Abstract
Circadian clocks are widespread endogenous mechanisms that control the temporal pattern of diverse biological processes, including gene transcription. KaiA is the positive element of the cyanobacterial clock because KaiA overexpression elevates transcription levels of clock components. Recently, we showed that the structure of KaiA is that of a domain-swapped homodimer. The N-terminal domain is a pseudo-receiver; thus, it is likely to be involved in signal transduction in the clock-resetting pathway. The C-terminal domain of KaiA is structurally novel and enhances the KaiC autokinase activity directly. Here, we report the NMR structure of the C-terminal domain of KaiA (ThKaiA180C) in complex with a KaiC-derived peptide from the cyanobacterium Thermosynechococcus elongatus BP-1. The protein–peptide interface is revealed to be different from a model that was proposed earlier, is stabilized by a combination of hydrophobic and electrostatic interactions, and includes many residues known to produce a circadian-period phenotype upon substitution. Although the structure of the monomeric subunit of ThKaiA180C is largely unchanged upon peptide binding, the intersubunit dimerization angle changes. It is proposed that modulation of the C-terminal KaiA domain dimerization angle regulates KaiA–KaiC interactions.
Evolutionarily divergent organisms possess mechanisms by which biological processes are regulated rhythmically with an ≈24-h period (1, 2). Such mechanisms, known as circadian clocks, have been found in multiple eukaryotic groups, including insects, plants, fungi, and mammals (3), as well as in a single prokaryotic group, cyanobacteria (4). The cyanobacterial circadian system is potentially the oldest (5) and simplest identified thus far. Three genes, kaiA, kaiB, and kaiC, are necessary for rhythmic transcription (4), whereas virtually all promoter elements of Synechococcus elongatus (PCC 7942), as well as those introduced from other organisms, are under circadian control (4, 6, 7). The KaiA, KaiB, and KaiC proteins associate in vitro (8) and in vivo in a circadian-oscillating manner (9), and amino acid substitutions in these proteins directly affect protein–protein interactions and the circadian period (4, 10, 11). Light resetting of the phase of the circadian clock is likely to proceed through a signal transduction cascade initiated by the sensory protein CikA (12).
Overexpression of KaiC attenuates global transcription levels (4, 7). In vitro studies show binding of KaiC to forked DNA (13); thus, KaiC has been proposed to interact directly with DNA in vivo and affect transcription levels by global chromatin remodeling (14). KaiC shares sequence similarity with the DnaB/RecA bacterial helicase superfamily (15), and recent electron microscopy studies show that KaiC forms double ring-shaped hexameric particles in the presence of ATP or nonhydrolysable ATP analogs (13, 16, 17), with an ≈20-Å-diameter pore present at the center (13, 17). KaiC has an autokinase activity as demonstrated in vitro (11, 14, 18) and in vivo (11, 14, 19). The equilibrium between phosphorylated and unphosphorylated KaiC oscillates with a circadian pattern (11), and the KaiC autokinase activity is enhanced by KaiA in vitro (11, 14, 18) and in vivo (11, 14). Regulation of the KaiC phosphorylation state by means of KaiA enhancement is believed to be important for the circadian mechanism (11, 14, 18, 19), yet no details of the KaiA–KaiC interaction have been available.
We recently showed that KaiA is a two-domain protein with an N-terminal pseudo-receiver and a structurally novel, KaiC-interacting C-terminal domain (17, 18). The C-terminal domain is sufficient to enhance the autokinase activity of KaiC in vitro, and we proposed (18) that protein–protein interactions at the N-terminal pseudo-receiver domain of KaiA function in signal-input to the clock. The putative N-terminal domain protein–protein interactions are thought to modulate KaiA C-terminal domain enhancement of the KaiC autokinase activity (18). The NMR structure of the dimeric C-terminal domain revealed a likely site for the KaiA–KaiC interaction at the groove between the two KaiA subunits (17). The recently completed x-ray structure of full-length KaiA from S. elongatus shows a domain-swapped homodimer where the N-terminal pseudo-receiver of one subunit interacts with the C-terminal effector domain of the other subunit (20). The interdomain interactions are relatively weak, which led to the hypothesis of two possible KaiA states in solution, “closed” and “open,” depending on the association or dissociation of the N- and C-terminal domains (20). In addition, the x-ray structure of KaiA from Anabaena sp. PCC 7120 (essentially an independent C-terminal KaiA domain) was recently solved (21). Individually, the x-ray structures of the monomeric subunits of KaiA are virtually identical to the NMR structures, although the dimerization angle of the C-terminal domain differs by ≈20° (20).
Here, we report the identification of a Thermosynechococcus elongatus BP-1 KaiC-derived peptide (residues 488–518, referred to as CIIABD) that interacts directly with the C-terminal domain of T. elongatus KaiA (residues 180–283, referred to as ThKaiA180C), and we present the NMR structure of the resulting ≈32-kDa complex. CIIABD binding on ThKaiA180C alters the ThKaiA180C dimerization angle, suggesting that KaiA–KaiC affinity can be modulated by changes in the dimerization geometry of the KaiA C-terminal domain. We propose that such changes in quaternary structure occur in vivo to regulate KaiC autokinase activity enhancement by KaiA and achieve clock-resetting, possibly by means of interactions of the N-terminal domain of KaiA with upstream signal transduction proteins (18).
Materials and Methods
Protein Preparation. Details of expression and purification of ThKaiA180C, as well as preparation of 13C-enriched, unenriched heterodimeric ThKaiA180C samples, have been described (17, 22). Segments of the T. elongatus KaiC gene that encode the desired peptide sequences were cloned in a pET32a+ vector (Novagen), thereby creating thioredoxin–polyHis–peptide fusions. The unmodified pET32a+-encoded thioredoxin fusion was used as control peptide for KaiA-binding experiments. Escherichia coli BL21(DE3) (Novagen) was transformed with the resulting plasmids and grown in Luria broth or in minimal medium containing 15N-enriched ammonium chloride and unenriched or 13C-enriched glucose (Cambridge Isotope Laboratories, Cambridge, MA) as sole nitrogen and carbon sources, respectively. Expression of peptide fusion constructs was induced by adding isopropyl-β-d-thiogalactopyranoside (Calbiochem) to a final concentration of 1 mM. The cells were harvested after 4 h, resuspended in a buffer containing 50 mM NaCl and 20 mM Tris·Cl (pH 7.4), and lysed by heat treatment at 60°C for 15 min. Cell lysates were separated by centrifugation, and the fusion peptide was purified from the supernatant fraction by metal affinity chromatography. The sample was buffer exchanged to 50 mM NaCl/20 mM Tris·Cl, pH 7.4, and cleaved by using enterokinase (Novagen). Cleavage by this enzyme results in the addition of three non-KaiC-derived residues (AMA) at the N terminus of the peptide. Peptides were isolated by reverse-phase chromatography and lyophilized. Peptide identity and purity were confirmed by matrix-assisted laser desorption ionization–time-of-flight spectroscopy, and quantification was carried out by amino acid analysis (Protein Chemistry Laboratory, Texas A&M University). NMR samples consisted of ≈1.2 mM ThKaiA180C/peptide in a 1:1 molar ratio. NMR buffer conditions were 20 mM NaCl/20 mM Na2HPO4, pH 7.07, at 23°C.
NMR Experiments. All experiments were performed at 50°C by using Inova 600- and 500-MHz spectrometers (Varian) equipped with triple-axis gradient probes. Sequential 13C, 15N, and 1H-backbone, side-chain, and stereospecific assignments of ThKaiA180C were performed as described in refs. 18 and 22. CIIABD peptide assignments were performed in a similar fashion by using a sample of uniformly 15N, 13C-enriched CIIABD peptide and unenriched ThKaiA180C. Interproton nuclear Overhauser effect (NOE) distance restraints were obtained by 13C, 13C-edited 4D NOESY (in a sample containing 13C-enriched ThKaiA180C and unenriched CIIABD peptide), 15N-edited 3D NOESY (15N-enriched ThKaiA180C and unenriched CIIABD peptide), 13C-edited, 12C-filtered 3D NOESY (13C-enriched ThKaiA180C and unenriched CIIABD peptide), 13C-edited, 12C-filtered 3D NOESY (heterodimeric 13C-enriched, unenriched ThKaiA180C and 13C-enriched CIIABD peptide), and 13C-filtered, 13C-edited 3D NOESY (unenriched ThKaiA180C and 13C-enriched CIIABD peptide) spectra. Hydrogen bond restraints were applied based on hydrogen-exchange protection data collected by NMR, as well as the existence of expected regular secondary structure NOEs (23). The isomerization state of all proline residues was determined as trans (23). Backbone dynamics measurements (15N T1, 15N T2 and 15N{—1H} NOE) were performed and analyzed as described in ref. 24. Spectrometer temperature was calibrated by using a methanol sample. Backbone and side-chain chemical shifts of ThKaiA180C and CIIABD have been deposited in the BioMagResBank (accession no. 6174).
NMR Structure Calculations. The ϕ and ψ dihedral angle values were derived from the TALOS database (25). For residues for which all 10 predictions lie in the same region of the Ramachandran plot, the restraint boundaries were set to two SDs. Residues with nine predictions in the same region of the Ramachandran plot were restrained within two SDs plus 10°. The x-plor-nih software package (26) was used for all stages of NMR structure calculations, which were performed in a manner identical to that described in ref. 17. Noncrystallographic symmetry was enforced between the two monomeric ThKaiA180C subunits and between the two CIIABD peptide chains throughout the calculation. However, to better sample the possible conformations of the intermolecular disulfide bond (C272 with C272′, where prime denotes the second subunit), C272 of ThKaiA180C was excluded from this symmetry term. In the final family of structures, this disulphide bridge shows variable geometry, and therefore, we cannot place it in one of the well defined classes of disulphide bonds (27). However, the Hβ2/3 and Cβ C272 resonances show significant broadening in all of the acquired spectra, indicating that in solution, the geometry of the disulphide bridge varies on a millisecond–microsecond timescale. The family of structures consists of the 25 lowest-energy structures that satisfy all experimental restraints (see Table 1, which is published as supporting information on the PNAS web site). The structures and structure-calculation restraints have been deposited in the Protein Data Bank (accession nos. 1SUY and 1SV1, for the average minimized structure and the 25-structure ensemble, respectively).
Figure Preparation and Structure Notes. Figures were prepared by using spock(28). The three N-terminal non-KaiA-derived residues of ThKaiA180C and, similarly, the three N-terminal non-KaiC-derived residues of CIIABD peptide are not shown.
Results
Identification of a KaiA Binding Site in KaiC. Despite the importance of KaiA–KaiC interactions in the circadian clock (11), only one study (29) has focused on identifying the KaiA binding site of KaiC. Taniguchi et al. (29) were able to locate two different regions of S. elongatus KaiC, residues 206–263 and 418–519, that interacted with KaiA in yeast two-hybrid and in vitro coimmobilization assays. From these results, and KaiC structure modeling based on the KaiC–helicase similarity, the authors predicted that the KaiA–KaiC interaction occurs at the interface between the predicted CI and CII domains of KaiC (8).
Based on sequence similarity and fold recognition (30), we expect KaiC to be composed of two RecA-like domains (31), spanning residues 19–240 and 261–472 of the T. elongatus KaiC sequence. These domains presumably occupy the two globular domains observed in the KaiC electron microscopy structures (13, 16, 17). The highly conserved residues 241–260 (90% sequence identity between the S. elongatus and T. elongatus KaiC) constitute a sequence linker between the two RecA-like domains and probably occupy the physical linker observed in the KaiC electron microscopy structure (17). The C-terminal residues 473–518 are highly conserved as well (see Fig. 6, which is published as supporting information on the PNAS web site). A small non-RecA-like N-terminal KaiC sequence was not considered here because of a lack of sequence conservation.
The peptide fragments corresponding to residues 241–260 and 473–518 of T. elongatus KaiC were expressed and purified as fusions to thioredoxin, as described in Materials and Methods. Binding to ThKaiA180C was tested by titrating unlabeled KaiC peptide fusions to 15N-enriched ThKaiA180C with stoichiometric ratios of 1:1 and by acquiring 1H, 15N heteronuclear single quantum correlation (HSQC) spectra. No change was observed in these spectra when the fusion of thioredoxin to KaiC residues 241–260 was used (data not shown). In contrast, the fusion construct of KaiC residues 473–518 caused dramatic changes in the 1H, 15N HSQC spectra of ThKaiA180C (see Fig. 7B, which is published as supporting information on the PNAS web site). Because no such changes are seen when titrating a control peptide fusion construct (see Fig. 7C) or the fusion of KaiC residues 241–260, we attribute this change to a specific interaction of ThKaiA180C with KaiC residues 473–518.
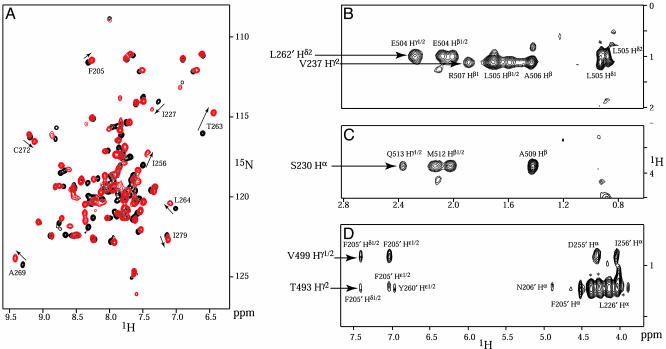
A shorter peptide fragment spanning residues 488–518 (CIIABD; see Fig. 6) with the thioredoxin tag removed was sufficient to produce similar changes as the larger constructs in the ThKaiA180C 1H, 15N HSQC spectra, whereas the quality of the spectra improved (Figs. 1A and 7B). Constructs with further N-terminal truncations (spanning residues 496–518) or C-terminal truncations (residues 488–511) showed decreased binding, as indicated by the higher stoichiometric ratios that are necessary to produce a change in the HSQC spectra. CIIABD is part of the second fragment identified by Taniguchi et al. (29) as interacting with KaiA, and it includes a site (T495) where an amino acid substitution is known to induce a clock-period phenotype (4). An HSQC spectrum of 15N-enriched ThKaiA180C in complex with CIIABD at a 1:1 ratio is shown in Fig. 1 A and is compared with an HSQC spectrum of free ThKaiA180C. Higher ratios of CIIABD to ThKaiA180C did not produce any further visible changes in the HSQC spectra. Substoichiometric titrations of CIIABD to ThKaiA180C produce HSQC spectra with gradual appearance and disappearance of the bound and free ThKaiA180C resonances, suggesting a slow timescale of binding. Only one set of resonances is observed in the 1H, 15N HSQC spectra of bound ThKaiA180C. Therefore, the complex is symmetric on the NMR timescale, which is consistent with two CIIABD peptides being bound to a single ThKaiA180C dimer.
Fig. 1.
Observation of ThKaiA180C–CIIABD interactions. (A) The 1H, 15N HSQC spectra of 15N-enriched ThKaiA180C, both free (black) or in a 1:1 complex with unenriched CIIABD peptide (red). Some of the ThKaiA180C residues that undergo significant chemical shift changes are labeled, and the changes are indicated by arrows. The residues that experience significant chemical-shift change are at the ThKaiA180C–CIIABD and ThKaiA180C dimerization interfaces. Strips from 3D 13C-edited, 12C-filtered NOESY spectra acquired on 1:1 complexes of 13C-enriched ThKaiA180C and unenriched CIIABD (B and C) and unenriched ThKaiA180C and 13C-enriched CIIABD (D) are also shown. Several NOESY crosspeaks of specific interactions between ThKaiA180C and CIIABD are labeled. *, Crosspeaks with a maximum in intensity on adjacent 13C planes.
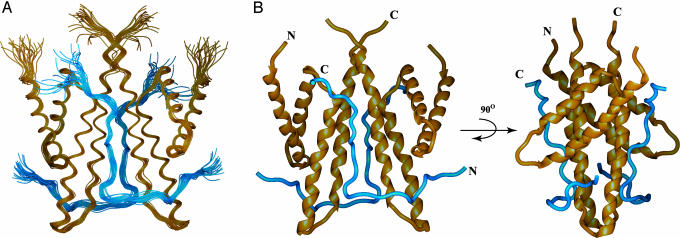
Description of the Structure. The solution structure of the ThKaiA180C dimer in complex with two CIIABD peptides was calculated from 2,420 experimental restraints per symmetry unit (see Table 1) by using the x-plor-nih software package (26). The ThKaiA180C–CIIABD interface is well defined, with 166 NOE-derived distance restraints connecting the protein and peptide. Fig. 1 B–D shows strips from the 13C-edited/12C-filtered NOESY spectra where some of these NOEs are identified. The precision of the final family of 25 structures (Fig. 2A) was calculated against the average structure for all ordered residues (defined residues of ThKaiA180C with 15N{—1H} NOE values >0.6 and CIIABD residues 490–513; see Fig. 8, which is published as supporting information on the PNAS web site) and is 0.33 ± 0.07 Å for the backbone and 0.76 ± 0.08 Å for heavy atoms of the ThKaiA180C dimer, and 0.47 ± 0.08 for the backbone and 0.89 ± 0.09 for heavy atoms of the complex (see Table 1).
Fig. 2.
Structure of the ThKaiA180C–CIIABD complex. Shown are the overlaid backbones of the 25-structure ensemble (PDB ID code 1SV1) (A) and the average minimized structure (PDB ID code 1SUY) in two mutually orthogonal views (B). ThKaiA180C backbones are shown in gold, and CIIABD is shown in blue.
The overall fold of ThKaiA180C of the complex is that of a right-turning four-helix bundle (32) with the four α-helices (α1–α4) organized in two antiparallel helix–loop–helix pairs. The two helix–loop–helix pairs pack together at a wide angle (≈50° on average). As noted in ref. 17, this wide angle of packing is unusual and places ThKaiA180C in the so-called X-class of four-helix bundles (33). ThKaiA180C dimerizes along the C-terminal half of the long α4 helix by using primarily coiled-coil hydrophobic interactions with α4′. Additional hydrophobic interactions, as well as an intersubunit salt bridge and two putative hydrogen bonds, are formed between α4 and the loop connecting α2′ and α3′, thereby stabilizing the dimer interface. Four of the amino acid substitutions of S. elongatus KaiA that are known to affect clock period can be mapped to identical or chemically similar residues in the ThKaiA180C hydrophobic core (10, 17), whereas three substitutions involve residues of the dimer interface. Substitutions of these residues likely destabilize the ThKaiA180C structure.
The two CIIABD peptides bound to a ThKaiA180C dimer have an extended L-shaped conformation, as shown in Fig. 2B. Starting at the N terminus, each peptide chain crosses the wide angle formed between the helix–loop–helix pairs of one ThKaiA180C subunit, forms a tight turn, and follows the groove between the two ThKaiA180C subunits (Figs. 2B and 3). The C terminus of each peptide chain interacts with the N terminus of α3′. Thus, it is clear that KaiA dimerization is necessary for CIIABD (and presumably KaiC) binding because each peptide chain makes extensive contacts with both ThKaiA180C subunits. It is worth noting that the observed arrangement of CIIABD and ThKaiA180C is different from the hypothesis in ref. 17. Our earlier model predicted that a KaiC-derived peptide would interact with ThKaiA180C by passing through the groove between the ThKaiA180C subunits. Instead, CIIABD lies along that groove. Further, CIIABD binding completely occludes the groove, as indicated by accessible surface area calculations of the entire complex (data not shown). Thus, we expect that additional KaiC interactions inside the ThKaiA180C groove are unlikely. The two symmetry-related CIIABD peptides could correspond to two KaiC molecules of the same hexameric KaiC particle or, alternatively, to two different hexameric KaiC particles in close proximity.
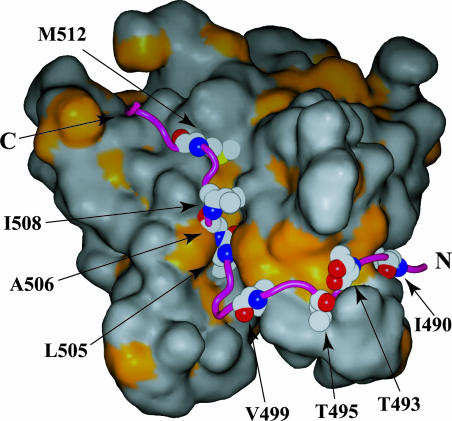
Fig. 3.
Hydrophobic interactions in the ThKaiA180C–CIIABD complex. An accessible surface area representation of ThKaiA180C is shown, with the area of exposed hydrophobic side chains shown in gold. CIIABD (purple) is anchored mainly on a series of hydrophobic patches through mostly conserved nonpolar residues (labeled). The CIIABD N and C termini are indicated.
Description of the ThKaiA180C–CIIABD Interface. CIIABD forms a combination of hydrophobic, electrostatic, and hydrogen bonding interactions with ThKaiA180C (see Fig. 6) that result in burial of ≈1,400 Å2 per peptide unit. An accessible surface area representation of ThKaiA180C, with the surface of hydrophobic side chains shown in gold, is shown in Fig. 3. The peptide (purple) lies along a series of mostly conserved hydrophobic patches created by the L202, F205, F243, I256, L258, L262, I265, L233′, V237′, and L257′ residues of ThKaiA180C (see Fig. 6). A total of eight hydrophobic side chains anchor CIIABD to that surface (I490, T493, T495, V499, L505, A506, I508, and M512). Several electrostatic interactions between ThKaiA180C and CIIABD can be inferred from the structure. Charge–charge interactions are seen between D500 of CIIABD and K248′ of ThKaiA180C: E501 with K248′, K502 with D241′, K502 with E238′, and K510 with D266. Also, a large number of putative hydrogen bonds between ThKaiA180C and CIIABD can be seen in many structures of the ensemble, which would increase the specificity of interaction. These hydrogen bonds occur between the C′ and N of I490 of CIIABD and Oε1 and Nε2 of Q246 of ThKaiA180C, respectively, as well as between C′ of P494 and Nε of R252; N and Oγ1 of T495 with C′ and N of S253; Nη2 of R496 and C′ of F205; Oε1 and Oε2 of E501 and C′ and Oγ of S244′; Nζ of K510 and Nδ1 of H270; Sδ of M511 and Oε2 of E273; and Nε2 of Q513 and Oε1 of E234′.
The importance of this peptide–protein interface for the correct function of the circadian clock is demonstrated by the significant number of clock-period phenotype amino acid substitutions that can be mapped here (4, 10). Six known substitutions are mapped to E238, D241, F243, S244, K248, and E273 of ThKaiA180C, which are residues that provide important interactions for peptide binding as revealed by the structure (see Fig. 6). A seventh ThKaiA180C amino acid with a clock-period phenotype substitution, T242, is placed near the flexible N terminus of CIIABD and is possibly implicated in full-length KaiC binding. In CIIABD, T495 is responsible for important hydrophobic and putative hydrogen bonding interactions to ThKaiA180C (see Fig. 6). The NMR structure suggests that substitution of these residues is likely to weaken the KaiA–KaiC interaction, thereby causing the observed lengthening of the clock period (4, 10).
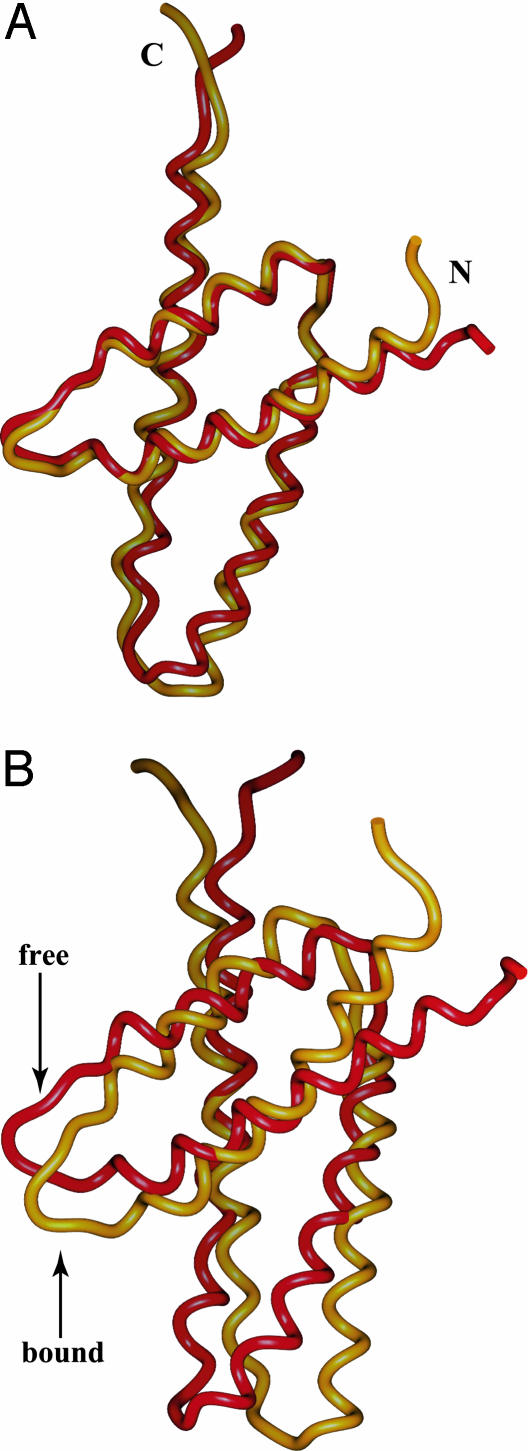
Comparison of Free and Bound ThKaiA180C. The ThKaiA180C monomeric subunit is relatively unchanged upon CIIABD binding and maintains the unusually wide angle of packing between the two helical pairs (≈50° in the bound and free forms) (17). The backbone rms deviation of the average minimized structures of free and bound ThKaiA180C is 1.27 Å over all ordered residues (inclusive, 187–208, 212–247, and 258–279) (17). The most important differences occur at the C-terminal part of α3 and the N-terminal part of α4, where CIIABD makes several contacts. Exclusion of that region (ThKaiA180C residues 241–247 and 258–260) improves the superposition to 0.94 Å. This case is shown in Fig. 4A.
Fig. 4.
Comparison of the structures of C-terminal KaiA domains. Shown here are the backbone representations of free ThKaiA180C (red) (17) and ThKaiA180C when in complex with CIIABD (gold). The N and C termini of chains are indicated. The monomeric subunits of the two domains are superimposed and shown in A. The backbone rms deviation is 0.94 Å for free vs. bound ThKaiA180C. (B) Superpositions of one of the subunits shows differing rotations around the dimerization interface of the second subunits. (For clarity, the superimposed subunits are not shown.)
The biggest change between free and bound ThKaiA180C is in the angle of dimerization. As shown in Fig. 4B, when the monomeric subunits of ThKaiA180C are superimposed, the second monomeric subunits show a relative rotation around the dimerization interface. This rotation is driven by insertion of CIIABD nonpolar side chains (L505 and A506) in the ThKaiA180C dimerization groove, where they form a hydrophobic cluster with side chains of residues at or near the ThKaiA180C dimerization interface (L233, H236, L264, and I265′). A total of eight NOESY crosspeaks were unambiguously identified from the 13C-edited, 12C-filtered NOESY spectra as connecting this cluster. In addition, ThKaiA180C residues at or near the dimer interface (I227, Y275, and I279), but relatively far in space from the ThKaiA180C–CIIABD interactions also show significant shifts in the 1H, 15N HSQC spectra upon CIIABD binding (Fig. 1 A), which also suggests changes of the quaternary dimer structure.
Discussion
The ThKaiA180C–CIIABD complex structure discussed here provides structural explanations for earlier mutant phenotypes (4, 10). It is important to note that the changes observed in the quaternary structure of the ThKaiA180C dimer upon binding CIIABD should be bidirectional; i.e., protein–KaiA interactions that influence the dimerization angle of the C-terminal domain of KaiA will thereby also alter the affinity of KaiA for KaiC. Thus, we propose that protein–protein interactions at the N-terminal pseudo-receiver domain of KaiA, possibly from a signal cascade originating at the sensory protein CikA (12), alter the dimerization angle of the C-terminal domain of KaiA. In this way, the KaiA–KaiC affinity and KaiA enhancement of the KaiC autokinase activity can be regulated, and the clock can be reset in response to environmental stimuli (Fig. 5). KaiA proteins from filamentous cyanobacteria, such as Anabaena sp. PCC 7120 and Nostoc punctiforme, do not include an N-terminal pseudoreceiver domain (18). It is possible that in these cases the putative signal cascade directly interacts with the KaiA C-terminal domain and suggests differences in the circadian clock resetting mechanism between filamentous and nonfilamentous cyanobacterial species. Previous KaiC autokinase activity assays show that full-length KaiA stimulates KaiC phosphorylation to a higher extent than the C terminus of KaiA alone (18). These results can be interpreted by our proposal because the dimerization angle was found to be different between the full-length KaiA structure and that of the free C-terminal KaiA domain in solution (20). However, the quaternary structural differences observed between the crystal and solution structures could also be induced by packing forces in the crystal.
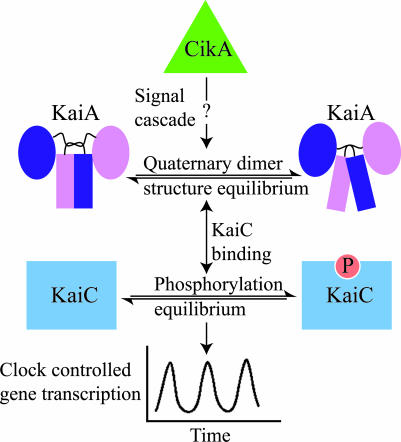
Fig. 5.
Putative mechanism for clock resetting. An upstream signal cascade, possibly originating from CikA (12), interacts with the N-terminal domain of KaiA and alters the quaternary dimer structure of the C-terminal domain. Changes in the dimerization angle of the KaiA C-terminal domain change its affinity for KaiC and, thus, change the KaiA-induced KaiC autokinase activity enhancement and affect the KaiC phosphorylation equilibrium. Perturbations in the KaiC phosphorylation equilibrium alter the circadian oscillator and possibly cause resetting.
Previous models of Kai protein complex formation can also be analyzed based on our results. Ye et al. (20) proposed a model in which the KaiA–KaiC interaction drives KaiA in a hypothesized open conformation that has high affinity for KaiB. Our results show that KaiC binding indeed changes the quaternary structure of the C-terminal KaiA domain. We expect that these changes can also influence the relative conformation of the two KaiA domains, possibly in a manner similar to that suggested by Ye et al. (20). Garces et al. (21) have proposed a different model of how KaiB may attenuate KaiA enhancement of KaiC phosphorylation, in which KaiA and KaiB use a similar concave surface (corresponding to the surface at the bottom of the KaiA particle, as shown in Fig. 3) for KaiC binding and, thus, compete for KaiC. However, the surface proposed earlier for KaiA (21) as interacting with CIIABD is perpendicular to that found here. Further, in the orientation proposed by Garces et al., KaiB does not feature the largely hydrophobic groove used by CIIABD for KaiA binding. Thus, our structural data do not provide evidence for competition between KaiA and KaiB for CIIABD binding.
It is important to note that earlier studies showed KaiA binding to both CI and CII subunit of KaiC by using in vitro or yeast two-hybrid assays (8, 29), yet no sequence homologous to CIIABD exists in CI. Thus, it is possible that an additional, different, KaiA-interacting fragment is present in CI. However, the ThKaiA180C–CIIABD interaction described here is extensive and can account for all of the KaiA C-terminal domain solvent exposed amino acid substitutions with a clock-period phenotype (4, 10). Therefore, we believe that interactions of KaiA with CI are secondary to those with the CII domain of KaiC, although it is likely that they will contribute to the specificity and affinity of the KaiA–KaiC complex. The stoichiometry of the ThKaiA180C–CIIABD complex is likely to be physiologically relevant. KaiC abundance in vivo is relatively high compared with KaiA (19), and therefore, we expect that a KaiA dimer will be bound to two KaiC protein molecules for at least some fraction of the circadian period. However, KaiA can fully enhance the KaiC autokinase activity even at relatively low KaiA/KaiC stoichiometries (34), suggesting that formation of the KaiA–KaiC complex is transient. Currently, it is not known whether the KaiC autokinase activity operates as cis or trans (transphosphorylation). Because a KaiA dimer can bind simultaneously to two KaiC-derived peptides, which could correspond to independent KaiC hexamers, it is conceivable that KaiA enhances KaiC autophosphorylation by acting as a linker module that temporarily brings together two different KaiC hexamers in order for transphosphorylation to occur.
Supplementary Material
Acknowledgments
We thank Dr. Susan S. Golden for critical reading of the manuscript, Dr. James C. Hu for useful discussions, and Dr. Karl Koshlap for technical assistance with the NMR instrumentation. We also thank the Protein Chemistry Laboratory for help with peptide identification and quantitation. This work was supported by National Institutes of Health Grant GM064576 (to A.C.L.). The NMR instrumentation in the Biomolecular NMR Laboratory at Texas A&M University was supported by National Science Foundation Grant DBI-9970232.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HSQC, heteronuclear single quantum correlation; NOE, nuclear Overhauser effect.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1SUY and 1SV1). Backbone and side-chain chemical shifts of ThKaiA180C and CIIABD have been deposited in the BioMagResBank (accession no. 6174).
References
- 1.Bünning, E. (1973) The Physiological Clock: Circadian Rhythms and Biological Chronometry (Springer, New York), 3rd Ed.
- 2.Dunlap, J. C., Loros, J. J. & DeCoursey, P. J. (2004) Chronobiology: Biological Timekeeping (Sinauer, Sunderland, MA).
- 3.Dunlap, J. C. (1999) Cell 96, 271–290. [DOI] [PubMed] [Google Scholar]
- 4.Ishiura, M., Kutsuna, S., Aoki, S., Iwasaki, H., Andersson, C. R., Tanabe, A., Golden, S. S., Johnson, C. H. & Kondo, T. (1998) Science 281, 1519–1523. [DOI] [PubMed] [Google Scholar]
- 5.Dvornyk, V., Vinogradova, O. & Nevo, E. (2003) Proc. Natl. Acad. Sci. USA 100, 2495–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu, Y., Tsinoremas, N. F., Johnson, C. H., Lebedeva, N. V., Golden, S. S., Ishiura, M. & Kondo, T. (1995) Genes Dev. 9, 1469–1478. [DOI] [PubMed] [Google Scholar]
- 7.Nakahira, Y., Katayama, M., Miyashita, H., Kutsuna, S., Iwasaki, H., Oyama, T. & Kondo, T. (2004) Proc. Natl. Acad. Sci. USA 101, 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwasaki, H., Taniguchi, Y., Ishiura, M. & Kondo, T. (1999) EMBO J. 18, 1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kageyama, H., Kondo, T. & Iwasaki, H. (2003) J. Biol. Chem. 278, 2388–2395. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura, H., Nakahira, Y., Imai, K., Tsuruhara, A., Kondo, H., Hayashi, H., Hirai, M., Saito, H. & Kondo, T. (2002) Microbiology 148, 2903–2909. [DOI] [PubMed] [Google Scholar]
- 11.Iwasaki, H., Nishiwaki, T., Kitayama, Y., Nakajima, M. & Kondo, T. (2002) Proc. Natl. Acad. Sci. USA 99, 15788–15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitz, O., Katayama, M., Williams, S. B., Kondo, T. & Golden, S. S. (2000) Science 289, 765–768. [DOI] [PubMed] [Google Scholar]
- 13.Mori, T., Saveliev, S. V., Xu, Y., Stafford, W. F., Cox, M. M., Inman, R. B. & Johnson, C. H. (2002) Proc. Natl. Acad. Sci. USA 99, 17203–17208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu, Y., Mori, T. & Johnson, C. H. (2003) EMBO J. 22, 2117–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leipe, D. D., Aravind, L., Grishin, N. V. & Koonin, E. V. (2000) Genome Res. 10, 5–16. [PubMed] [Google Scholar]
- 16.Hayashi, F., Suzuki, H., Iwase, R., Uzumaki, T., Miyake, A., Shen, J. R., Imada, K., Furukawa, Y., Yonekura, K., Namba, K. & Ishiura, M. (2003) Genes Cells 8, 287–296. [DOI] [PubMed] [Google Scholar]
- 17.Vakonakis, I., Sun, J., Wu, T., Holzenburg, A., Golden, S. S. & LiWang, A. C. (2004) Proc. Natl. Acad. Sci. USA 101, 1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams, S. B., Vakonakis, I., Golden, S. S. & LiWang, A. C. (2002) Proc. Natl. Acad. Sci. USA 99, 15357–15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitayama, Y., Iwasaki, H., Nishiwaki, T. & Kondo, T. (2003) EMBO J. 22, 2127–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye, S., Vakonakis, I., Ioerger, T. R., LiWang, A. C. & Sacchettini, J. C. (2004) J. Biol. Chem. 279, 20511–20518. [DOI] [PubMed] [Google Scholar]
- 21.Garces, R. G., Wu, N., Gillon, W. & Pai, E. F. (2004) EMBO J. 23, 1688–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vakonakis, I. & LiWang, A. C. (2004) J. Biomol. NMR 28, 403–404. [DOI] [PubMed] [Google Scholar]
- 23.Wüthrich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 24.LiWang, A. C., Cao, J. J., Zheng, H., Lu, Z., Peiper, S. C. & LiWang, P. J. (1999) Biochemistry 38, 442–453. [DOI] [PubMed] [Google Scholar]
- 25.Cornilescu, G., Delaglio, F. & Bax, A. (1999) J. Biomol. NMR 13, 289–302. [DOI] [PubMed] [Google Scholar]
- 26.Schwieters, C. D., Kuszewski, J. J., Tjandra, N. & Marius Clore, G. (2003) J. Magn. Reson. 160, 65–73. [DOI] [PubMed] [Google Scholar]
- 27.Richardson, J. S. (1981) Adv. Protein Chem. 34, 167–339. [DOI] [PubMed] [Google Scholar]
- 28.Christopher, J. A. (1999) spock, The Structural Properties Observation and Calculation Kit (Center for Macromolecular Design, Texas A&M University, College Station, TX).
- 29.Taniguchi, Y., Yamaguchi, A., Hijikata, A., Iwasaki, H., Kamagata, K., Ishiura, M., Go, M. & Kondo, T. (2001) FEBS Lett. 496, 86–90. [DOI] [PubMed] [Google Scholar]
- 30.Kelly, L. A., MacCallum, R. M. & Sternberg, M. J. E. (2000) J. Mol. Biol. 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 31.Story, R. M. & Steitz, T. A. (1992) Nature 355, 374–376. [DOI] [PubMed] [Google Scholar]
- 32.Kamtekar, S. & Hecht, M. H. (1995) FASEB J. 9, 1013–1022. [DOI] [PubMed] [Google Scholar]
- 33.Harris, N. L., Presnell, S. R. & Cohen, F. E. (1994) J. Mol. Biol. 236, 1356–1368. [DOI] [PubMed] [Google Scholar]
- 34.Hayashi, F., Ito, H., Fujita, M., Iwase, R., Uzumaki, T. & Ishiura, M. (2004) Biochem. Biophys. Res. Commun. 316, 195–202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.