Abstract
An ability to rationally design complex networks from the bottom up can offer valuable quantitative model systems for use in gaining a deeper appreciation for the principles governing the self-organization and functional characteristics of complex systems. We report herein the de novo design, graph prediction, experimental analysis, and characterization of simple self-organized, nonlinear molecular networks. Our approach makes use of the sequence-dependant auto- and cross-catalytic functional characteristics of template-directed peptide fragment condensation reactions in neutral aqueous solutions. Starting with an array of 81 sequence similar 32-residue coiled-coil peptides, we estimated the relative stability difference between all plausible A2B-type coiled-coil ensembles and used this information to predict the auto- and cross-catalysis pathways and the resulting plausible network motif and connectivities. Similar to most complex systems, the generated graph displays clustered nodes with an overall hierarchical architecture. To test the validity of the design principles used, nine nodes composing a main segment of the graph were experimentally analyzed for their capacity in establishing the predicted network connectivity. The resulting self-organized chemical network is shown to display 25 directed edges in good agreement with the graph analysis estimations. Moreover, we show that by varying the system parameters (presence or absence of certain substrates or templates), its operating network motif can be altered, even to the extremes of turning pathways on or off. We suggest that this approach can be expanded for the construction of large-scale networks, offering a means to study and to understand better the emergent, collective behaviors of networks.
Networks appear in numerous aspects of the world we live in, from the large-scale ecological systems, social networks, and World Wide Web, to the microscopic biochemical networks of living cells (1–15). Recent breakthroughs in graph-theoretic analysis have provided a revealing global view of the architectural features of complex networks. Statistical analyses suggest that most complex networks, including metabolic and proteomic networks, have scale-free topology (1–5). Unlike regular or random network topologies, scale-free networks exhibit both relatively short average distances between any two nodes and high clustering coefficients by having a few highly connected nodes. For instance, in biological networks some proteins act as hubs to engage in a large number of interactions with other proteins, whereas the majority of proteins seem to behave as links and partake in only one or a few interactions. This top-down view of complex systems provides key boundary conditions on network topology and functional properties but gives relatively few details and only a static view of the system (6, 11). On the other hand, from the bottom-up perspective, it is often possible to gather detailed information about the properties of the individual components of a network. For instance, molecular biology and biochemical sciences have and continue to provide a wealth of detailed information about the functional characteristics of biomolecules and their interaction diagrams (13, 15–17). Yet our current inadequate understanding of the influence, transfer, and processing of molecular-level information into the overall dynamic macroscopic behavior of a living cell remains as a major impediment in the bottom-up approach to systems biology. Moreover, the overall collective behavior of a given self-organized network, its “emergent” characteristics, does not depend solely on the properties of its individual components but also on the complex web of dynamic nonlinear interactions and information transfer processes between its components. Therefore, to better understand the underlying principles and factors contributing to and influencing complex system characteristics, various aspects of complex networks need to be addressed, quantitatively analyzed, and rigorously modeled (6, 11). These include all levels of organization and hierarchy; from the properties of individual components, their interaction selectivity, strength, dynamics, and information transfer capacity, to local network motifs and the large-scale graph architecture and behavior. Furthermore, it is imperative that the studies provide sufficient quantitative detail that could be used to arrive at experimentally testable hypotheses. However, a significant limitation hampering progress toward these goals stems from the fact that most existing complex systems are seldom amenable to deliberate and systematic manipulation or network reorganization (2, 17). This constraint is evident particularly in the context of the highly evolved isotropic biological networks where substantial mutations, protein deletions, or network tampering can often result in the disruption of vital cellular functions. Therefore, as an alternative possibility, we reasoned that rationally designed synthetic self-organized molecular systems might provide useful model networks for the study and better understanding of complex system behavior (18–23). In this account we describe the rationale, de novo design, graph estimation, and analyses of a relatively simple synthetic molecular system that seem to bear many of the basic properties one commonly ascribes to complex self-organized networks.
Methods
Assessment of Autocatalytic and Cross-Catalytic Efficiencies. An aqueous mixture was prepared containing equimolar amounts of N and the studied Ei, reducing agent tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 4-acetamidobenzoic acid (ABA) as the internal standard, and the desired amount of seeded template. Under these acidic conditions, the ligation reaction is inhibited. After equilibrating for ≈30 min, the reactions were initiated by the addition of 3-(N-morpholino)propanesulfonic acid (Mops) buffer to give initial reaction conditions as described in the text and figure legends. Aliquots (15 μl) were removed at various time points, immediately quenched in 3% trifluoroacetic acid in water, and stored frozen before RP-HPLC analysis. Ligation experiments that involved two electrophiles and the nucleophile (see Fig. 8a) were performed in the same manner. All experiments were repeated at least twice.
Fig. 8.
Establishing catalytic pathways in isolated subnetworks. (a) The cross-catalytic rate of production of T1 (filled symbols) and T4 (open symbols) over time are shown in reaction mixtures containing E1 and E4 (90 μM each) and N (200 μM), in the absence (• and ○), or the presence of 20 ± 2 μM initial concentration of T1 (▪ and □), T4 (♦ and ⋄), or both T1 and T4 (▴ and ▵). (b) Production of T7 in reaction mixtures containing E7 and N (80 μM each), in the absence (•), or the presence of 25 ± 2 μM initial concentration of T3 (○), T4 (□), T5 (⋄), T8 (▵), or all four templates together (6.5 ± 1 μM each, ▪). All reactions were performed with 5 mM TCEP as the reducing agent in 100 mM Mops (pH 7.2) at 22°C with ABA as internal standard. Curves are shown to guide the eye.
Probing the Network Connectivity. Acidic stock solutions containing the common nucleophile N, all nine electrophiles E1–9 (or eight electrophiles for experiments shown in Fig. 6), TCEP, and ABA were seeded with one of the templates Ti, or with water. Reactions were initiated by adding Mops buffer to give initial reaction conditions as described in the text and figure legends. Aliquots were removed at various time points and placed in a solution of Mops buffer (85 μl, 200 mM, pH 7.50) saturated with 6 M guanidinium hydrochloride to allow for any intermediate thiolester to rearrange into product. After 20 min, this mixture was quenched with 10 μl of 10% trifluoroacetic acid in water and stored frozen. RP-HPLC analysis by using Zorbax c-18 column with the following solvent gradient (solvent A, 99% H2O/1% CH3CN/0.1% trifluoroacetic acid; solvent B, 10% H2O/90% CH3CN/0.07% trifluoroacetic acid): 0% B for 2 min, 0 → 20% B for 1 min, 20% → 34% B for 5 min, and 34% → 50% B for 25 min. The identities of all peptide peaks in the chromatogram were determined by matrix-assisted laser desorption ionization–time-of-flight or HPLC-sonic spray ionization MS and by direct coinjection and retention time comparisons with authentic samples.
Fig. 6.
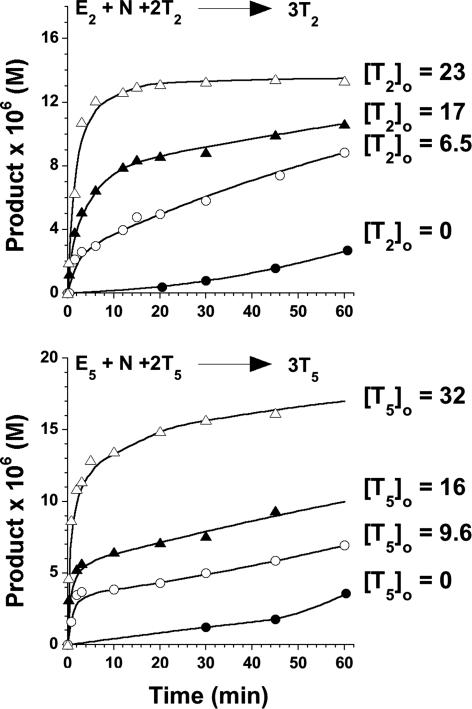
The autocatalytic rates of T2 and T5 production over time. For each template a set of reactions were performed by using 100 μM Ei, 100 μM N, 5 mM TCEP, 100 mM Mops (pH 7.2) in the presence or absence of various initial concentration (μM) of [Ti]0 as indicated. Curves are shown to guide the eye. See the supporting information (Fig. 11, which is published as supporting information on the PNAS web site) for similar plots for T1, T3, T4, T6, T7, T8, and T9.
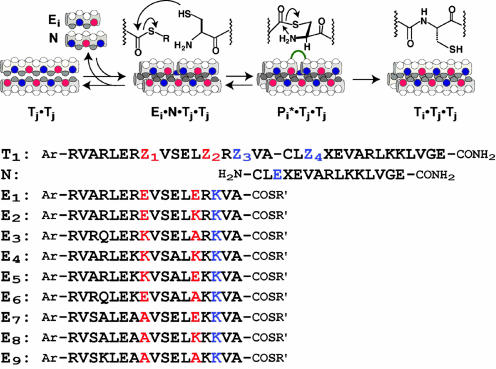
Design Rationale. In constructing the molecular network, we have used template-directed peptide fragment condensation reactions (24–27) (Fig. 1) as a common circuit design element (7) to considerably simplify the de novo design, graph prediction, and experimental analyses. The approach is therefore fundamentally rooted in the functional consequences of the sequence-dependant characteristics of coiled-coil interactions. It has been shown previously that appropriate amino acid substitutions at the coiled-coil recognition interface can alter the aggregate stabilities of substrates, reactive intermediates, and products, and thereby influence the template-directed replication, cross-catalytic selectivity, and efficiencies (28–32). Moreover, we have established recently that peptide sequences that allow facile interconversion between parallel dimeric and trimeric coiled-coils can use dimeric coiled-coils as the productive templates to enhance considerably substrate selectivity, catalytic efficiency, and turnover (31). Although functional coiled-coil sequences can be chosen in principle from a rather large sequence space, in the present study we selected to assess a relatively small peptide array composed of eighty-one 32-residue coiled-coils. The peptide sequences were derived from a recently characterized autocatalytic coiled-coil (31) by substituting glutamic acid (E), alanine (A), and lysine (K) residues at four e and g heptad positions (g8, e13, g15, and e20) that line a portion of the coiled-coil recognition interface. It is well known that amino acids at these positions can participate in g ⇔ e′ (i, i′ + 5) interhelical side-chain–side-chain interactions and significantly influence the stability of the coiled-coil ensembles. A simple scoring algorithm was then devised, based on the following rationale, assumptions, and empirical data, to assess whether a given coiled-coil sequence is likely to partake in network formation by functioning as a competent template for the production of itself and/or other sequences.
Fig. 1.
Schematic representation of the template-directed peptide fragment ligation and the peptide sequences used in this study. The electrophilic peptide fragment Ei bearing a C-terminal thiolester moiety and the nucleophilic peptide N possessing an N-terminal cysteine residue can preorganize on the complementary peptide template Tj to form a coiled-coil ternary (not shown) or quaternary (depicted) complex. The productive juxtaposition and enhanced effective concentration of the reactive functionalities facilitate the Kent ligation process (35), proceeding through the transthiolester intermediate followed by intramolecular aminolysis, to give product Ti. The reaction is autocatalytic when Ti = Tj and cross-catalytic when Ti ≠ Tj. The array of 81 peptides used in the theoretical network analysis were generated from the template sequence T by amino acid substitutions (Glu, Ala, and Lys) at positions Z1-4. The nine experimentally analyzed peptide sequences T1–9 were derived by fragment coupling of N with the corresponding electrophilic fragments E1–9. To facilitate product analysis, additional amino acid substitutions were introduced at the solvent exposed residues on the coiled coil surface (heptad positions, b, c, and f) to give mass differentiation and distinct RP-HPLC retention times. Ar, ABA; X, Lys-ABA; R′, ethanesulfonic acid.
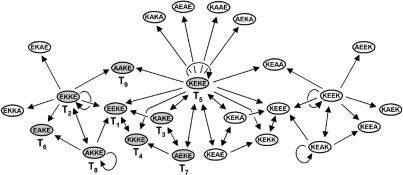
Scoring Analysis. In previous studies we had demonstrated a direct correlation between the apparent stability of productive coiled-coil intermediates (substrate–template complexes) and the corresponding initial rates of template-directed product formation (24, 25). Furthermore, in reaction mixtures where two or more templates were competing for common substrates, the observed selectivity in template-directed fragment condensation reactions could be rationalized based on the relative stability differences (–ΔΔG) between the competing intermediate and product ensembles (28, 30, 31). These observations suggest that with respect to more complex reaction mixtures it might be possible to determine, a priori, the network of governing template-directed pathways by estimating the differences in the stability (–ΔΔG) of all plausible template-product ensembles—by analogy to the corresponding intermediate complexes—assuming that by using sequence similar coiled-coils, the functional complexes have similar aggregation state, folding, and structural organizations. For estimating the relative stability of the coiled-coil species in our system, we have made use of the experimental data from a recent biophysical study in which the coupling energies (–ΔΔGint) for amino acid side-chain pairs at the interhelical g ⇔ e′ (i, i′+ 5) positions have been measured (33). Assuming homodimeric peptides as templates (see above), we calculated the change in stability (–ΔΔG) for all possible A3- and A2B-type parallel trimeric products (81 × 81) versus the reference (–ΔΔG = 0) homotrimeric coiled-coil having only alanine residues at positions 8, 13, 15, and 20. The –ΔΔG scores were calculated by summing the values for the six pairwise cross-strand g ⇔ e′ (i, i′+ 5) interactions. The calculated values range from –3.6 to 8.4 ± 0.6 kcal·mol–1, from the least stable homotrimeric structure having Lysg ⇔ Lyse' interactions to the most favorable coiled-coil with Lysg ⇔ Glue′ pairs at all the amino acid substitution sites, respectively. However, although the estimated –ΔΔG values can be used to sort the order of stability of all plausible coiled-coil complexes, in the absence of a validated correlation factor, the magnitude of these values cannot be used to forecast the efficiency of the corresponding template-directed pathways. In lieu of that, we assigned and ranked a given sequence as capable of template-directed ligation (of another or itself) if the resulting trimeric product–template complex had a threshold value of –ΔΔG ≥ 5.6 ± 0.2 kcal·mol–1. The threshold value was approximated by applying the same scoring analysis to previously reported similar examples of kinetically well characterized template-directed ligases and replicases (25, 31), and based on the empirical observations that neutralization of two complementary interhelical Lysg ⇔ Glue′ interactions [–ΔΔG of ≈2.8 ± 0.2 kcal·mol–1 (33)] can often lead to a dramatically reduced rate of template-directed product formation. Analysis of the plausible template-directed pathways in the coiled-coil matrix by using the threshold value of –ΔΔG ≥ 5.6 resulted in a nonrandom graph with 25 nodes joined by 53 directed edges, including five autocatalytic pathways (Fig. 2). This graph is clustered and hierarchical, and it possesses a mirror symmetry that is due to peptides with different permutations of residues along the recognition interface, but it offers the same combinations of interacting residues. Interestingly, the experimental studies detailed below seem to indicate that the theoretical approach above can offer, to a first approximation, a useful guide for designing relatively complex molecular networks.
Fig. 2.
Calculated graph architecture illustrating formation of a self-organized peptide network composed of 25 nodes joined by 53 vector edges obtained by using threshold –ΔΔG ≥ 5.6 kcal/mol. Each node represents a template or product sequence identified by the nomenclature that denotes the amino acid residues of a given sequence at the varied positions 8, 13, 15, and 20, respectively. The directed edges (arrows) signify template-assisted ligation pathways pointing from the dimeric template to the product (curved arrows indicate autocatalysis). Nodes highlighted in grey (and named T1 through T9) represent those sequences evaluated experimentally for their ability to form that portion of the graph.
Experimental Results and Discussion
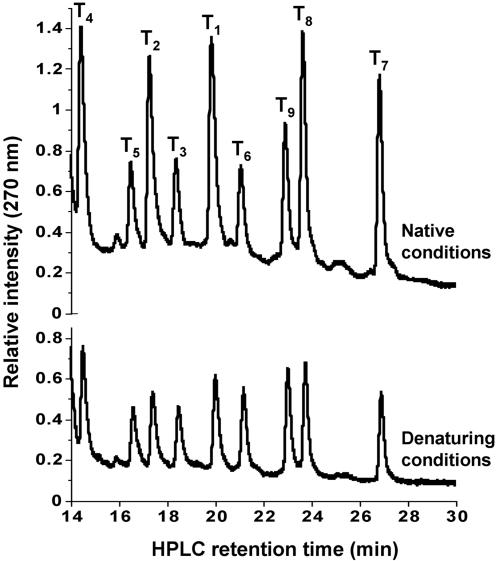
The analysis above suggests that even relatively few peptides can potentially provide a rich graph architecture. But how accurate are these predictions? Do the graph predictions represent a static view of the entire networking potential? How does the dynamic aspect of a self-organized network, in the present case the reaction rates and template-directed selectivities, manifest themselves? To address these questions, we chose to evaluate experimentally the nine nodes (highlighted in Fig. 2, with sequences defined in Fig. 1) that constitute the main segment of the graph, including the apparent branching nodes T5 (KEKE) and T2 (EKKE). These nine nodes are connected theoretically by 20 cross-catalytic and 3 autocatalytic edges. We used the experimental approaches described below to uncover these connections and their directions between nodes and to probe the strength of their connectivity in the peptide network under several different reaction conditions (system inputs). The network was probed by reacting all nine electrophilic peptide fragments, E1–9 (50 ± 5 μM each), and a substoichiometric amount of the nucleophilic peptide fragment, N (300 μM), in a single reaction vessel. Quantitative analytical RP-HPLC analysis was used to follow the rate of production of all nine species simultaneously in a given reaction mixture. Products were identified by matrix-assisted laser desorption ionization-time-of-flight or HPLC-sonic spray ionization MS and by direct coinjection and HPLC retention-time comparisons with authentic samples. Product formation, T1–9, over time was measured under both native (100 mM Mops, pH 7.2) and denaturing (3 M guanidine hydrochloride, 100 mM Mops, pH 7.2) conditions. Under the native reaction conditions, all nine products were formed but in different yields, with T1, T2, T4, T7, and T8 being the most abundant (Fig. 3). However, under denaturing conditions, all nine products were again formed but at a slower rate and in approximately equal yield to each other. These studies confirm the expectations that it is the assembled, three-dimensional structures in the reaction mechanism that give rise to template-assisted ligation selectivities and specificities, not some inherent differences in reactivity among the peptide fragments.
Fig. 3.
RP-HPLC chromatograms of the network reaction (N + E1–9 → T1–9) under native and denaturing reaction conditions at 24 h reaction time. The two reactions were carried out in parallel under similar initial reaction conditions: 50 ± 5 μM each E1–9, 300 μM N, 5 mM TCEP, 100 mM Mops (pH 7.2) at 22°C in the presence or absence of 3 M guanidine hydrochoride. For clarity only the product region of the chromatogram is shown.
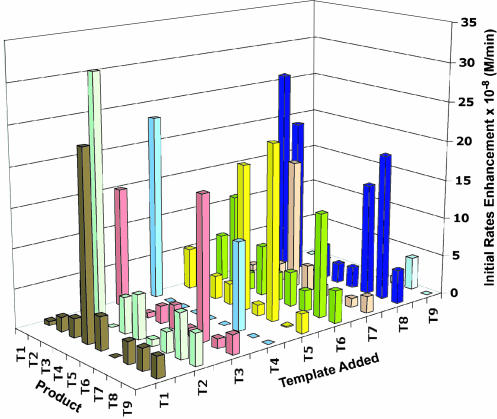
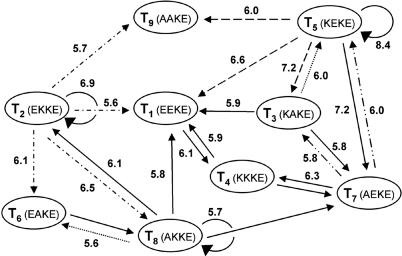
To determine the network connectivity, the network reaction under native conditions was repeated nine more times, each time seeded with ≈30 μM concentration of only one of the templates T1–9. Monitoring product formation over time shows that, in the initial stages of the reaction, each added template enhances the formation of not all but only certain products relative to the unseeded background reaction (Fig. 4 and Fig. 9, which is published as supporting information on the PNAS web site). Moreover, these initial rate enhancements allow for an overall increased yield of all products in the mixture as noted at later points in the course of the reaction. The observed product enhancements in the seeded reactions represent operating auto- and cross-catalytic pathways within the network. Fig. 5 illustrates the resulting network motif having 8 nodes and 14 directed edges depicted by black solid arrows. In agreement with the scoring analysis, the experimental network shows that T2, T5, and T8 enhance their own rates of production. This network, however, lacks 11 directed edges and one node as compared with the theoretical predictions (Fig. 2). Moreover, three observed connections were not predicted; these pathways have scores of 5.2–5.5 kcal·mol–1, just below the chosen threshold of the theoretical graph (5.6 kcal·mol–1), and therefore not displayed. Is the source of apparent lack of close similarity between the observed and predicted network architectures due to the dynamic nature of the system? After all, by analogy to the more complex biological networks, it is quite reasonable that nodal interdependencies are sensitive to the system inputs (concentration of reactants, presence or absence of templates, and time allowed for reaction), thereby under a given reaction condition the network can overemphasize or underrepresent (switching on or off) certain pathways. The following studies support this hypothesis and serve to uncover the latent edges.
Fig. 4.
The effects of templates on the auto- and cross-catalytic initial rates enhancement of product formation. Initial rates of product formation were derived from the amounts of product formed in time in nine network reactions (N + E1–9 → T1–9) carried out under native conditions (50 ± 5 μM each E1–9, 300 μM N, 5 mM TCEP, 100 mM Mops, pH 7.2) each containing an initial concentration (28 ± 3 μM) of only one template Ti. The data shown are average of at least two sets. Estimated errors in measuring the concentrations of products formed in the background reactions and cross-catalytic pathways are on the order of ≤0.5 μM and for autocatalytic pathways ≤1.0 μM. Because of the minor variations in the initial concentrations of added templates between different sets of experiments (±10%), only the initial rates of product formation that are enhanced by ≥2.0-fold over the background (unseeded) reactions (≥100% enhancements) are considered to be significant (see Fig. 10, which is published as supporting information on the PNAS web site) and used to generate the experimental network diagram illustrated in Fig. 5.
Fig. 5.
The experimentally derived network architecture. The arrows (edges) designate template-assisted ligation pathways pointing from the template to the product. The graph represents compilation of the results of four studies (see text). Solid arrows were generated from data shown in Fig. 4, and dashed and dotted arrows were from data in Fig. 7. Numbers along the edges are the estimated –ΔΔG scoring values (kcal·mol–1) for the specified template-directed pathways.
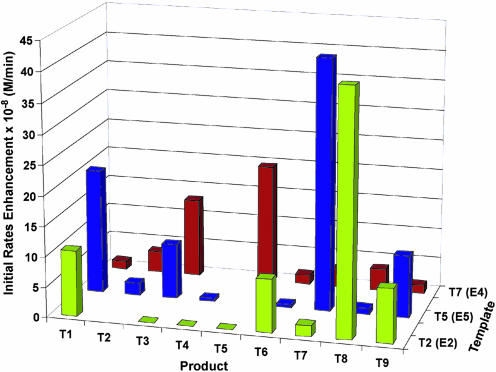
To gage the contribution of autocatalysis to the network self-organization, the inherent autocatalytic efficiencies for all nine peptides T1–9 were determined by reacting each electrophile individually with an equimolar amount of the nucleophile (100 μM each) under native reaction conditions, both in the absence and presence of varying initial concentrations of the corresponding template Ti (Fig. 6). These data were fitted to the template-assisted ligation model (25) to determine various kinetic parameters (see Fig. 12, which is published on the PNAS web site), providing the following series for autocatalytic efficiency, Krel: T5 > T2 > T8 ≈ T3 > T7 ≈ T6, with T9, T1, and T4 essentially autocatalytically infertile (Table 1). This series agrees well with the scoring analysis and ranking of the autocatalytic pathways. To a first approximation, the strong autocatalyic efficiency of T2 and T5 seem to offer an explanation for why the eight expected edges emanating from these nodes (T5 ⇒ T1, T3, T4, T9; and T2 ⇒ T1, T6, T8, T9) were not observed in the complex reaction mixture. It suggests that because of the high autocatalytic efficiencies of T2 and T5, these two templates were engaged in their own synthesis and thus were not available for cross-catalyzing weaker pathways, the exception being that of the T5 ⇒ T7 edge, which was also predicted to have a high cross-catalytic efficiency (score = 7.2 kcal·mol–1). The network analysis described by the studies recapitulated in Fig. 4 is apparently not sufficient for revealing the entire repertoire of operable reaction pathways. Therefore, to verify the inherent networking capacity of T2 and T5, their autocatalytic pathways were turned off and the effect on network architecture was studied (Fig. 7). According to the graph analysis, T2 and T5 constitute the two main branching out nodes (signal distribution points) in the graph. The networking capacity of T5 was established by analyzing the rates of product formation in reaction mixtures made up of the nucleophilic fragment (300 μM), and all electrophiles but E5 (50 ± 5 μM each) in the presence or absence of T5 (25 μM) under native conditions. Under these reaction conditions T5 displays a strong propensity for the cross-catalytic production of T7, as was also observed in the previous network reaction that included E5. However, by turning off the T5 autocatalysis the network reorganizes to unveil three of the four latent cross-catalytic pathways predicted by the algorithm. Their relative cross-catalytic efficiencies are T5 ⇒ {T1 ≥ T3 ≈ T9}, represented by the dashed arrows in Fig. 5. Similarly, using reaction mixtures made up of the nucleophilic fragment N (300 μM) and all electrophiles but E2 (50 ± 5 μM each) in the presence or absence of T2 (25 μM), the predicted branching out node capacity of T2 was established by the four observed pathways T2 ⇒ {T8 > T6 ≈ T1 ≈ T9} (dash-dot arrows in Fig. 5). Two directed edges that were predicted to emanate from T7 are also lacking in the experimental network. Their absence was hypothesized to be due to the strong, highly favorable connectivity from T7 to T4, making T7 unavailable for weaker cross-catalytic pathways. Accordingly, to uncover these and any other connections directed from T7, the network reaction seeded with T7 was repeated but with the exclusion of substrate E4. This experiment (Fig. 7) confirmed the two additional cross-catalytic pathways that were predicted; T7 ⇒ {T3 ≈ T5} (dash-dot arrows in Fig. 5). The final missing directed connectivities of T3 ⇒ T5 and T8 ⇒ T6 were established by isolated cross-catalysis reactions (dotted arrows in Fig. 5). Therefore, altogether these studies establish that the nine peptide-nodes can participate in the formation of a synthetic, self-organized network composed of 25 edges in good agreement with the estimated graph architecture.
Table 1. Relative autocatalytic efficiency of the nine experimental nodes in isolated reactions.
| Product | k1 (kuncat), M-1·s-1 | Km, μM | k6 (kcat), s-1 | k6/Km, M-1·s-1 | Krel* | Score† |
|---|---|---|---|---|---|---|
| T5 | 0.08 | 130 | 0.09 | 690 | 69 | 8.4 |
| T2 | 0.10 | 320 | 0.09 | 280 | 22 | 6.9 |
| T8 | 0.09 | 860 | 0.10 | 120 | 11 | 5.7 |
| T3 | 0.09 | 990 | 0.08 | 80 | 7.1 | 4.8 |
| T7 | 0.10 | 970 | 0.03 | 31 | 2.5 | 4.8 |
| T6 | 0.13 | 1,000 | 0.04 | 40 | 2.5 | 4.5 |
| T9 | 0.10 | 1,000 | 0.02 | 20 | 1.6 | 4.2 |
| T1 | 0.08 | 970 | 0.01 | 10 | 1.0 | 3.0 |
| T4 | <1‡ | 2.4 |
¶The kinetic parameters obtained from fitting the rates of product formation to template-assisted ligation reaction model (25) (see Fig. 12) using the program simfit (34).
Krel equal to the [(k6/Km)/k1] for each reaction divided by [(k6/Km)/k1] of the slowest measurable reaction (to make T1).
Values are in kcal/mol, calculated by using the scoring method described in the text for each homotrimer catalyst-product complex.
No intermediate species was detected.
Fig. 7.
The effects of T2, T5, and T7 on the initial rates of product formation. Each reaction mixture contained 300 μM N, 50 ± 5 μM for each of eight of nine electrophilic fragments Ei (the excluded electrophilic fragment is indicated in parentheses on the template axis), 25 ± 2 μM initial concentration of Ti (indentified on the template axis), 5 mM TCEP, 100 mM Mops (pH 7.2).
It is instructive to use the experimentally derived network architecture and to gage retrospectively how sensitive the accuracy of the network predictions can be with respect to the parameters used in the scoring analysis. The thermodynamic data (33) used in the scoring analysis has an estimated error of ±0.2 kcal·mol–1 in the selected range of –ΔΔG = 5.6–8.4 kcal·mol–1. By using the corresponding threshold values of 5.8 to 5.4 kcal·mol–1 (5.6 ± 0.2), the estimated number of edges in the graph would range from 19 to 28, respectively, thus reducing the accuracy of the network predictions to ≈80%. Therefore, although the simple scoring analysis used in the present study clearly can be a useful guide for selecting appropriate peptide sequences for the experimental design and evaluations, more rigorous calculation methods are needed to better enhance the reliability and general applicability of the graph predictions.
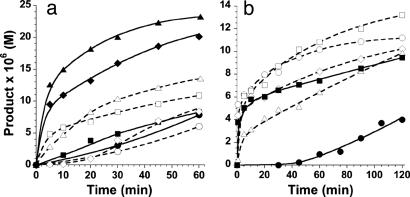
The functional analyses above provide an exciting perspective into the dynamic properties of a network that are not readily apparent when examining the overall graph structure or studying the individual components of the network. For example, despite the fact that T5 is inherently the most efficient autocatalytic species (Table 1), in the complex reaction mixture five other peptides are produced in larger amounts (Fig. 3). The greater abundance of the two other autocatalytic species T2 and T8 is attributed to their tendency for mutual cross-catalysis (Fig. 5). The autocatalytically infertile T1 and T4, and also T7 (a weak autocatalyst), are among the most rapidly produced species in the reaction mixture. Inspection of the network architecture reveals a small reciprocal and interdependent subnetwork composed of T1 ⇔ T4 ⇔ T7. We have also experimentally validated the T1 ⇔ T4 reciprocal catalysis pathway by seeding a reaction mixture that contained E1, E4 (90 μM each), and N (200 μM) by either one or both templates T1 and T4 (20 μM each) (Fig. 8a). Significant rate enhancements due to the cross-catalytic pathways were observed with negligible contributions from autocatalysis. The T1 ⇔ T4 ⇔ T7 network is also fed by seven other pathways ensuring their important roles in network trafficking. T1 and T7 serve as signal integration nodes receiving inputs from five other nodes (T2, T3, T4, T5, and T8) and outputs in a reciprocal manner into T4. This hypothesis was also verified in isolated experiments by monitoring the cross-catalytic production of T7 (from E7 and N, 80 μM each) by either T3, T4, T5, or T8 (25 μM) or in a reaction mixture that included all four templates (6.5 μM each) (Fig. 8b). Therefore, even this relatively simple molecular network displays some of the elementary dynamic characteristics of larger and more complex systems and as such could provide a potentially useful and general synthetic model for quantitative analyses of network self-organization at various levels of hierarchy.
The studies presented here highlight a synthetic chemical approach toward the rational de novo design of complex self-organized molecular systems. Even the relatively small molecular network described seems to mimic some of the graph architectural and basic dynamic features commonly associated with much larger complex systems. Similar to biological networks, our synthetic network possesses directed edges and is characterized by reaction rates. The rate of formation of various nodes is significantly different in isolation than in the context of the network. Different pathways operate under different sets of system inputs. The preponderance of certain nodes seems to depend on the integrated inputs from other nodes. Moreover, because of the common circuit design element used, it seems plausible that a variety of other networks, including more complex systems or networks possessing desired topological features, could be selected from the available large coiled-coil sequence space. Furthermore, since the functional characteristics of each network component can be estimated and/or experimentally assessed, the approach may also provide more accurate data facilitating various mathematical approaches used to model network behavior. It is our hope that the built-in dynamic features of the synthetic network influenced by the underlying graph architecture, myriad of reaction rates, and template-directed selectivities, would provide a rich model system for the study and better understanding of complex system behavior.
Supplementary Material
Acknowledgments
This work was supported by National Aeronautics and Space Administration Astrobiology Institute Grant NCC2-1055. Research fellowships were received from the Human Frontier Science Program (to G.A.) and the Natural Sciences and Engineering Research Council (Canada) (to R.J.).
Abbreviations: TCEP, tris(2-carboxyethyl)phosphine hydrochloride; ABA, 4-acetamidobenzoic acid.
References
- 1.Watts, D. J. & Strogatz, S. H. (1998) Nature 393, 440–442. [DOI] [PubMed] [Google Scholar]
- 2.Strogatz, S. H. (2001) Nature 410, 268–276. [DOI] [PubMed] [Google Scholar]
- 3.Jeong, H., Tombor, B., Albert, R., Oltval, Z. N. & Barabasi, A. L. (2000) Nature 407, 651–654. [DOI] [PubMed] [Google Scholar]
- 4.Ravasz, E., Somera, A. L., Mongru, D. A., Oltvai, Z. N. & Barabasi, A. L. (2002) Science 297, 1551–1555. [DOI] [PubMed] [Google Scholar]
- 5.Girvan, M. & Newman, M. E. J. (2002) Proc. Natl. Acad. Sci. USA 99, 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bray, D. (2003) Science 301, 1864–1865. [DOI] [PubMed] [Google Scholar]
- 7.Alon, U. (2003) Science 301, 1866–1867. [DOI] [PubMed] [Google Scholar]
- 8.Wuchty, S. (2001) Mol. Biol. Evol. 18, 1694–1702. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg, D. S. & Roth, F. P. (2003) Proc. Natl. Acad. Sci. USA 100, 4372–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kauffman, S. (1993) Origin of Order: Self Organization and Selection in Evolution (Oxford Univ. Press, New York).
- 11.Alm, E. & Arkin, A. P. (2003) Curr. Opin. Struct. Biol. 13, 193–202. [DOI] [PubMed] [Google Scholar]
- 12.Mering, C. v., Krause, R., Snel, B., Cornell, M., Oliver, S., Fields, S. & Bork, P. (2002) Nature 417, 399–403. [DOI] [PubMed] [Google Scholar]
- 13.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298, 799–804. [DOI] [PubMed] [Google Scholar]
- 14.Shen-Orr, S., Milo, R., Mangan, S. & Alon, U. (2002) Nat. Genet. 31, 64–68. [DOI] [PubMed] [Google Scholar]
- 15.Bhalla, U. S. & Iyengar, R. (1998) Science 283, 381–387. [DOI] [PubMed] [Google Scholar]
- 16.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 17.Ideker, T., Thorsson, V., Ranish, J., Christmas, R., Buhler, J., Eng, J., Bumgarner, R., Goodlett, D., Aebersold, R. & Hood, L. (2001) Science 292, 929–934. [DOI] [PubMed] [Google Scholar]
- 18.Elowitz, M. & Leibler, S. (2000) Nature 403, 335–338. [DOI] [PubMed] [Google Scholar]
- 19.Gardner, T., Cantor, C. & Collins, J. (2000) Nature 403, 339–342. [DOI] [PubMed] [Google Scholar]
- 20.Becskei, A. & Serrano, L. (2000) Nature 405, 590–593. [DOI] [PubMed] [Google Scholar]
- 21.Arkin, A., Ross, J. & McAdams, H. (1998) Genetics 149, 1633–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guet, C., Elowitz, M., Hsing, W. & Leibler, S. (2002) Science 296, 1466–1470. [DOI] [PubMed] [Google Scholar]
- 23.Dueber, J. E., Yeh, B. J., Chak, K. & Lim, W. A. (2003) Science 301, 1904–1908. [DOI] [PubMed] [Google Scholar]
- 24.Severin, K., Lee, D. H., J., K. A. & Ghadiri, M. R. (1997) Nature 389, 706–709. [DOI] [PubMed] [Google Scholar]
- 25.Kennan, A. J., Haridas, V., Severin, K., Lee, D. H. & Ghadiri, M. R. (2001) J. Am. Chem. Soc. 123, 1797–1803. [DOI] [PubMed] [Google Scholar]
- 26.Lee, D. H., Granja, J. R., Martinez, J. A., Severin, K. & Ghadiri, M. R. (1996) Nature 382, 525–528. [DOI] [PubMed] [Google Scholar]
- 27.Severin, K., Lee, D. H., Martinez, J. A. & Ghadiri, M. R. (1997) Chem. Eur. J. 3, 1017–1024. [Google Scholar]
- 28.Severin, K., Lee, D. H., Martinez, J. A., Vieth, M. & Ghadiri, M. R. (1998) Angew. Chem. Int. Ed. Engl. 37, 126–128. [Google Scholar]
- 29.Lee, D. H., Severin, K., Yokobayashi, Y. & Ghadiri, M. R. (1997) Nature 390, 591–594. [DOI] [PubMed] [Google Scholar]
- 30.Saghatelian, A., Yokobayashi, Y., Soltani, K. & Ghadiri, M. R. (2001) Nature 409, 797–801. [DOI] [PubMed] [Google Scholar]
- 31.Yokobayashi, Y. (2001) in Catalytic and Auto-Catalytic Peptide Networks (The Scripps Research Institute, La Jolla, CA).
- 32.Yao, S., Ghosh, I., Zutshi, R. & Chmielewski, J. (1998) Nature 396, 447–450. [DOI] [PubMed] [Google Scholar]
- 33.Krylov, D., Barchi, J. & Vinson, C. (1998) J. Mol. Biol. 279, 959–972. [DOI] [PubMed] [Google Scholar]
- 34.Sievers, D. & von Kiedrowski, G. (1994) Nature 369, 221–224. [DOI] [PubMed] [Google Scholar]
- 35.Dawson, P. E., Muir, T. W., Clark-Lewis, I. & Kent, S. B. H. (1994) Science 266, 776–779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.