatchSurfer2 with the holo form of the target and 1st (LCS) and 3rd (BS) with the model despite the conformational difference of the model to the holo form (Figure 7D). Close inspection found that the compound docks to the pocket in the opposite direction when the model was used (lower panels in Figure 7D) relative to the pose when the holo form was used (upper panels); however, because this active compound is almost symmetric, both termini of the compound can form hydrogen-bonds in both orientations between terminal hydroxyl groups and residues in the pocket (patches in red). Also hydrophobic interaction in the middle of the compound (the patch in orange) can clearly be formed in either orientation. In contrast to PL-PatchSurfer2, AutoDock Vina and DOCK6 suffer from the structural difference of the model. ZINC01686128, which was ranked 5th by AutoDock Vina on the holo structure, was ranked 115th on the model, because one terminal of the compound could not form hydrogen bonds (Figure 7E). As for DOCK6, ZINC03615416 was the top-scoring active compound when the holo structure was used. However, with the model it was ranked very low, 1393th, because a favorable pose was not found within the pocket and the ligand was placed outside of the binding site (Figure 7F).
Results on the WOMBAT dataset
Finally, we tested the four VS methods on the WOMBAT dataset. The WOMBAT dataset has more diverse active compounds than DUD, addressing observations that VS benchmark result is influenced by the diversity of targets and ligands in a dataset32,57,58. The results are summarized in Table 5 and results for individual targets are given in Supporting Information Table S13. Pairwise p-values are provided in Supporting Information Table S14.
Table 5.
Virtual screening results on the WOMBAT dataset.
| EF1% | EF5% | EF10% | AUC | BEDROC | |
|---|---|---|---|---|---|
| PL-PS2 (LCS) | 6.41 | 2.70 | 1.60 | 0.313 | 0.177 |
| PL-PS2 (BS) | 6.47 | 2.54 | 1.61 | 0.329 | 0.226 |
| AutoDock Vina | 5.37 | 2.88 | 2.22 | 0.558 | 0.197 |
| DOCK6 | 5.56 | 2.39 | 1.63 | 0.419 | 0.155 |
| ROCS | 4.70 | 2.13 | 1.57 | 0.434 | 0.147 |
On this dataset, PL-PatchSurfer2 (BS) again showed the highest performance among the three SBVS methods in terms of EF1% and BEDROC. The performances of the methods in all metrics were worse than DUD, although it is difficult to make direct comparison because the compound sets are different from DUD. Interestingly, ROCS performed worst in all metrics except for AUC even though it performed best on the DUD benchmark. This may be reflecting the fact that active compounds in WOMBAT are on average more dissimilar to the cognate ligand than DUD (the average dissimilarity is 0.645 in DUD and 0.757 in WOMBAT), which in general makes it more difficult to find actives from a cognate ligand using LBVS methods13,59. It needs to be mentioned that, however, these differences are not statistically significant: After applying Bonferroni correction, no programs are distinguishable at p < 0.1 level, in terms of EF1%. For AUC, AutoDock Vina is statistically significant at p < 0.1, and for BEDROC, only AutoDock Vina and ROCS are distinguishable with each other.
Computational Time
Table 6 reports computational time for the four methods. It is the average time taken to screen the whole compound library of over the 25 DUD targets. PL-PatchSurfer2 is 22 times and 18 times faster than Autodock Vina and DOCK6, respectively, and on the same scale with ROCS, an LBVS program.
Table 6.
Average computational time for screening the DUD dataset.
| Program | Computation Time (hours) |
|---|---|
| PL-PatchSurfer2 | 2.64 |
| AutoDock Vina | 58.81 |
| DOCK6 | 48.03 |
| ROCS | 1.38 |
The values do not include the time for computing compound conformations, patches of a target and compounds. All the computations were processed on a Linux machine with Intel i7–3820 3.60 GHz CPU and 64 GB RAM.
CONCLUSION
We introduced and benchmarked PL-PatchSurfer2, which evaluates protein-ligand interactions based on compatibility of local molecular surfaces. A molecular surface is divided into a number of overlapping patches, which are described by four physicochemical properties, shape, electrostatic potential, hydrophobicity, and hydrogen bonding. The features are further converted to 3DZD, a 72-dimensional vector. Representing and comparing the features by 3DZD has two technical advantages: 1) the descriptor is rotationally invariant and 2) comparison can be fast because Euclidean distance comparison of vectors is fast12,20,23–28.
The thorough benchmark study performed in this work has indicated that PL-PatchSurfer2 performs better in general than the two existing SBVS methods, AutoDock Vina and DOCK6, and particularly outperforms the two methods when apo form or template-based models of targets were used. ROCS, an LBVS method, showed comparable performance to PL-PatchSurfer2 on DUD, but PL-PatchSurfer2 consistently showed a higher value than ROCS on all the metrics when applied to the WOMBAT dataset, which has more diverse active compounds. These results indicate that the unique surface patch representation imparts an interesting strength to PL-PatchSurfer2, i.e. increased tolerance to conformation change of targets. Taking advantage of this unique strength, PL-PatchSurfer2 can substantially extend the capability of virtual screening by providing more accurate predictions in difficult cases, such as scaffold hopping or cases where only apo form of targets or template-based models of targets are available. Sufficient performance on template-based models is worth attention because it can substantially extend the target space where virtual screening is applicable. To further improve the performance, it may be effective to perform ensemble docking, where multiple conformations are used also for a target protein, in addition to ligand conformation ensemble, to cover receptor structural flexibility60,61.
Supplementary Material
Acknowledgments
Funding Sources
This work is partly supported by a grant from the Lilly Research Award Program, the National Institute of General Medical Sciences of the National Institutes of Health (R01GM097528), and the National Science Foundation (IIS1319551, DBI1262189, IOS1127027).
We thank Hyunho Chun, Purdue University, for the discussion about statistical analysis of the virtual screening results.
ABBREVIATIONS
- LBVS
ligand-based virtual screening
- SBVS
structure-based virtual screening
- 3DZD
three-dimensional Zernike descriptor
- AUC
area-under receiver operating characteristic curve
- VS
virtual screening
- BS
Boltzmann-weighted ligand score
- LCS
lowest conformer score
Footnotes
Author Contributions
WHS participated in designing the research, coded the programs, conducted the experiments, and wrote the paper. CC participated in coding the programs. JW participated in designing the research and in interpreting the results. D.K. conceived the study, participated in its design and coordination and wrote the paper. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Results of training and testing of weights in Eq. 5; The targets in the DUD dataset with their apo structures and templates used for building their computational models; Targets from the WOMBAT dataset used in this study; Pairwise p-values for comparison between PL-PatchSurfer1 and PL-PatchSurfer2; Results for individual targets in the DUD benchmark dataset; Performance of the Boltzmann-weighted ligand score with different combinations of parameters; Pairwise p-values from student’s t-test for the performance on the DUD dataset; Virtual screening results for individual targets in the apo structure dataset; Pairwise p-values from student’s t-test for the apo form targets; Virtual screening results for individual targets in the template-based model dataset; Pairwise p-values for the template-based model dataset; Virtual screening results of individual template-based models that were constructed with templates of two different range of sequence identity; Virtual screening results for individual targets in the WOMBAT dataset; Pairwise p-values for the WOMBAT set; Interaction pattern between RXRα and its cognate ligand; and Performance difference to holo and apo form targets.
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Walters WP, Stahl MT, Murcko MA. Virtual Screening—An Overview. Drug Discov Today. 1998;3:160–178. [Google Scholar]
- 2.Vidović D, Busby SA, Griffin PR, Schürer SC. A Combined Ligand- and Structure-Based Virtual Screening Protocol Identifies Submicromolar PPARγ Partial Agonists. ChemMedChem. 2011:94–103. doi: 10.1002/cmdc.201000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langmead CJ, Andrews SP, Congreve M, Errey JC, Hurrell E, Marshall FH, Mason JS, Richardson CM, Robertson N, Zhukov A, Weir M. Identification of Novel Adenosine A2A Receptor Antagonists by Virtual Screening. J Med Chem. 2012;55:1904–1909. doi: 10.1021/jm201455y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fazi R, Tintori C, Brai A, Botta L, Selvaraj M, Garbelli A, Maga G, Botta M. Homology Model-Based Virtual Screening for the Identification of Human Helicase DDX3 Inhibitors. J Chem Info Model. 2015;55:2443–2454. doi: 10.1021/acs.jcim.5b00419. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz J, Awale M, Reymond JL. SMIfp (SMILES fingerprint) Chemical Space for Virtual Screening and Visualization of Large Databases of Organic Molecules. J Chem Info Model. 2013;53:1979–1989. doi: 10.1021/ci400206h. [DOI] [PubMed] [Google Scholar]
- 6.Durant JL, Leland BA, Henry DR, Nourse JG. Reoptimization of MDL Keys for Use in Drug Discovery. J Chem Inf Comput Sci. 2002;42:1273–1280. doi: 10.1021/ci010132r. [DOI] [PubMed] [Google Scholar]
- 7.Raymond JW, Gardiner EJ, Willett P. RASCAL: Calculation of Graph Similarity using Maximum Common Edge Subgraphs. Comput J. 2002;45:631–644. doi: 10.1021/ci010381f. [DOI] [PubMed] [Google Scholar]
- 8.Bender A, Mussa HY, Glen RC. Similarity Searching of Chemical Databases Using Atom Environment Descriptors (MOLPRINT 2D): Evaluation of Performance. JChem Inf Comput Sci. 2004;44:1708–1718. doi: 10.1021/ci0498719. [DOI] [PubMed] [Google Scholar]
- 9.Hattori M, Okuno Y, Goto S, Kanehisa M. Development of a Chemical Structure Comparison Method for Integrated Analysis of Chemical and Genomic Information in the Metabolic Pathways. J Am Chem Soc. 2003;125:11853–11865. doi: 10.1021/ja036030u. [DOI] [PubMed] [Google Scholar]
- 10.Ballester PJ, Richards WG. Ultrafast Shape Recognition to Search Compound Databases for Similar Molecular Shapes. J Comput Chem. 2007;28:1711–1723. doi: 10.1002/jcc.20681. [DOI] [PubMed] [Google Scholar]
- 11.Hawkins PCD, Skillman AG, Nicholls A. Comparison of Shape-Matching and Docking as Virtual Screening Tools. J Med Chem. 2007;50:74–82. doi: 10.1021/jm0603365. [DOI] [PubMed] [Google Scholar]
- 12.Shin WH, Zhu X, Bures MG, Kihara D. Three-Dimensional Compound Comparison Methods and Their Application in Drug Discovery. Molecules. 2015;20:12841–12962. doi: 10.3390/molecules200712841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Korff M, Freyss J, Sander T. Comparison of Ligand- and Structure-Based Virtual Screening on the DUD Data Set. J Chem Info Model. 2009;49:209–231. doi: 10.1021/ci800303k. [DOI] [PubMed] [Google Scholar]
- 14.Shin WH, Kim JK, Kim DS, Seok C. GalaxyDock2: Protein–Ligand Docking Using Beta-Complex and Global Optimization. J Comput Chem. 2013;34:2647–2656. doi: 10.1002/jcc.23438. [DOI] [PubMed] [Google Scholar]
- 15.Jain AN. Surflex-Dock 2.1: Robust Performance from Ligand Energetic Modeling, Ring Flexibility, and Knowledge-Based Search. J Comput-Aided Mol Des. 2007;21:281–306. doi: 10.1007/s10822-007-9114-2. [DOI] [PubMed] [Google Scholar]
- 16.Trott O, Olson AJ. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen WJ, Balius TE, Mukherjee S, Brozell SR, Moustakus DT, Lang PT, Case DA, Kuntz ID, Rizzo RC. DOCK 6: Impact of New Features and Current Docking Performance. J Comput Chem. 2015;36:1132–1156. doi: 10.1002/jcc.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leach AR, Gillet VJ, Lewis RA, Taylor R. Three-Dimensional Pharmacophore Methods in Drug Discovery. J Med Chem. 2010;53:539–558. doi: 10.1021/jm900817u. [DOI] [PubMed] [Google Scholar]
- 19.Wolber G, Langer T. LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. J Chem Info Model. 2005;45:160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 20.Hu B, Zhu X, Monloe L, Bures MG, Kihara D. PL-PatchSurfer: A Novel Molecular Local Surface-Based Method for Exploring Protein-Ligand Interactions. Int J Mol Sci. 2014;15:15122–15145. doi: 10.3390/ijms150915122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canterakis N. 3D Zernike Moments and Zernike Affine Invariants for 3D Image Analysis and Recognition. Proceedings of 11th Scandinavian Conference on Image Analysis; 1999. pp. 85–93. [Google Scholar]
- 22.Novotni M, Klein R. 3D Zernike Descriptors for Content Based Shape Retrieval. Proceedings of eighth ACM symposium on Solid modeling and applications; 2003. pp. 216–225. [Google Scholar]
- 23.Chikhi R, Sael L, Kihara D. Real-Time Ligand Binding Pocket Database Search Using Local Surface Descriptors. Proteins. 2010;78:1007–2028. doi: 10.1002/prot.22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sael L, Kihara D. Binding Ligand Prediction for Proteins Using Partial Matching of Local Surface Patches. Int J Mol Sci. 2010;11:5009–5026. doi: 10.3390/ijms11125009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sael L, Kihara D. Detecting Local Ligand-Binding Site Similarity in Nonhomologous Proteins by Surface Patch Comparison. Proteins. 2012;80:1177–1185. doi: 10.1002/prot.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu X, Xiong Y, Kihara D. Large-Scale Binding Ligand Prediction by Improved Patch-Based Method Patch-Surfer2. 0. Bioinformatics. 2015;31:707–713. doi: 10.1093/bioinformatics/btu724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esquivel-Rodriguez J, Xiong Y, Han X, Gang S, Christoffer C, Kihara D. Navigating 3D Electron Microscopy Maps with EM-SURFER. BMC Bioinformatics. 2015;16:181. doi: 10.1186/s12859-015-0580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong Y, Esquivel-Rodriguez J, Sael L, Kihara D. 3D-SURFER 2.0. Web Platform for Real-Time Search and Characterization of Protein Surfaces. Methods in Molecular Biology. 2014;1137:105–117. doi: 10.1007/978-1-4939-0366-5_8. [DOI] [PubMed] [Google Scholar]
- 29.Truchon JF, Bayly CI. Evaluating Virtual Screening Methods: Good and Bad Metrics for the “Early Recognition” Problem. J Chem Info Model. 2007;47:488–508. doi: 10.1021/ci600426e. [DOI] [PubMed] [Google Scholar]
- 30.Zhao W, Hevener KE, White SW, Lee RE, Boyett JM. A Statistical Framework to Evaluate Virtual Screening. BMC Bioinformatics. 2009;10:255. doi: 10.1186/1471-2105-10-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang N, Shoichet BK, Irwin JJ. Benchmarking Sets for Molecular Docking. J Med Chem. 2006;49:6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Good AC, Oprea TI. Optimization of CAMD Techniques 3. Virtual Screening Enrichment Studies: A Help or Hindrance in Tool Selection? J Comput-Aided Mol Des. 2008;22:169–178. doi: 10.1007/s10822-007-9167-2. [DOI] [PubMed] [Google Scholar]
- 33.Hawkins PCD, Skillman AG, Warren GL, Ellingson BA, Stahl MT. Conformer Generation with OMEGA: Algorithm and Validation Using High Quality Structures from the Protein Databank and Cambridge Structural Database. J Chem Info Model. 2010;50:572–584. doi: 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kellogg GE, Semus SF, Abraham DJ. HINT: A New Method of Empirical Hydrophobic Field Calculation for CoMFA. J Comput–Aided Mol Des. 1991;5:545–552. doi: 10.1007/BF00135313. [DOI] [PubMed] [Google Scholar]
- 36.Heiden W, Moeckel G, Brickmann J. A New Approach to Analysis and Display of Local Lipophilicity/Hydrophilicity Mapped on Molecular Surfaces. J Comput-Aided Mol Des. 1993;7:503–514. doi: 10.1007/BF00124359. [DOI] [PubMed] [Google Scholar]
- 37.Cheng T, Zhao Y, Li X, Lin F, Xu Y, Zhang X, Li Y, Wang R. Computation of Octanol–Water Partition Coefficients by Guiding an Additive Model with Knowledge. J Chem Info Model. 2007;47:2140–2148. doi: 10.1021/ci700257y. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Liu Z, Li Z, Han L, Liu Z, Zhao Z, Wang L. Comparative Assessment of Scoring Functions on an Updated Benchmark: 1. Compilation of the Test Set. J Chem Info Model. 2014;54:1700–1716. doi: 10.1021/ci500080q. [DOI] [PubMed] [Google Scholar]
- 39.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WTM, Mortenson PN, Murray CW. Diverse, High-Quality Test Set for the Validation of Protein–Ligand Docking Performance. J Chem Info Model. 2007;50:726–740. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 40.Paulsen JL, Anderson AC. Scoring Ensembles of Docked Protein:Ligand Interactions for Virtual Lead Optimization. J Chem Info Model. 2009;49:2813–2819. doi: 10.1021/ci9003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vosmeer CR, Pool R, van Stee MF, Perić-Hassler L, Vermeulen NPE, Geerke DP. Towards Automated Binding Affinity Prediction Using an Iterative Linear Interaction Energy Approach. Int J Mol Sci. 2014;15:798–816. doi: 10.3390/ijms15010798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan H, Irwin JJ, Webb BM, Klebe G, Shoichet BK, Sali A. Molecular Docking Screens Using Comparative Models of Proteins. J Chem Info Model. 2009;49:2512–2527. doi: 10.1021/ci9003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bordogna A, Pandini A, Bonati L. Predicting the Accuracy of Protein–Ligand Docking on Homology Models. J Comput Chem. 2011;32:81–98. doi: 10.1002/jcc.21601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cavasotto CN, Phatak SS. Homology Modeling in Drug Discovery: Current Trends and Applications. Drug Discov Today. 2009;14:676–683. doi: 10.1016/j.drudis.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Databank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Šali A, Blundell TL. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Skolnick J. TM-align: A Protein Structure Alignment Algorithm Based on the TM-Score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 50.Geppert H, Vogt M, Bajorath J. Current Trends in Ligand-Based Virtual Screening: Molecular Representations, Data Mining Methods, New Application Areas, and Performance Evaluation. J Chem Info Model. 2010;50:205–216. doi: 10.1021/ci900419k. [DOI] [PubMed] [Google Scholar]
- 51.Jain AN. Effects of Protein Conformation in Docking: Improved Pose Prediction through Protein Pocket Adaptation. J Comput-Aided Mol Des. 2009;23:355–374. doi: 10.1007/s10822-009-9266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cavasotto CN, Abagyan R. Protein Flexibility in Ligand Docking and Virtual Screening to Protein Kinases. J Mol Biol. 2004;337:209–225. doi: 10.1016/j.jmb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 53.Xu M, Yu L, Wan B, Yu L, Huang Q. Predicting Inactive Conformations of Protein Kinases Using Active Structures: Conformational Selection of Type-II Inhibitors. PLoS One. 2011;6:e22644. doi: 10.1371/journal.pone.0022644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kufareva I, Katritch V, Stevens RC, Abagyan R Participants of. GPCR Dock 2013. Advances in GPCR Modeling Evaluated by the GPCR Dock 2013 Assessment: Meeting New Challenges. Structure. 2014;22:1120–1139. doi: 10.1016/j.str.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engström O, Öhman L, Greene GL, Gustafsson J-Å, Carlquist M. Molecular Basis of Agonism and Antagonism in the Oestrogen Receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 56.Steirand K, Raney M. Drawing PDB: Protein-Ligand Complexes in Two Dimensions. ACS Med Chem Lett. 2010;1:540–545. doi: 10.1021/ml100164p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J Med Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cross JB, Thompson DC, Rai BK, Baber JC, Fan KY, Hu Y, Humblet C. Comparison of Several Molecular Docking Programs: Pose Prediction and Virtual Screening Accuracy. J Chem Info Model. 2009;49:1455–1474. doi: 10.1021/ci900056c. [DOI] [PubMed] [Google Scholar]
- 59.Venkatraman V, Perez-Nueno VI, Mavridis L, Ritchie DW. Comprehensive Comparison of Ligand-Based Virtual Screening Tools Against the DUD Data set Reveals Limitations of Current 3D Methods. J Chem Info Model. 2010;50:2079–2093. doi: 10.1021/ci100263p. [DOI] [PubMed] [Google Scholar]
- 60.Huang SY, Zou X. Ensemble Docking of Multiple Protein Structures: Considering Protein Structural Variations in Molecular Docking. Proteins. 2007;66:399–421. doi: 10.1002/prot.21214. [DOI] [PubMed] [Google Scholar]
- 61.Campbell AJ, Lamb ML, Joseph-McCathy D. Ensemble-Based Docking Using Biased Molecular Dynamics. J Chem Inf Model. 2014;54:2127–2138. doi: 10.1021/ci400729j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.