Abstract
Iron-regulatory protein 1 (IRP1) is a dual-function protein with mutually exclusive roles as a posttranscriptional regulator of animal-cell iron metabolism or as the cytosolic isoform of the iron–sulfur enzyme aconitase (c-acon). Much effort has focused on the role of IRP1 in posttranscriptional gene regulation and in factors that influence its interconversion with c-acon, but little is known about the metabolic function and regulation of c-acon. The role of PKC-dependent phosphorylation of S711 on IRP1/c-acon function was examined. Phosphorylation state-specific antibodies revealed that S711 is phosphorylated by PKC in vitro and in human embryonic kidney cells treated with a PKC activator. In aco1 yeast, the phosphomimetic mutants S711D and S711E exhibited severely impaired aconitase function, whereas S711A and S711T were unaffected relative to the WT protein. Aconitase activity in yeast extracts displayed a similar pattern when assayed for capacity to convert citrate to isocitrate: WT, S711A, and S711T were active, but S711D and S711E activity was undetectable. In contrast, when measured by the conversion of isocitrate to cis-aconitate, S711D and S711E displayed substantial activity, indicating that phosphorylation impairs the citrate but not isocitrate mode of aconitase function. This possibility was confirmed in vivo by demonstrating that S711D and S711E specifically antagonized the requirement for isocitrate in two metabolic scenarios. Iron-responsive element RNA-binding affinity was unaffected by S711 mutations. Our results show that S711 is a target of phosphorylation capable of conferring distinct effects on c-acon function potentially dictating changes in cytosolic citrate/isocitrate metabolism.
It is critical that organisms balance the metabolic requirement for iron with its potential toxicity. Vertebrates possess an elegant system to sense cellular iron status, regulate the uptake and metabolic fate of iron, and thereby maintain iron homeostasis. Iron-regulatory protein 1 (IRP1) and IRP2 are sensory components of this critical regulatory network that promote increased iron uptake when cells are iron depleted and storage of iron when cells are iron overloaded (1, 2). IRPs control mRNA fate by binding to iron-responsive elements (IRE) in the untranslated regions of mRNA encoding proteins that control iron homeostasis. The RNA-binding activity of IRP can be regulated by multiple factors. Iron promotes the loss of IRP1 RNA-binding activity through insertion of a [4Fe–4S] cluster into the protein, converting it to the cytosolic isoform of the tricarboxylic acid (TCA) cycle enzyme aconitase (c-acon). Solvent accessibility of the Fe–S cluster in aconitases allows for perturbants to promote loss of the cluster from c-acon, converting it to IRP1 (1, 2). Consequently, reactive species such as NO and H2O2 are able to promote removal of the Fe–S cluster, favoring generation of IRP1 from c-acon, although H2O2 may do so through direct and indirect mechanisms (reviewed in refs. 1 and 2).
Whereas much is known about the role of IRP1 as an RNA-binding protein, relatively little is understood about its function in intermediary metabolism as the cytosolic aconitase. Aconitases are ancient enzymes that are present in organisms from eubacteria to humans and are members of a superfamily of dehydratases that require an [4Fe–4S] iron–sulfur cluster to perform their varied enzymatic roles (3). Aconitases catalyze two separate reactions, the conversion of citrate to cis-aconitate and cis-aconitate to isocitrate, and the binding of all three compounds to the Fe–S cluster is required for catalysis. The nature of the overall aconitase reaction is such that the intermediate cis-aconitate must dissociate from the enzyme and bind in a topologically different mode for the next reaction to proceed (4).
Three families of aconitases are known: the acnB family of prokaryotes, the eukaryotic mitochondrial aconitase (m-acon) family, and the acnA/IRP family found in both prokaryotes and eukaryotes (3). In mammals, there are individual mitochondrial and cytosolic aconitases that are members of the latter two gene families. Interest has focused on the potential differing roles of specific aconitases, and in Escherichia coli, acnB is the major aconitase expressed during logarithmic-phase growth, whereas acnA is induced during the stationary phase or in response to oxidative stress (5, 6). The evidence suggests a potential role of both enzymes in iron metabolism. Like IRP1, some bacterial aconitases are RNA-binding proteins when devoid of their Fe–S cluster (7, 8). Despite the discovery of c-acon decades ago, the physiological role in mammalian systems remains ill defined.
We have demonstrated that IRP1 and IRP2 also are regulated by PKC-dependent phosphorylation (9–13). Treatment of HL-60 cells with a PKC activator increases IRP1 phosphorylation and RNA-binding activity, with a concurrent increase in transferrin receptor mRNA level (9, 10). Two PKC phosphorylation sites, S138 and S711, have been proposed in IRP1 (9). It was also demonstrated that phosphomimetic mutants of S138 exhibit a cluster-instability phenotype, suggesting that phosphorylation alters the set point for iron regulation of IRP1 (11, 12). Finally, we (S.L.C. and R.S.E., unpublished data) and others (14) have found that S138 phosphomimetic mutants of IRP1 undergo iron-dependent degradation.
Relatively less is known about the role of S711 in IRP1 phosphoregulation. S711 is located near R699, which is required for citrate/isocitrate binding, and R728, R732, and other residues involved in RNA binding (15), suggesting that phosphorylation might target one or both of these functions. Interestingly, the S711 region is highly conserved in bacterial acnA enzymes and in IRP1/c-acon in species from Drosophila to humans, whereas in IRP2, which is not an aconitase, this region also is well conserved with the exception that residue 711 is an Ala. Hence, defining the consequence of S711 phosphorylation on aconitase function may provide insight into uncharacterized roles of aconitases in multiple systems.
In this report, a study of the second proposed site of PKC-dependent phosphorylation of IRP1, S711, is described. These studies reveal that S711 of IRP1 is phosphorylated by PKC in vitro and in vivo in human embryonic kidney (HEK) cells. Aconitase activity of S711 phosphomimetic mutants is preferentially inhibited in the citrate mode of c-acon function. Interestingly, substitution of phosphomimetic amino acids for S711 failed to affect the RNA-binding affinity of IRP1. These results show that S711 is a target of PKC-dependent phosphorylation, which confers distinct effects on c-acon function potentially capable of dictating changes in cytosolic citrate and isocitrate metabolism.
Experimental Procedures
Yeast Strains and Culture Conditions. The aconitase-deficient aco1 strain 0615d (MATa,ura3-52, trp1-Δ63, his3-Δ200, aco1, ade2) was used for the analysis of the aconitase function of IRP1 (16). Transformed 0615d cells were maintained in synthetic complete (SC) medium with 2% dextrose lacking uracil (SD-ura). All cultures were grown at 30°C. Δzwf1 and Δzwf1/Δidp2 strains that contain either a kanamycin-resistance or ura3 gene inserted into the glucose-6-phosphate dehydrogenase (ZWF1) and cytosolic NADP-linked isocitrate dehydrogenase (IDP2) genes, respectively, of the parental haploid strain S173-6B (MATa leu2-3,112 his3-1 ura3-52 trp 1-289) (17) were used. Δzwf1cells transformed with pYAD (16) plasmids expressing IRP1 were grown on SD-ura plus kanamycin. Δzwf1/Δidp2 cells were maintained on SD-ura plus kanamycin. To determine whether IRP1 mutants antagonized the requirement for isocitrate, transformed Δzwf1 cells were grown in SC-ura plus 2% dextrose, 2% glycerol, or 2% acetate with or without H2O2.
Site-Directed Mutagenesis of IRP1. The S711 codon was mutated to GCA, GAT, GAA, or ACC to code for A, D, E, and T, respectively, with QuikChange (Stratagene) or a PCR method (18). DNA sequencing confirmed the mutation.
Yeast Extract Preparation and Aconitase Assays. Yeast extracts were from mid-logarithmic-phase cells grown in SC-ura (11). Protein concentration was determined by using bicinchoninic acid (Pierce). Activity assays were performed against a blank with all assay components but substrate. Citrate-to-isocitrate conversion was measured by coupling to yeast Idp (GE Healthcare, Piscataway, NJ) and monitoring NADPH production in 0.1 M Tris-Cl (pH 8.0) containing 1.0 mM each of MgCl2, NADP+, and disodium citrate, and 0.4 unit of Idp (19). Isocitrate-to-cis-aconitate conversion was monitored directly at 240 nm in 0.1 M Tris-Cl (pH 8.0)/10 mM d-isocitrate (20 mM dl-isocitrate) (20). A unit of activity (U) is 1 μmol of product formed per min at 25°C.
RNA-Binding Assays. IRE RNA-binding analysis was performed by gel shift using 0.2–1.0 μg of cell extracts (11). Kd determination was made by RNA saturation analysis under equilibrium conditions using an RNA concentration range from 5.0 pM to 2.0 nM and through curve fitting using prism 4.0 software (GraphPad, San Diego). Bmax determination used 1 nM RNA.
Immunoblotting. Lysate protein (5 μg) was blotted as described in ref. 10. Antibodies produced against either IRP1 or the c-myc epitope at the C terminus of IRP1, horseradish peroxidase-coupled second antibody, and a chemiluminescence kit (SuperSignal, Pierce) were used. Blots were quantified by densitometry.
Antibodies. Antibodies against IRP1 were described in ref. 9. Rabbit phosphospecific antibodies against REFNSYG(p)S711RRGNDA, where (p)S denotes phosphoserine, were made and affinity-purified (BioSource International, Camarillo, CA).
Tissue Culture. HEK 293 cells expressing IRP1 were produced by using the Flp-In T-REx system (Invitrogen).
The results are representative of at least three experiments and are reported as mean ± SEM unless noted otherwise.
Results
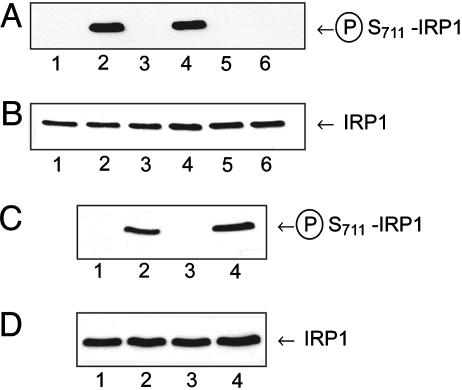
S711 Is a PKC Phosphorylation Site in IRP1. Our laboratory has demonstrated that IRP1 is an efficient substrate for PKC and that a peptide fragment of the S711 region is phosphorylated by PKC (9). To demonstrate that S711 is a phosphorylation site, phosphorylation state-specific antibodies against S711 were generated. These antibodies recognize purified recombinant WT IRP1 or the S138A mutant of IRP1 when they have been phosphorylated by PKC (Fig. 1A, lanes 2 and 4) but fail to recognize nonphosphorylated WT or S138A IRP1 (Fig. 1A, lanes 1 and 3). The S711A mutant of IRP1 was not recognized whether it was PKC phosphorylated or not (Fig. 1A, lanes 5 and 6). Reprobing of the blot from Fig. 1A with an antibody against the entire IRP1 protein showed equal loading of all samples (Fig. 1B). The S711-phosphospecific antibody also was used to probe the IRP1 phosphorylation state in HEK 293 cells expressing WT IRP1 and treated with or without phorbol 12-myristate 13-acetate (PMA) for 1 or 3 h. In cells treated with diluent only, the S711-phosphospecific antibody failed to recognize IRP1 (Fig. 1C, lanes 1 and 3). In contrast, a strong band was observed at Mr ≈100,000 1 h after treatment of the cells with 1 μM PMA, which increased in intensity by 1.7-fold after 3 h of PMA treatment (Fig. 1C, lanes 2 and 4). PMA did not alter the IRP1 protein level (Fig. 1D, lanes 1–4).
Fig. 1.
Serine-711 of IRP1 is phosphorylated in vitro and in vivo. (A and B) Purified recombinant WT (lanes 1 and 2), S138A (lanes 3 and 4), or S711A (lanes 5 and 6) rabbit IRP1 (56 ng of protein) were treated without (lanes 1, 3, and 5) or with (lanes 2, 4, and 6) PKC in the presence of 158 μg/ml phosphatidylserine, and 0.3 mM unlabeled ATP for 2 h (9). (A) Immunoblot with S711-phosphospecific antibody. (B) Reprobing of immunoblot in A with anti-rat liver IRP1 antibody (9) after stripping the membrane. (C and D) HEK 293 cells expressing WT IRP1 were treated with diluent (lanes 1 and 3) or 1 μM PMA for 1 or 3 h (lanes 2 and 4). Cells were lysed (9), and lysate protein (50 μg) was immunoblotted. (C) Immunoblot with S711-phosphospecific antibody. (D) Same blot as in C, stripped and reprobed with nonphosphospecific antibody against S138 region (9).
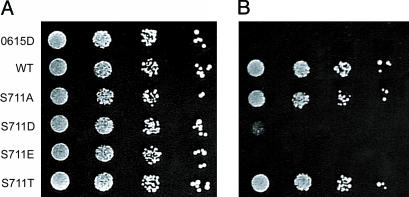
Phosphomimetic Mutations Introduced at S711 of IRP1 Impair Its Function as an Aconitase in aco1 Yeast. To investigate the role of phosphorylation of S711 on the ability of IRP1 to function as an aconitase, S711 was mutated to either aspartate (S711D) or glutamate (S711E) to mimic the effects of phosphorylation or to either nonphosphorylatable alanine (S711A) or threonine (S711T) as a further control, and the mutants were examined for their ability to rescue aconitase-deficient (aco1) yeast from glutamate auxotrophy. Aconitase-deficient (aco1) yeast cells are auxotrophic for glutamate presumably because TCA cycle flux is blocked and they cannot produce sufficient α-ketoglutarate (16, 21). When transformed with an aconitase gene, such as that encoding IRP1, aco1 yeast are rescued from glutamate auxotrophy (Fig. 2 A and B) (16). Both S711A and S711T supported aconitase-dependent growth as well as did the WT protein. However, the growth of yeast expressing the phosphomimetic S711D protein was significantly impaired in the absence of glutamate (Fig. 2B). Cells expressing S711E failed to grow in the absence of glutamate in the time frame of this experiment. In aco1 cells, the expression levels of S711A, S711D, and S711T protein were all 75–80% of the WT protein, whereas S711E was expressed at 44% of WT (see Fig. 3C). Hence, substitution of a phosphomimetic amino acid for S711 selectively reduces the ability of IRP1/c-acon to support aconitase-dependent growth.
Fig. 2.
A phosphomimetic mutant impairs aconitase function in a yeast complementation assay. The aco1 yeast strain 0615d was or was not transformed with plasmids encoding WT or S711 IRP1 mutants. Individual colonies were grown to mid-logarithmic phase and washed, and then serial 10-fold dilutions were spotted onto SD-ura plates with glutamate (A) or without glutamate (B) for 5 days (11).
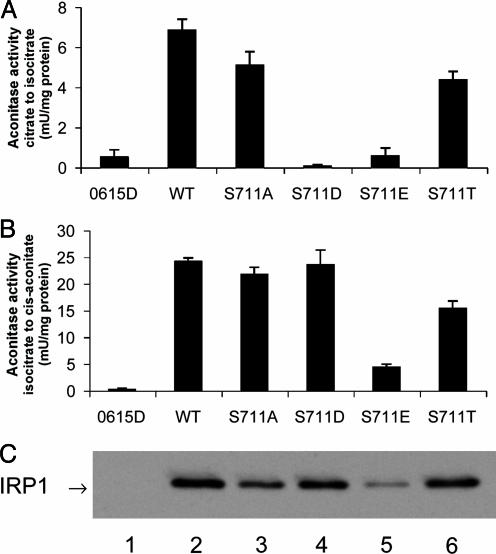
Fig. 3.
Selective inhibition of the citrate-to-isocitrate aconitase reaction by phosphomimetic mutation of S711. Extracts of transformed or nontransformed aco1 cells grown to mid-logarithmic phase (11) were assayed for aconitase activities. (A) Conversion of citrate to isocitrate (19). (B) Conversion isocitrate to cis-aconitate (20). (C) IRP1 level in extracts (5 μg of protein) determined by immunoblotting with an antibody against the myc-epitope tag on IRP1. Lane 1, nontransformed; lane 2, WT; lane 3, S711A; lane 4, S711D; lane 5, S711E; lane 6, S711T.
Rate of Conversion of Citrate to Isocitrate in Extracts of Yeast Expressing WT and S711 Mutants Mimics Growth Response. The ability of extracts from yeast grown in liquid culture to convert citrate to isocitrate was determined. Extracts of nontransformed aco1 yeast contained negligible activity (0.53 ± 0.36 mU/mg of protein) (Fig. 3A). When WT IRP1 was expressed, the aconitase activity was 6.84 ± 0.55 mU/mg of protein. S711A IRP1 displayed an activity of 5.09 ± 0.68 mU/mg of protein, which was similar to that of WT IRP1. Cells expressing S711T converted citrate to isocitrate with an activity of 4.36 ± 0.42 mU/mg of protein, which was 64% of WT activity. In extracts of cells expressing S711D (0.07 ± 0.07 mU/mg of protein) or S711E (0.56 ± 0.40 mU/mg of protein), aconitase activity was not detectable. Thus, S711D and S711E are severely impaired in the ability to convert citrate to isocitrate, consistent with their reduced ability to support glutamate-dependent growth of aco1 yeast.
S711 Phosphomimetic Mutants Actively Convert Isocitrate to cis-Aconitate. Given the proximity of S711 to residue R699, which, on the basis of sequence identity with m-acon, is required for substrate binding, it seemed relevant also to determine the ability of the WT and S711 mutants to convert isocitrate to the intermediate cis-aconitate. Furthermore, studies on the equivalent Arg residue of m-acon have shown that mutating this residue can greatly reduce enzymatic activity in general but preferentially impairs the citrate-to-isocitrate reaction (22).
Activity for the isocitrate-to-cis-aconitate reaction was not detected in extracts of nontransformed cells (0.19 ± 0.19 mU/mg of protein) (Fig. 3B). Extracts from cells expressing WT IRP1 had an activity of 24.2 ± 0.67 mU/mg of protein. Extracts containing S711A IRP1 had an activity of 21.7 ± 1.4 mU/mg of protein. Extracts from cells expressing S711T had a rate of 15.4 ± 1.4 mU/mg of protein that was 64% of the WT rate. In contrast to what was observed for the citrate-to-isocitrate reaction, both phosphomimetic mutants exhibited a rate of conversion of isocitrate to cis-aconitate that was equivalent to (S711D) or somewhat lower than (S711E) that observed for the WT protein. S711D and S711E converted isocitrate to cis-aconitate at a rate of 23.5 ± 2.8 and 4.4 ± 0.6 mU/mg of protein, respectively. The level of expression of WT and S711 IRP1 mutants expressed in aco1 cells grown in liquid culture was then determined. A representative immunoblot is shown in Fig. 3C. Analyzing three separate transformants (mean ± SEM), we found that S711A (77 ± 4%), S711D (79 ± 8%), and S711T (75 ± 11%) were expressed at a similar level, whereas S711E (44 ± 11%) was noticeably lower when expressed as a percent of WT IRP1 expression. Hence, when normalized to WT IRP1 protein level, the activity in converting isocitrate to cis-aconitate was 24.2, 28.2, 29.7, 10.0, and 20.5 mU/mg of protein for the WT, S711A, S711D, S711E, and S711T proteins, respectively.
The aconitase activity measurements were expressed as the ratio of the rates of conversion of isocitrate to cis-aconitase relative to the conversion of citrate to isocitrate. Purified native c-acon has a ratio of 2.8, and m-acon has a ratio of 3.8 (23). WT, S711A, and S711T IRP1 expressed in yeast had ratios of 3.8 ± 0.3, 4.7 ± 0.1 and 4.2 ± 0.8 (mean ± SEM, n = 3), respectively. To determine whether activity in converting citrate to isocitrate on the part of the S711D and S711E mutants could be detected, the Fe–S cluster in purified recombinant protein was reconstituted with iron and sulfide (11). After reconstitution, specific activities for WT, S138D, and S138E were 8.3, 1.1, and 0.4 U/mg of protein, respectively, for the conversion of citrate to isocitrate; the activity in converting isocitrate to cis-aconitate was 22.7, 26.5, and 8.5 U/mg of protein, respectively. Under these conditions, the ratio of the rates for isocitrate to cis-aconitate relative to the rate of conversion of citrate to isocitrate was 24:1 and 21:1 for S711D and S711E, respectively, and 2.7 for WT protein. Taken together, these results indicate that the replacement of S711 with a phosphomimetic amino acid selectively and dramatically impairs the ability of c-acon to convert citrate to isocitrate.
S711 Phosphomimetic Mutants of IRP1/c-acon Antagonize the Requirement for Cytosolic Isocitrate. The observations described above suggested that alterations of c-acon activity induced by phosphorylation of S711 may influence isocitrate metabolism. In yeast (25) and mammalian cells (26), Idp2p is important in protecting cells against oxidative stress, and mammalian Idp2p generates NADPH that is necessary for redox maintenance (24). Hence, it was reasoned that the S711D and S711E mutants of c-acon might impair the ability of yeast to survive oxidative stress by competing against Idp2, a key source of NADPH, for isocitrate.
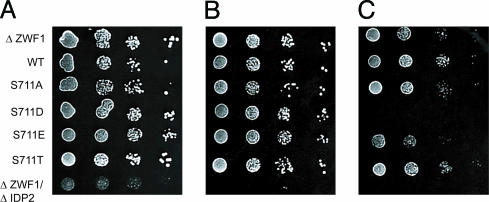
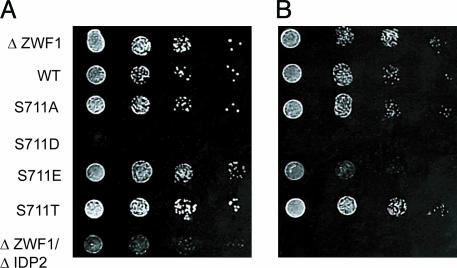
To test this hypothesis, we used Saccharomyces cerevisiae strains that are compromised in their ability to handle oxidative stress: Δzwf1, which lack glucose-6-phosphate dehydrogenase (G6PD), and Δzwf1/Δidp2, which lack G6PD and Idp2. Δzwf1/Δidp2 cells cannot survive in the presence of oxidant stress such as that induced by H2O2, because they are defective in the capacity to generate cytosolic NADPH (25). A single mutation in either gene is nonlethal when cells are exposed to oxidative stress. However, in Δzwf1 cells, Idp2 is essential for survival in response to H2O2 and other oxidants (25). Hence, we tested the ability of phosphomimetic S711D and S711E proteins to antagonize Idp2 activity and mimic the Δzwf1/Δidp2 double-mutant phenotype in Δzwf1 cells. When plated onto media with glucose as the sole carbon source, the nontransformed Δzwf1 cells, the Δzwf1 cells expressing WT or the S711 mutants of IRP1, and the Δzwf1Δidp2 cells all grew identically (results not shown). To induce oxidative stress, the cells were grown with glycerol as the carbon source, with or without H2O2. Neither the WT nor any of the S711-mutant proteins antagonized the ability of Δzwf1 cells to survive when grown on glycerol in the presence of low (0.5 mM) H2O2 (Fig. 4 A and B). The reduced ability of the Δzwf1/Δidp2 double mutant to grow was used as a positive control for the presence of oxidative stress when cells are grown on a nonfermentable carbon source. However, when Δzwf1 cells were plated on media containing a higher concentration of H2O2 (1.0 mM) (Fig. 4C), the cells expressing S711E and particularly S711D were either impaired or unable to grow, whereas the cells expressing WT, S711A, or S711T were unaffected. Interestingly, the cells expressing S711D showed the most dramatic effect, and this is the mutant that displayed both a high ratio of isocitrate-to-cis-aconitate activity relative to the citrate-to-isocitrate activity and also displayed an absolute rate of conversion of isocitrate to cis-aconitate that was similar to that observed for the WT protein. Hence, these results demonstrate that the S711 phosphomimetic mutants are specifically capable of antagonizing the requirement for cytosolic isocitrate.
Fig. 4.
Expression of S711 phosphomimetic mutants increases susceptibility to oxidative stress. The yeast double mutant Δzwf1/Δidp2 cannot grow in the presence of H2O2 (17). Δzwf1 cells were or were not transformed with plasmids expressing WT or S711 IRP1 mutants to assess their ability to antagonize Idp2p activity through competition for isocitrate. Cells were grown in SD plus kanamycin media to mid-logarithmic phase and washed, and then serial 10-fold dilutions were spotted onto SC plus glycerol plates without (A) (11) or with the addition of 0.5 mM H2O2 (B) or 1 m MH2O2 (C). Immunoblotting gave relative levels (shown in parentheses) of IRP1 in Δzwf1 extracts from cells grown in liquid culture: WT (1), S711A (0.7), S711D (0.4), S711E (1.1), and S711T (1.5) (results not shown).
The ability of WT and S711 mutants of c-acon/IRP1 to influence yeast growth when acetate is the carbon source was examined. In this situation, isocitrate is required to fuel the glyoxylate cycle. Δzwf1 cells expressing the S711D mutant of IRP1 failed to grow on acetate (Fig. 5A). The Δzwf1/Δidp2 double mutant was able to grow on acetate plates, in contrast to cells expressing the S711D mutant, showing that the observed growth defect is not solely because of an oxidative stress response but is specific to the requirement for isocitrate in certain metabolic pathways. When Δzwf1 cells were grown in the presence of acetate plus 0.5 mM H2O2, which should further enhance the need for isocitrate, expression of S711E antagonized cell growth relative to that observed for cells expressing the WT, S711A, or S711T proteins (Fig. 5B). Collectively, these results show that the direct impact of phosphomimetic S711 mutants on isocitrate usage can result in a discernible in vivo effect on cellular metabolism.
Fig. 5.
Expression of S711 phosphomimetic mutants antagonizes glyoxylate cycle function. Δzwf1 cells transformed or not transformed with plasmids expressing WT and S711 mutants were assayed for their ability to grow when acetate was the carbon source. Cells were grown to mid-logarithmic phase in SD with appropriate selections and washed, and then serial 10-fold dilutions were spotted on SC-media containing 2% acetate without (A) or with (B) 0.5 mM H2O2.
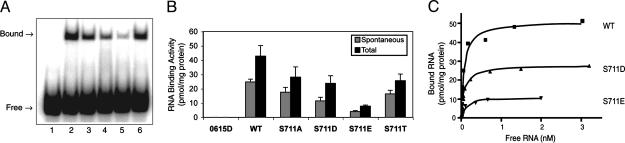
RNA-Binding Activity of WT and S711 Mutants of IRP1. Extracts containing WT IRP1 had an IRE-binding activity of 24.7 ± 1.9 pmol/mg of protein (Fig. 6 A and B). S711A and S711T mutants had RNA-binding activities of 17.5 ± 3.4 and 16.4 ± 2.5 pmol/mg of protein, respectively. Because the abundance of both S711A and S711T proteins was reduced by about 20%, compared with WT IRP1, it is apparent that these proteins bound the ferritin IRE with a similar capacity per unit of total IRP1 protein. RNA-binding activity of the phosphomimetic mutants S711D and S711E was 11.5 ± 2.5 and 4.0 ± 0.6 pmol/mg of protein, respectively. When normalized to IRP1 protein level (see Fig. 3C), the RNA-binding activity of S711D and S711E was estimated to be 15 and 9 pmol/mg of protein, which is lower than that observed for WT, S711A, or S711T IRP1. Because these differences in spontaneous IRE-binding activity could be the result of a difference in the distribution of IRP1 between its RNA-binding and aconitase forms, the extracts were treated with 2% 2-mercaptoethanol to estimate total binding activity. After 2-mercaptoethanol, the RNA-binding activity of S711A, S711D, and S711T was 70–80% of WT IRP1 when normalized to IRP1 protein level, whereas S711E was about 40% of WT IRP1, indicating that a portion of S711E was not active for RNA binding.
Fig. 6.
IRE RNA-binding activity in yeast extracts. RNA-binding activity was determined by gel-shift assay. (A) IRE RNA-binding activity was determined in yeast extracts as follows: lane 1, nontransformed; lane 2, WT IRP; lane 3, S711A; lane 4, S711D; lane 5, S711E; and lane 6, S711T. 1 nM RNA was used. (B) RNA-binding activity for transformants expressing WT or the S711 mutants of IRP1 plus nontransformed cells (n = 3 independent colonies). The spontaneous (gray bars) or 2% 2-mercaptoethanol-induced total (black bars) IRE RNA-binding activity was determined. (C) An RNA saturation analysis for extracts containing WT, S711D, or S711E IRP1.
IRP1 has been reported to bind to the ferritin IRE with a Kd ranging from 20 to 100 pM (13). As in our previous work (11), WT IRP1 expressed in yeast bound the rat L-ferritin IRE with an affinity of 34 ± 9pM(n = 3), whereas S711D and S711E exhibited a Kd of 98 ± 46 pM (n = 3) and 73 ± 20 pM (n = 2), respectively (Fig. 6C). Hence, the S711 phosphomimetic mutants retain the ability to bind the IRE with high affinity.
Discussion
Previous work from our laboratory has established that IRP1 can be phosphorylated by PKC in vitro and that there are two major phosphorylation sites (9). Ser-138 and Ser-711 were identified as putative sites of PKC action in IRP1 on the basis of the ability of the kinase to phosphorylate peptide fragments of these regions of IRP1 (9). In the study reported here, phosphorylation state-specific antibodies that recognize IRP1 when it is phosphorylated at S711 were used. By using recombinant WT and phosphorylation-site mutants of IRP1, bovine brain PKC was shown to specifically phosphorylate S711 in vitro. Furthermore, S711 was phosphorylated when HEK 293 cells expressing WT IRP1 were treated with PMA. These findings demonstrate that S711 is a phosphorylation site in vivo and set the stage for our structure/function studies of this phosphorylation site.
Initially, aco1 yeast was used as a means to determine the extent to which insertion of phosphomimetic amino acids for S711 affected the ability of IRP1 to serve as an aconitase in vivo. In this context, the S711A and S711T mutants along with WT c-acon rescued aco1 yeast from glutamate auxotrophy. In contrast, the two phosphomimetic mutants S711D and S711E were very poor or not able, respectively, to rescue aco1 cells, supporting the concept that phosphorylation at S711 impairs the ability of c-acon to convert citrate to isocitrate. This finding was further supported when the ability of yeast extracts expressing WT or S711 mutants of IRP1 to convert citrate to isocitrate was determined. Aconitase activity in yeast extracts measured in this manner was not detectable for the two phosphomimetic proteins, although a low level of activity was observed when purified protein was reconstituted with iron and sulfide. Hence, measurements of the ability of WT and S711 mutants to convert citrate to isocitrate reflected the growth results with aco1 yeast.
Given the fact that some active site mutants of m-acon display differential effects on specific reactions of m-acon (22), although greatly reducing overall activity, the ability of the S711 mutants to convert isocitrate to cis-aconitate also was examined. Interestingly, the phosphomimetic mutants, especially S711D, retained significant activity when their capacity to convert isocitrate to cis-aconitate was determined. In fact, the two phosphomimetic mutants display a ratio of rates for conversion of isocitrate to cis-aconitate relative to the citrate-to-isocitrate reaction that is on the order of 7-fold higher than WT, S711A, or S711T. Importantly, the fact that S711T behaved like WT whereas S711D displayed a very different ratio suggests that side-chain charge has a greater role than side-chain size in influencing aconitase function in this regard. To confirm these findings, the ability of the S711 phosphomimetic mutants to compete with other enzymes for isocitrate was determined in two metabolic scenarios where isocitrate is required for cell viability: (i) the use of isocitrate to generate cytosolic reducing equivalents during oxidative stress, and (ii) the requirement for isocitrate to fuel the glyoxylate cycle. In both scenarios, both phosphomimetic mutants antagonized growth of yeast requiring increased levels of cytosolic isocitrate, and the phenotype observed was directly related to the overall activity of each mutant. In other words, cells expressing S711D displayed a greater velocity of converting isocitrate to cis-aconitate and also displayed a greater antagonism of the requirement for cytosolic isocitrate. Hence, these results suggest that phosphorylation of IRP1 at S711 alters cytosolic citrate and isocitrate metabolism.
Enzymatic, spectroscopic, and crystallographic analyses of macon provide insight into how phosphorylation of IRP1 might alter specific aspects of the reaction mechanism of c-acon. First, m-acon catalyzes the second and third steps in the TCA cycle, conversion of citrate to cis-aconitate and conversion of cis-aconitate to isocitrate. The aconitase mechanism requires stereospecific trans removal of water from the Cα and Cβ carbons of citrate or isocitrate and then trans addition of water across the double bond of cis-aconitate (4). From a variety of enzymological data, it was inferred that after dehydration of citrate or isocitrate, the cis-aconitate formed must dissociate from the enzyme, rotate 180°, and rebind for the hydration to occur trans relative to the dehydration reaction and using the proton retained by the enzyme (reviewed in ref. 4). Second, the [4Fe–4S] cluster of aconitases contains one iron atom that directly binds substrate. The manner of this binding results in the different placement of the acetyl moiety of the Cγ of citrate and isocitrate in the active site (reviewed in ref. 4). This mechanism dictates that cis-aconitate must bind to the enzyme in two ways, one referred to as the citrate mode and the other as the isocitrate mode, depending on the direction of the reaction. Support for this dual binding mode comes from crystallographic studies of aconitase with various natural substrates and transition-state analogs or with sulfate bound as a model of substrate-free enzyme (27). Third, the crystallographic investigations demonstrate that there are significant structural rearrangements of the active site cleft, including formation of different salt bridges, as well as some surface-exposed regions, depending on the substrate bound (27). How might these factors contribute to the selective effect of S711 mutation on the citrate mode of c-acon activity?
It is noteworthy that S711 is modeled to be near the entrance to the cleft in c-acon and is in close proximity to R699 which, compared with m-acon, is one of several residues required for citrate or isocitrate binding. In addition, there are conformational changes in surface regions of m-acon transmitted from the active site on binding of substrate (27). One way in which phosphorylation at S711 may selectively inhibit the citrate conversion to isocitrate might involve altered formation or elimination of salt bridges in the cleft that impair binding or release of cis-aconitate in the citrate mode or perhaps reduce the ability to displace citrate from the enzyme. Release of cis-aconitate is the rate-limiting step for m-acon (28). The work described in Fig. 3A used 1 mM citrate, which argues against a mere Km effect of the S711 mutations because the Km of c-acon for citrate is 1.5 μM (M. C. Kennedy, personal communication). Taken together, the results reported herein suggest that phosphorylation selectively impairs one mode of c-acon enzymatic action and provide a unique example of how phosphorylation may alter enzyme function.
Whereas there is evidence that a number of TCA cycle enzymes, including m-acon, are phosphoproteins, there are few examples where the consequences of phosphorylation are clearly understood (29). A well defined example is the case of E. coli isocitrate dehydrogenase (30), where phosphorylation at the active site residue S113 inactivates the enzyme when acetate is the sole carbon source and isocitrate flux through the TCA cycle is decreased to provide for increased flux through the glyoxylate cycle. Our results support the concept that phosphorylation of S711 selectively impairs the citrate mode of c-acon action in mammalian cells and that the metabolic consequences in the cytosol might include enhanced accumulation of citrate and/or a depletion of isocitrate and possibly NADPH. It will be of interest to determine whether S711 phosphorylation reduces cytosolic isocitrate levels and impairs the defense against oxidative stress in mammalian cells as the S711 phosphomimetic mutants do in yeast.
The demand for cytosolic citrate might increase in iron deficiency as an adaptive response to alter fuel metabolism in the face of impaired muscle mitochondrial function. Iron deficiency can lead to high liver and serum triglycerides (reviewed in ref. 31). The reasons for this effect are unclear but may relate to reduced muscle use of fatty acids as a fuel because of impaired muscle mitochondrial function. Increased accumulation and storage of triglyceride might represent a means to store fuel in anticipation of future iron sufficiency and proper muscle mitochondrial function (31). Alternatively, the increased requirement for glucose as a fuel source for iron-deficient muscle might require phosphorylation of liver c-acon to enhance liver gluconeogenic capacity (31). Recent studies suggest there is increased citrate export from iron-deficient liver mitochondria (K. Ross and R.S.E., unpublished observations). In terms of a more direct use in iron metabolism, accumulation of citrate might expand its use as an iron carrier, possibly for enhancing mitochondrial uptake of iron when mammalian cells are iron deficient. On a similar theme, and given the conservation of the S711 region in bacterial acnA enzymes, it is tempting to speculate that if S711 phosphorylation occurs in E. coli, then citrate accumulation and export could be of use in iron uptake by the ferric citrate transport system (32). In this regard, it is of interest to note that acnA is regulated by Fur, a key transcription factor in E. coli iron homeostasis (5, 6). A role for E. coli aconitases in iron metabolism has been suggested, but this role is focused on the effects of loss of the Fe–S cluster primarily in acnB (5, 6). It will be of interest to determine whether S711 phosphorylation is altered as a function of iron status.
What are the relative roles of phosphorylation of S138 and S711 in controlling IRP1/c-acon function in mammalian cells? Our work described here supports the hypothesis that phosphorylation of S711 preferentially affects the aconitase function of IRP1. In contrast, previous work from our laboratory (9–13) indicates that although S138 phosphorylation greatly destabilizes the Fe–S cluster in IRP1 and consequently indirectly alters c-acon activity, the primary effect of S138 phosphorylation is to alter the set point and mechanism of iron regulation of the RNA-binding activity. Hence, it appears that phosphorylation of S138 or S711 by PKC or other kinases may serve to dictate the use of IRP1/c-acon in cellular metabolism and regulation. Future studies should be directed at determining the effects of IRP1 phosphorylation at these sites, the factors that induce changes in S138/711 phosphorylation, their kinetics of in vivo phosphorylation, and which phenotype predominates when both sites are phosphorylated.
Acknowledgments
We thank L. McAlister-Henn for providing yeast strains, the W. Walden and E. Craig labs for advice on yeast work, the P. Bertics lab for PKC, D. Stout for insightful suggestions, and T. Carpenter for technical assistance. This study was supported in part by National Institutes of Health Grant DK47219 and U.S. Department of Agriculture Grant 01-35200-10683. J.S.P. and S.L.C. were supported by National Institutes of Health Grant T32 DK07665, and A.V. was supported by U.S. Department of Agriculture Hatch Project No. 3951.
Abbreviations: c-acon, cytosolic aconitase; Idp2, cytosolic NADP-linked isocitrate dehydrogenase; IRP, iron-regulatory protein; IRE, iron-responsive element; m-acon, mitochondrial aconitase; PMA, phorbol 12-myristate 13-acetate; U, enzyme activity unit.
References
- 1.Eisenstein, R. S. (2000) Annu. Rev. Nutr. 20, 627–662. [DOI] [PubMed] [Google Scholar]
- 2.Cairo, G., Recalcati, S., Pietrangelo, A. & Minotti, G. (2002) Free Radical Biol. Med. 32, 1237–1243. [DOI] [PubMed] [Google Scholar]
- 3.Gruer, M. J., Artymiuk, P. J. & Guest, J. R. (1997) Trends Biochem. Sci. 22, 3–6. [DOI] [PubMed] [Google Scholar]
- 4.Beinert, H., Kennedy, M. C. & Stout, C. D. (1996) Chem. Rev. (Washington, D.C.) 96, 2335–2373. [DOI] [PubMed] [Google Scholar]
- 5.Jordan, P. A., Tang, Y., Bradbury, A. J., Thomson, A. J. & Guest, J. R. (1999) Biochem. J. 344, 739–746. [PMC free article] [PubMed] [Google Scholar]
- 6.Varghese, S., Tang, Y. & Imlay, J. A. (2003) J. Bacteriol. 185, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alen, C. & Sonenshein, A. L. (1999) Proc. Natl. Acad. Sci. USA 96, 10412–10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang, Y. & Guest, J. R. (1999) Microbiology 145, 3069–3079. [DOI] [PubMed] [Google Scholar]
- 9.Eisenstein, R. S., Tuazon, P. T., Schalinske, K. L., Anderson, S. A. & Traugh, J. A. (1993) J. Biol. Chem. 268, 27363–27370. [PubMed] [Google Scholar]
- 10.Schalinske, K. L. & Eisenstein, R. S. (1996) J. Biol. Chem. 271, 7168–7176. [DOI] [PubMed] [Google Scholar]
- 11.Brown, N. M., Anderson, S. A., Steffen, D. W., Carpenter, T. B., Kennedy, M. C., Walden, W. E. & Eisenstein, R. S. (1998) Proc. Natl. Acad. Sci. USA 95, 15235–15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, N. M., Kennedy, M. C., Antholine, W. E., Eisenstein, R. S. & Walden, W. E. (2002) J. Biol. Chem. 277, 7246–7254. [DOI] [PubMed] [Google Scholar]
- 13.Schalinske, K. L., Anderson, S. A., Tuazon, P. T., Chen, O. S. & Eisenstein, R. S. (1997) Biochemistry 36, 3950–3958. [DOI] [PubMed] [Google Scholar]
- 14.Fillebeen, C., Chahine, D., Caltagirone, A., Segel, P. & Pantopoulos, K. (2003) Mol. Cell. Biol. 23, 6973–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaldy, P., Menotti, E., Moret, R. & Kuhn, L. C. (1999) EMBO J. 18, 6073–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narahari, J., Ma, R., Wang, M. & Walden, W. E. (2000) J. Biol. Chem. 275, 16227–16234. [DOI] [PubMed] [Google Scholar]
- 17.Minard, K. I., Jennings, G. T., Loftus, T. M., Xuan, D. & McAlister-Henn, L. (1998) J. Biol. Chem. 273, 31486–31493. [DOI] [PubMed] [Google Scholar]
- 18.Marini, F., Naeem, A. & Lapeyre, J. N. (1993) Nucleic Acids Res. 21, 2277–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose, I. A. & O'Connell, E. L. (1967) J. Biol. Chem. 242, 1870–1879. [PubMed] [Google Scholar]
- 20.Kennedy, M. C., Emptage, M. H., Dryer, J.-L. & Beinert, H. (1983) J. Biol. Chem. 258, 11098–11105. [PubMed] [Google Scholar]
- 21.Ogur, M., Coker, L. & Ogur, S. (1964) Biochem. Biophys. Res. Commun. 14, 193–197. [DOI] [PubMed] [Google Scholar]
- 22.Zheng, L., Kennedy, M. C., Beinert, H. & Zalkin, H. (1992) J. Biol. Chem. 267, 7895–7903. [PubMed] [Google Scholar]
- 23.Kennedy, M. C., Mende-Mueller, L., Blondin, G. A. & Beinert, H. (1992) Proc. Natl. Acad. Sci. USA 89, 11730–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickinson, D. A. & Forman, H. J. (2002) Ann. N.Y. Acad. Sci. 973, 488–504. [DOI] [PubMed] [Google Scholar]
- 25.Minard, K. I. & McAlister-Henn, L. (2001) Free Radical Biol. Med. 31, 832–843. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S. M., Koh, H. J., Park, D. C., Song, B. J., Huh, T. L. & Park, J. W. (2002) Free Radical Biol. Med. 32, 1185–1196. [DOI] [PubMed] [Google Scholar]
- 27.Lauble, H. & Stout, C. D. (1995) Proteins 22, 1–11. [DOI] [PubMed] [Google Scholar]
- 28.Schloss, J. V., Emptage, M. H. & Cleland, W. W. (1984) Biochemistry 23, 4572–4580. [DOI] [PubMed] [Google Scholar]
- 29.Bykova, N. V., Egsgaard, H. & Moller, I. M. (2003) FEBS Lett. 540, 141–146. [DOI] [PubMed] [Google Scholar]
- 30.Hurley, J. H., Dean, A. M., Sohl, J. L., Koshland, D. E. & Stroud, R. M. (1990) Science 249, 1012–1016. [DOI] [PubMed] [Google Scholar]
- 31.Ross, K. L. & Eisenstein, R. S. (2003) J. Nutr. 133, 1510S–1516S [DOI] [PubMed] [Google Scholar]
- 32.Braun, V. & Killman, H. (1999) Trends Biochem. Sci. 24, 104–109. [DOI] [PubMed] [Google Scholar]