Abstract
Background
Identifying core microbiota is an important step for understanding the key components of microbial communities. Traditional approach that identifies core taxa at the OTU level ignores potential ecological coherence of higher rank taxa. There is a need to develop software that can systematically identify core taxa at and above the species level.
Results
Here we developed PhyloCore, an application that uses a phylogeny-based algorithm to identify core taxa at the proper taxonomic levels. It incorporates a number of features that users can set according to their needs. Using multiple gut microbiota as test cases, we demonstrate that PhyloCore is more powerful and flexible than OTU-based approaches.
Conclusions
PhyloCore is a flexible and fast application that identifies core taxa at proper taxonomic levels, making it useful to sequence-based microbial ecology studies. The software is freely available at http://wolbachia.biology.virginia.edu/WuLab/Software.html
Keywords: Core taxa, microbiota, OTU
1 Introduction
Core microbiota are defined as members shared by most microbial assemblages from similar habitats. It has been suggested that core taxa may play important roles in the function of the community (Shade and Handelsman, 2011). Thus, identifying core is a very useful step in microbial ecology studies. It provides valuable insights into what ‘healthy’ microbiota look like for a particular habitat and may also help in identifying keystone species in the community. The traditional approach used to identify core taxa is to find OTUs (operational taxonomic units) that are present in a large proportion of samples (Caporaso et al., 2011; Huse et al., 2012; Martínez et al., 2013). However, this method can only identify core taxa at the OTU level and ignores potential phylogenetic redundancy in microbial communities where multiple closely related OTUs are present.
Since closely related bacterial taxa can be ecologically interchangeable (Harvey and Pagel, 1991), it may be useful to consider phylogenetic relationships when identifying core taxa. Recent studies suggest that there is ecological coherence (members of a taxonomic group share common ecological traits that distinguish them from members of other taxonomic groups) among bacterial taxonomic ranks higher than the species (OTU) level (Lozupone and Knight, 2007; 2005; Philippot et al., 2010; 2009). These findings suggest that microbial cores might exist at taxonomic levels higher than species. Therefore, it is not surprising that traditional core identification method often fails to detect core OTUs. Higher taxonomic level cores have been identified in previous studies (Benson et al., 2010; Zhang et al., 2015). However, the cores were identified either manually or using in-house scripts that are not available to the public. There is a need to develop software that can systematically identify core taxa at and above the species level. Here we present PhyloCore, an application that uses a phylogeny-based algorithm to identify core taxa at the proper taxonomic levels.
2 Material and methods
2.1 Algorithm
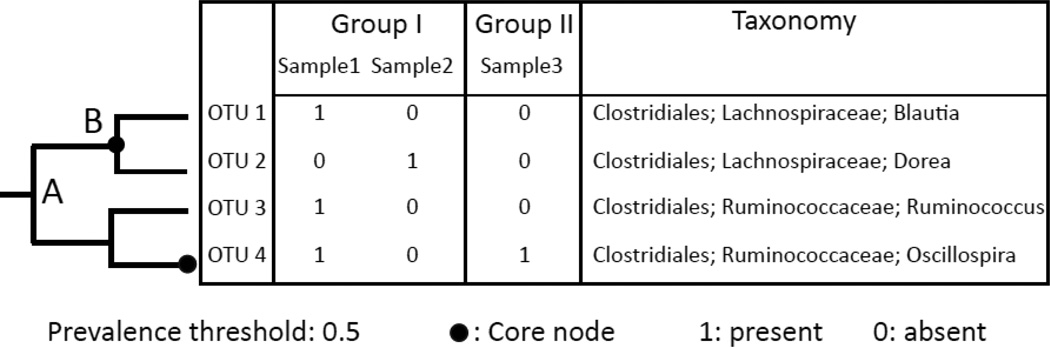
PhyloCore takes an OTU table that describes the presence/absence of each OTU in all samples and their taxonomic assignments, and optionally a phylogenetic tree of all OTUs. When a phylogenetic tree of all OTUs is provided, PhyloCore will use it to infer relationships between OTUs. In the absence of an OTU tree, PhyloCore will construct a tree using taxonomic information in the OTU table. OTU table and tree can be generated from 16S rRNA (or SSU rRNA) sequence data using a microbial ecology analysis software such as QIIME (Caporaso et al., 2010). To identify the core taxa, PhyloCore starts at the root node and traverses the whole tree in breadth-first order. For each internal node, PhyloCore calculates a prevalence value, defined as the cumulative presence of all its descendant OTUs. For example, in Figure 1 the prevalence of the internal node B is 2/3, because its two descendant OTUs (OTU 1 and OTU 2) appear in 2 out of 3 samples. For each leaf node or OTU, (e.g., OTU 4 in Fig. 1), PhyloCore calculates its prevalence value in all samples.
Figure 1.
A diagram illustrating the criteria of core node identification. A phylogenetic tree of all OTUs and an OTU table describing the presence/absence each OTU are required as input. Samples 1–3 represent microbial communities (e.g. gut microbiota of different hosts). Group I and II represent categories of microbial community (e.g. from male or female hosts). Using a prevalence threshold of 0.5, internal node B and OTU 4 are identified as core nodes.
PhyloCore initially stores any node i that has a prevalence value greater than a user supplied threshold as a core taxon. However, if PhyloCore finds a descendant of node i (e.g., node j) that also passes the threshold, then node i is replaced by node j in the stored core node list. For example, using a 0.5 threshold, node A is initially defined as a core node. However, when PhyloCore moves down the lineage and finds that node B also qualifies, node A is replaced by node B in the core node list. This will guarantee that for a particular lineage only the core at the lowest possible taxonomic level is identified, assuming that cores at lower taxonomic levels are more informative than those at higher levels in revealing the functions of the core that are important for the community.
Once a list of core nodes is identified, PhyloCore assigns taxonomy to each core node. If the core is an OTU, its taxonomy is used directly. For an internal core node, it is done by finding the lowest common taxonomy of all its descendant OTUs. Take node B for example, the lowest common taxonomy of OTU 1 and OTU 2 is family Lachnospiraceae. Therefore, Lachnospiraceae is assigned to node B.
Dataset can be imbalanced when sampling among groups is uneven. For example, in Figure 1 group I has two samples while group II has only one sample. This will result in a bias towards group I if all samples are treated equally. In this case, weighted core taxa can be identified. The prevalence of each node Pweighted will be calculated based on the formula below, giving each group (but not each sample) the same weight:
where Pi is the prevalence in i’th group, and N is the total number of groups.
2.2 Features
PhyloCore is coded in both Perl and Python. It allows user to specify:
A prevalence threshold. A node is considered a core node if its prevalence is above the threshold.
An abundance threshold. OTUs with abundances lower than the threshold in a sample will be considered absent.
A sample ID list (with or without group information). Only samples in the list will be used in core identification. The group information, if provided, will be used to identify the weighted core nodes. This feature enables users to identify cores in a subset of the population (e.g., a specific group).
3 Results and Discussion
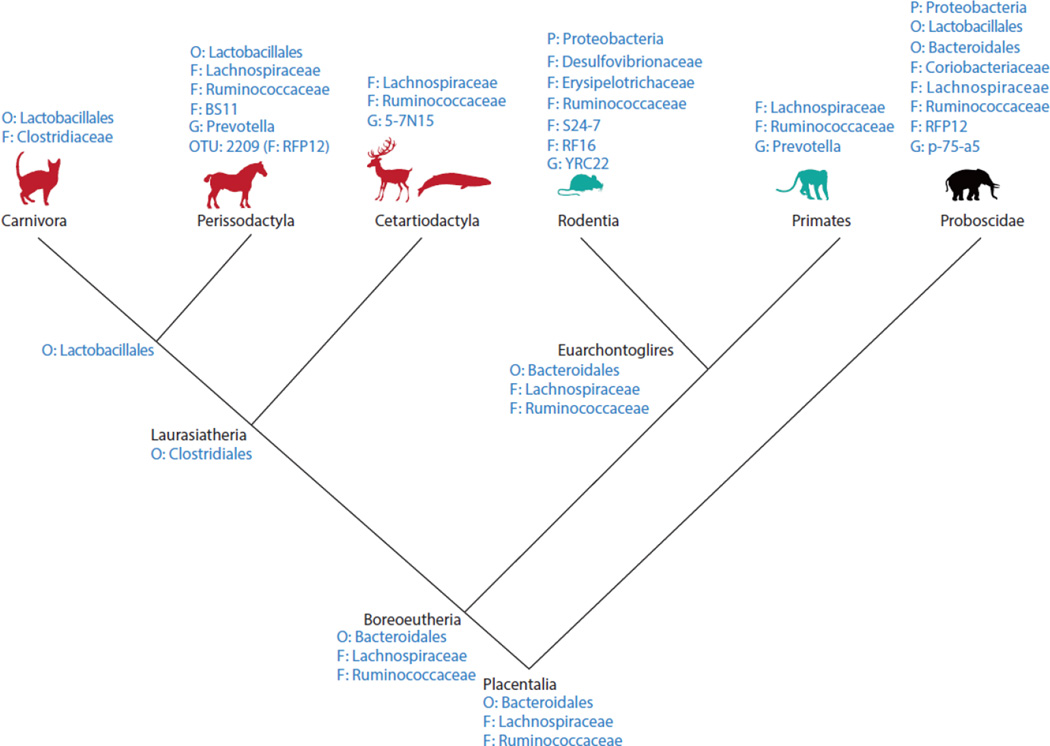
To demonstrate the use of PhyloCore, we first identified the core gut microbiota in mammals. The dataset contains 16S rRNA gene sequenced from 85 individuals belonging to 6 mammalian orders (Ley et al., 2008). Using QIIME, a 16S rRNA tree and an OTU table were generated and used as input files to PhyloCore. Prevalence threshold was set at 0.8 and samples were weighted. PhyloCore identified many core gut microbiota among different mammalian lineages (Figure 2). In comparison, traditional OTU-based core identification (as implemented in QIIME package) found only one OTU-level core (OTU 2209: family RFP12) in Perissodactyla, which was identified by PhyloCore as well. We found one order-level (Bacteroidales) and two family-level (Ruminococcaceae and Lachnospiraceae) core taxa that are shared among the mammals studied, suggesting that these taxa were likely present in the last common ancestor of mammals and are important in the codiversification of the gut microbiota and the mammalian hosts. As expected, the core hierarchy generally parallels the hierarchy of the host species phylogeny. In other words, if a microbial taxon a is a core of a host taxon b, then descendants of a are also cores of the descendants of b. For example, Order Bacteroidales is a core for Euarchontoglires. Genera YRC22 and Prevotella within Bacteroidales are cores of Rodentia and Primates, respectively (Figure 2). The parallelism in hierarchy should be perfect when the core is defined as being present in all samples (i.e., using a prevalence threshold of 1.0). Because a prevalence threshold of 0.8 was used here, the core hierarchy breaks down in some lineages. For example, Family Ruminococcaceae was identified as a core for Placentalia, but neither Ruminococcaceae nor its descendants were cores for Carnivora (Figure 2).
Figure 2.
Core microbial taxa identified in different mammalian lineages using a prevalence cutoff of 0.8. P: phylum, O: order, F: family, G: genus. The mammalian tree topology was derived from Reis et al. (2012). A 16S rRNA tree and an OTU table made with QIIME were used as input files to PhyloCore.
We also tested PhyloCore on 16S rRNA sequences from two human microbiome studies (Caporaso et al., 2011; Yatsunenko et al., 2012). The Yatsunenko et al. dataset contained 528 samples collected from healthy children and adults from Amazonas of Venezuela, rural Malawi and US metropolitan areas. The Caporaso et al. dataset encompassed 467 samples collected from two individual over 1 year period. 16S rRNA trees and OTU tables generated by QIIME were used as input files to PhyloCore. Yatsunenko et al. dataset contained 45,595 OTUs, and it took PhyloCore (Python version) 4 minutes to run on a Macbook (1.6 GHz Intel Core i5 processor and 8 GB of memory). At the 0.9 prevalence cutoff, QIIME found two OTU-level core taxa (OTU 1: Dorea and OTU 5: Blautia). Besides these two OTUs, PhyloCore identified additional core gut microbiota at higher taxonomic levels: one order (Bacteroidales), two families (Coriobacteriaceae and Veillonellaceae) and two genera (Faecalibacterium and Streptococcus). Caporaso et al. dataset contained 4,926 OTUs, and it took PhyloCore 25 seconds to run on the same computer. At the 0.9 prevalence cutoff, QIIME found 15 OTU-level core taxa (12 Bacteroides, 1 Roseburia, 1 Phascolarctobacterium and 1 unclassified Ruminococcaceae). In comparison, PhyloCore identified two more genus-level (Faecalibacterium and Blautia) core taxa in addition to the 15 core OTUs.
The confidence of core identification depends on the sample size. For example, using the same prevalence threshold, we would place more confidence in core taxa that are identified from 1,000 samples than those identified from 10 samples. It is therefore important that sufficient number of samples are included in the study. As a general guideline, the smaller the number of samples used in a study, the more stringent prevalence threshold should be applied. Ultimately, the users should decide what is a proper prevalence threshold for a given sample size. At the minimum, the users should report the sample size and the prevalence threshold used in the core analysis.
Based on the premise that ecological coherence can exist at higher taxonomic levels, we think it is useful to identify core microbiota at and above the species level. However, it is worth pointing out that by no means it implies that members of core taxa identified by PhyloCore all have the same functions, as many studies have demonstrated that closely related bacterial species do not have completely overlapping functions (Cordero et al., 2012; Youngblut et al., 2013). Instead, it suggests that functions shared by core taxa might be important for the function of the community. Although the correlation between 16S rRNA tree and ecological functions is not perfect, many studies have demonstrated an overall strong correlation that should be useful in predicting species function from phylogeny (Langille et al., 2013; Snel et al., 1999; Zaneveld et al., 2010).
4 Conclusion
We have developed PhyloCore, an application that uses phylogeny to identify core taxa in microbial communities. It has been suggested that core microbiota exist at the functional rather than the taxonomic level (Consortium, 2013; Turnbaugh et al., 2009). However, these two alternative hypotheses are not mutually exclusive. It is conceivable that core functions are shared by the phylogenetic core taxa and therefore they represent the two aspects of the same microbiota core. Having PhyloCore will help us further test this theory.
Supplementary Material
Highlights.
PhyloCore is an application that uses a phylogeny-based algorithm to identify core taxa in microbial communities.
PhyloCore systematically identify core taxa at the proper taxonomic levels.
Users can set prevalence/abundance thresholds and identify core in subgroups according to their needs.
Acknowledgments
This work has been supported by the National Institutes of Health [5R01GM108501 to MW].
Abbreviations
- OTU
operational taxonomic unit
- SSU rRNA
small subunit ribosomal RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Availability and requirements
Project name: PhyloCore
Project home page: http://wolbachia.biology.virginia.edu/WuLab/Software.html
Operating system(s): Unix/Linux (Perl and Python versions); Mac OS (Python version)
Programming language: Perl or Python 2
Other requirements: Bioperl 1.5.2 or later, or Biopython and Numpy
License: GNU GPL
Any restrictions to use by non-academics: None
Conflict of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
MW and TR conceived of the project and designed the software. TR implemented and tested the software. TR drafted the manuscript. MW revised the manuscript and supervised the work. All authors read and approved the final manuscript.
References
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh JI, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh JI, Knights D, Gajer P, Ravel J, Fierer N, Gordon JI, Knight R. Moving pictures of the human microbiome. Genome Biology. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium THMP. Structure, function and diversity of the healthy human microbiome. Nature. 2013;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero OX, Wildschutte H, Kirkup B, Proehl S, Ngo L, Hussain F, Le Roux F, Mincer T, Polz MF. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science. 2012;337:1228–1231. doi: 10.1126/science.1219385. [DOI] [PubMed] [Google Scholar]
- Huse SM, Ye Y, Zhou Y, Fodor AA. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS ONE. 2012;7:e34242. doi: 10.1371/journal.pone.0034242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone CA, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of Mammals and Their Gut Microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I, Muller CE, Walter J. Long-Term Temporal Analysis of the Human Fecal Microbiota Revealed a Stable Core of Dominant Bacterial Species. PLoS ONE. 2013;8:e69621-12. doi: 10.1371/journal.pone.0069621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L, Andersson SGE, Battin TJ, Prosser JI, Schimel JP, Whitman WB, Hallin S. The ecological coherence of high bacterial taxonomic ranks. Nature Reviews Microbiology. 2010;8:523–529. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- Philippot L, Bru D, Saby NPA, Cuhel J, Arrouays D, Simek M, Hallin S. Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environ Microbiol. 2009;11:3096–3104. doi: 10.1111/j.1462-2920.2009.02014.x. [DOI] [PubMed] [Google Scholar]
- Reis dos M, Inoue J, Hasegawa M, Asher RJ, Donoghue PCJ, Yang Z. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc. Biol. Sci. 2012;279:3491–3500. doi: 10.1098/rspb.2012.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Handelsman J. Beyond the Venn diagram: the hunt for a core microbiome. Environ Microbiol. 2011;14:4–12. doi: 10.1111/j.1462-2920.2011.02585.x. [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nature Genetics. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber CL, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblut ND, Shade A, Read JS, McMahon KD, Whitaker RJ. Lineage-specific responses of microbial communities to environmental change. Appl. Environ. Microbiol. 2013;79:39–47. doi: 10.1128/AEM.02226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneveld JR, Lozupone CA, Gordon JI, Knight R. Ribosomal RNA diversity predicts genome diversity in gut bacteria and their relatives. Nucleic Acids Research. 2010;38:3869–3879. doi: 10.1093/nar/gkq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Guo Z, Xue Z, Sun Z, Zhang M, Wang L, Wang G, Wang F, Xu J, Cao H, Xu H, Lv Q, Zhong Z, Chen Y, Qimuge S, Menghe B, Zheng Y, Zhao L, Chen W, Zhang H. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. The ISME Journal. 2015:1–12. doi: 10.1038/ismej.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.