An article by Cortez et al. (1) in a recent issue of PNAS makes several important connections between minichromosome maintenance (MCM) proteins and the DNA damage response. The authors establish that two MCM subunits, MCM2 and MCM3, are substrates for ATM- and Rad3-related (ATR) and ataxia-telangiectasia-mutated (ATM) checkpoint kinases, respectively. They also identify one of the phosphorylation sites on each MCM subunit. In addition, Cortez et al. show that decreasing the level of MCM7 protein, an ATR-interacting protein (ATRIP)-interacting subunit, causes an S-phase checkpoint defect as seen by radio-resistant DNA synthesis.
Initiation of DNA Replication
DNA replication starts at discrete origins marked by the origin recognition complex (ORC)-dependent assembly of a pre-replicative complex (pre-RC) (2, 3). Subsequent to ORC binding, Cdc6, Cdt1, and the MCM proteins assemble. Eukaryotes contain six related MCM proteins (MCM2–MCM7). MCM proteins are members of the AAA+ family (4) and display ATPase activity. Only the MCM467 subcomplex displays in vitro DNA helicase activity (5). MCM proteins and their associated ATPase activities are all essential for DNA replication (6) and are required for replication fork progression (7). Although it has not yet been conclusively established that MCM proteins are the replicative helicase unwinding DNA at the replication fork, they are the best candidates for this activity (8). The MCM-containing pre-RC is converted into an initiation complex after the concerted activity of S-phase kinases Cdk2/CycE and Cdc7/Dbf4 (3). Although the essential targets of Cdk2 and Cdc7 protein kinases on the pre-RC have not been identified, one consequence of their action is loading of Cdc45, an essential protein involved in DNA polymerase loading (9).
Several signal transduction pathways prevent duplication of damaged DNA, thus preserving genome integrity throughout S-phase (10). These cell-cycle checkpoint pathways can be activated in a replication-dependent or independent manner. They are triggered by a variety of genotoxic stresses, including ionizing radiation and radiomimetic drugs, which generate DNA double-strand breaks (DSBs), and UV irradiation and inhibitors of DNA replication, which generate single-stranded DNA (ssDNA). These aberrant DNA structures also arise as a consequence of unperturbed DNA replication (11). Two protein kinases are critical for the response to DNA damage: ATM (the protein mutated in the cancer-prone disease ataxia-telangiectasia) in the case of DSBs (12) and ATR in the case of ssDNA intermediates (13, 14). How these pathways target the S-phase kinases Cdk2 and Cdc7 to prevent origin firing has been extensively documented.
In addition to blocking origin firing, it has been postulated that checkpoints could directly affect progression of DNA replication forks; however, no biochemical evidence supported this idea. Because MCM proteins are implicated in origin firing as well as the elongation step of DNA replication, targeting MCMs by the DNA damage response has the potential to block both initiation and progression. In contrast, inhibition of S-phase kinases blocks only unfired origins from initiating because the kinases are dispensable postinitiation. The finding by Cortez et al. (1) that MCM proteins are substrates for ATM and ATR suggests a mechanism by which ongoing DNA replication could be modulated after origin firing.
Phosphorylation of MCM Proteins by ATM and ATR
Cortez et al. first identified MCM3 as a substrate of checkpoint kinases by using an elegant affinity purification scheme. Because ATM and ATR phosphorylate “SQ” sites, the investigators raised antibodies specific for phospho-SQ and used them to purify and identify ATM and ATR substrates from cells that were subjected to genotoxic stress.
What are the consequence of MCM phosphorylation by ATM and ATR? The authors show that phosphorylated MCM2 and MCM3 are found both chromatin-bound and in the cytosol, suggesting that ATM and ATR phosphorylation does not prevent chromatin assembly of MCM complexes. It remains to be tested whether phosphorylation of MCM inhibits the subsequent loading of Cdc45, thus preventing the activation of the pre-RC into an initiation complex. MCM2 or MCM3 phosphorylation, or both, could affect the ATPase or DNA helicase activities of the MCM heterohexameric complex. MCM2 is phosphorylated by ATR at S108, a residue conserved in vertebrates, but absent in yeast, suggesting a role for this modification in organisms with large genomes for which the mechanism of origin selection is not defined. This residue is located outside the conserved modules found in proteins of the AAA+ family. Therefore, phosphorylation at this site could have a regulatory role on the activity of the MCM hexamer rather than directly affecting the ATPase activity of MCM2 subunit. However, an argument against a direct inhibitory role of MCM2 phosphorylation by ATR on helicase function is the fact that aphidicolin treatment, which uncouples DNA polymerases from the helicase, leads to the generation of extensive regions of ssDNA (15), suggesting that the replicative helicase is not directly inhibited in these conditions. Additionally, phosphorylation of MCM2 in the N-terminal region could affect its known interactions with the transcription machinery (16, 17). MCM3 is phosphorylated by ATM at S535 located between the modules corresponding to box VII and VII′ of AAA+ protein (4). It is difficult to predict whether phosphorylation at this site will affect MCM3 ATPase activity because it is located on a stretch of amino acids present only in MCM3 proteins but absent in the other members of the MCM family (18).
To date, in vitro DNA helicase activity has been described for the MCM467 subcomplex exclusively. However, all six MCMs are essential for life in yeast and for DNA replication in Xenopus extracts (6, 19). If the in vivo DNA helicase activity is contributed exclusively by the MCM4, -6, and -7 subunits, it will be important to determine how MCM2, MCM3, and MCM5 cooperate with MCM467 subcomplex activity. In summary, considering the excess MCM protein on the chromatin (20) and without direct evidence of phosphorylation on the active MCM complex at origins, further investigation will be needed to assess the biochemical and biological consequences of MCM2 and MCM3 modifications by ATM and ATR.
MCM7 Interaction with the ATR Partner, ATRIP
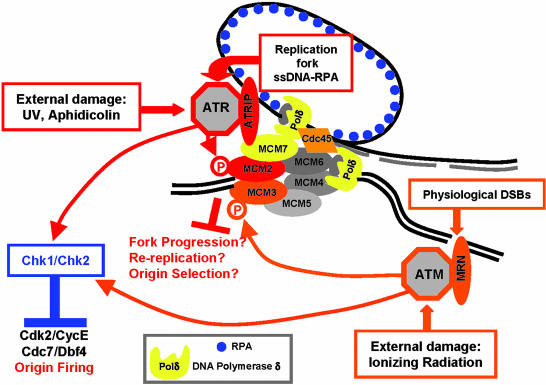
This article provides further compelling evidence for a direct connection between MCM and the DNA damage response by demonstrating the interaction between MCM7 and ATRIP (1). MCM7 also interacts with Rb, E6, and rad17 (21–24) and therefore could act as a platform for interactions with cell-cycle and checkpoint proteins. Indeed, MCM function is cell-cycle-regulated to ensure that the genome is replicated only once per cell cycle. Tethering active ATR to MCM complexes might be a way to locally prevent re-replication. ATRIP is required for ATR signaling in response to ssDNA-RPA intermediates (13). ATR/ATRIP coupled to MCM proteins may monitor and regulate the extent of ssDNA-RPA intermediates generated during DNA replication and after DNA damage (Fig. 1). The rate of DNA unwinding by the helicase is a limiting step for fork progression. Checkpoint activation by the presence of damage containing ssDNA or DSBs could therefore be used to slow down helicase progression before the DNA polymerase encounters the damage thus preventing replication fork collapse. Indeed, ATR-dependent phosphorylation of MCM2 is observed in S-phase in undamaged cells (1) suggesting a monitoring role in the absence of external damage. This is consistent with the observation that ATR is active during normal DNA replication and regulates the timing of origin firing (15).
Fig. 1.
A schematic view of the signaling pathways inhibiting DNA replication. ssDNA-RPA intermediates and DSBs arise as a consequence of external insults (irradiation and polymerase inhibitors) or during normal replication. These aberrant DNA structures trigger the activation of ATR and ATM protein kinases. In turn, these protein kinases inhibit origin firing through the Chk1/Chk2-dependent down-regulation of Cdk2 and Cdc7 protein kinases. MCM7 interaction with ATRIP could tether ATR protein kinase to the replication fork, providing a way to locally regulate ATR response to ssDNA. In addition to activating the canonical checkpoint pathways leading to Cdk2 and Cdc7 down-regulation, ATM and ATR directly phosphorylate MCM2 and MCM3. Although the consequences of these modifications are not yet known, they could modulate the activity of MCM proteins and either halt fork progression, prevent re-replication, or participate in origin selection.
Intriguingly, this manuscript shows that decreasing the amount of MCM7 protein to a level that does not significantly affect DNA replication leads to an intraS phase checkpoint defect and radio-resistant DNA synthesis after ionizing radiation. This phenotype establishes that MCM complex is a physiologically relevant ATM target for the DNA damage response. Furthermore, this observation is reminiscent of temperature-sensitive alleles of mcm mutant yeasts, that can progress through S-phase, yet display chromosome instability and accumulate DNA damage (21). Therefore, chromatin-bound MCM that are not present at the origin, or cytosolic MCM, might have a function unrelated to origin firing.
Finally, the data presented in this paper could provide an explanation for the “MCM paradox”: the fact that MCM proteins are in vast excess compared to the number of fired origins (20, 25). Excess MCM proteins might not be important for DNA replication but essential for checkpoint function and genome stability. Conversely, modulation of MCM activity by ATM and ATR in response to replication intermediates such as ssDNA-replication protein A (RPA) might provide a means for origin selection by inactivation of the excess MCM complexes on the chromatin.
Acknowledgments
This work was supported by National Institutes of Health Grant RO1 CA92245 and by American Cancer Society Grant RSG CCG-103367 (to J.G.).
See companion article on page 10078 in issue 27 of volume 101.
References
- 1.Cortez, D., Glick, G. & Elledge, S. J. (2004) Proc. Natl. Acad. Sci. USA 101, 10078–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, S. P. & Dutta, A. (2002) Annu. Rev. Biochem. 71, 333–374. [DOI] [PubMed] [Google Scholar]
- 3.Kelly, T. J. & Brown, G. W. (2000) Annu. Rev. Biochem. 69, 829–880. [DOI] [PubMed] [Google Scholar]
- 4.Neuwald, A. F., Aravind, L., Spouge, J. L. & Koonin, E. V. (1999) Genome Res. 9, 27–43. [PubMed] [Google Scholar]
- 5.Ishimi, Y. (1997) J. Biol. Chem. 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- 6.Forsburg, S. L. (2004) Microbiol. Mol. Biol. Rev. 68, 109–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labib, K., Tercero, J. A. & Diffley, J. F. (2000) Science 288, 1643–1647. [DOI] [PubMed] [Google Scholar]
- 8.Labib, K. & Diffley, J. F. (2001) Curr. Opin. Genet. Dev. 11, 64–70. [DOI] [PubMed] [Google Scholar]
- 9.Mimura, S. & Takisawa, H. (1998) EMBO J. 17, 5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou, B. B. & Elledge, S. J. (2000) Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]
- 11.Shechter, D., Costanzo, V. & Gautier, J. (April 9, 2004) DNA Repair, 10.1016/j.dnarep.2004.03.020. [DOI] [PubMed]
- 12.Shiloh, Y. (2003) Nat. Rev. Cancer 3, 155–168. [DOI] [PubMed] [Google Scholar]
- 13.Zou, L. & Elledge, S. J. (2003) Science 300, 1542–1548. [DOI] [PubMed] [Google Scholar]
- 14.Costanzo, V., Shechter, D., Lupardus, P. J., Cimprich, K. A., Gottesman, M. & Gautier, J. (2003) Mol. Cell 11, 203–213. [DOI] [PubMed] [Google Scholar]
- 15.Shechter, D., Costanzo, V. & Gautier, J. (2004) Nat. Cell Biol 6, 648–655. [DOI] [PubMed] [Google Scholar]
- 16.Holland, L., Gauthier, L., Bell-Rogers, P. & Yankulov, K. (2002) Eur. J. Biochem. 269, 5192–5202. [DOI] [PubMed] [Google Scholar]
- 17.Yankulov, K., Todorov, I., Romanowski, P., Licatalosi, D., Cilli, K., McCracken, S., Laskey, R. & Bentley, D. L. (1999) Mol. Cell. Biol. 19, 6154–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gozuacik, D., Chami, M., Lagorce, D., Faivre, J., Murakami, Y., Poch, O., Biermann, E., Knippers, R., Brechot, C. & Paterlini-Brechot, P. (2003) Nucleic Acids Res. 31, 570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tye, B. K. (1999) Annu. Rev. Biochem. 68, 649–686. [DOI] [PubMed] [Google Scholar]
- 20.Edwards, M. C., Tutter, A. V., Cvetic, C., Gilbert, C. H., Prokhorova, T. A. & Walter, J. C. (2002) J. Biol. Chem. 277, 33049–33057. [DOI] [PubMed] [Google Scholar]
- 21.Bailis, J. M. & Forsburg, S. L. (2004) Curr. Opin. Genet. Dev. 14, 17–21. [DOI] [PubMed] [Google Scholar]
- 22.Gladden, A. B. & Diehl, J. A. (2003) J. Biol. Chem. 278, 9754–9760. [DOI] [PubMed] [Google Scholar]
- 23.Kukimoto, I., Aihara, S., Yoshiike, K. & Kanda, T. (1998) Biochem. Biophys. Res. Commun. 249, 258–262. [DOI] [PubMed] [Google Scholar]
- 24.Sterner, J. M., Dew-Knight, S., Musahl, C., Kornbluth, S. & Horowitz, J. M. (1998) Mol. Cell. Biol. 18, 2748–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyrien, O., Marheineke, K. & Goldar, A. (2003) Bioessays 25, 116–125. [DOI] [PubMed] [Google Scholar]