Abstract
The Mildew resistance Locus O (MLO) family is unique to plants, containing genes that were initially identified as a susceptibility factor to powdery mildew pathogens. However, little is known about the roles and functional diversity of this family in rice, a model crop plant. The rice genome has 12 potential MLO family members. To achieve systematic functional assignments, we performed a phylogenomic analysis by integrating meta-expression data obtained from public sources of microarray data and real-time expression data into a phylogenic tree. Subsequently, we identified 12 MLO genes with various tissue-preferred patterns, including leaf, root, pollen, and ubiquitous expression. This suggested their functional diversity for morphological agronomic traits. We also used these integrated transcriptome data within a phylogenetic context to estimate the functional redundancy or specificity among OsMLO family members. Here, OsMLO12 showed preferential expression in mature pollen; OsMLO4, in the root tips; OsMLO10, throughout the roots except at the tips; and OsMLO8, expression preferential to the leaves and trinucleate pollen. Of particular interest to us was the diurnal expression of OsMLO1, OsMLO3, and OsMLO8, which indicated that they are potentially significant in responses to environmental changes. In osdxr mutants that show defects in the light response, OsMLO1, OsMLO3, OsMLO8, and four calmodulin genes were down-regulated. This finding provides insight into the novel functions of MLO proteins associated with the light-responsive methylerythritol 4-phosphate pathway. In addition, abiotic stress meta-expression data and real-time expression analysis implied that four and five MLO genes in rice are associated with responses to heat and cold stress, respectively. Upregulation of OsMLO3 by Magnaporthe oryzae infection further suggested that this gene participates in the response to pathogens. Our analysis has produced fundamental information that will enhance future studies of the diverse developmental or physiological phenomena mediated by the MLO family in this model plant system.
Keywords: abiotic stress, meta-expression analysis, MLO, rice, phylogenomic analysis
Introduction
The heritable significance of the Mildew resistance Locus O (MLO) family of seven-transmembrane proteins was first recognized in barley (Hordeum vulgare), where its loss of function caused resistance response against the powdery mildew pathogen Blumeria graminis f. sp. hordei (Bgh) (Jorgensen, 1992; Büschges et al., 1997). The MLO family is believed to be unique to plants and green algae (Devoto et al., 1999; Kim et al., 2002b). MLO gene family members in 22 land plant species including nine dicots and six monocots have been identified and the phylogenetic classification was performed (Kusch et al., 2016).
The MLO genes were originally found in the monocot barley. Homozygous mutant alleles of HvMLO are resistant to well-known fungal powdery mildew pathogens in that crop (Jorgensen, 1992; Piffanelli et al., 2002). Its expression can be regulated by many biotic and abiotic effectors, including Bgh fungus, leaf wounding, and the herbicide paraquat (Piffanelli et al., 2002). However, functional characterization of this family in dicots has been relatively limited. In Arabidopsis, AtMLO2 (one of 15 AtMLO members), has a role in conferring partial resistance to the powdery mildew pathogens Golovinomyces orontii and G. cichoracearum. Furthermore, a triple mutant of three paralogs—AtMLO2, AtMLO6, and AtMLO12—shows complete resistance to a fungal pathogen whereas single mutants of either atmlo6 or atmlo12 and the atmlo6/atmlo12 double mutant exhibit a phenotype with a percentage of host cell entry that is almost the same as that of wild-type (WT) plants (Consonni et al., 2006). This finding indicates that an unequal genetic redundancy might exist with respect to the modulation of defense between AtMLO paralogous genes. Therefore, further analyses remain to identify the most predominant member among those members which might facilitate the functional characterization through loss of function approaches.
In addition to the roles that MLO have in defenses against powdery mildew disease, they might also participate in morphological and developmental processes, e.g., root thigmomorphogenesis (Chen et al., 2009; Bidzinski et al., 2014), pollen tube reception in the ovary (Kessler et al., 2010), and pollen hydration (Yi et al., 2014). Shared genetic functions between closely linked family members within the same phylogenetic clade have also been reported but not yet clearly explained. For example, atmlo4 and atmlo11 mutant seedlings display a spiral-like root phenotype on minimal and mildly acidic media when they contact a hard surface. However, another close member of the same clade, AtMLO14, does not seem to be involved in this phenomenon (Chen et al., 2009). Although it is difficult to gain a complete understanding of the functional complexity within this gene family, it can be estimated by using gene expression patterns integrated into the phylogenetic tree context.
Since the completion of the rice genome annotation project, Liu and Zhu (2008) have identified and isolated 12 MLO genes in rice. However, that investigation focused primarily on their evolution, divergence, and structural features, and little was done to obtain clues about their predicted functions. In fact, the phenotype of only one of those 12 OsMLOs has been described. Nevertheless, because MLO genes in other species seem to be involved in many physiological processes, it is possible that we might be able to discover new phenotypes by evaluating expression profiles in rice as well.
Systematic assignments facilitate the functional identification of individual members within a gene family. Phylogenomic approaches that integrate diverse transcriptome data within the phylogenetic tree context are advantageous when conducting functional studies. For example, we have performed genome-wide phylogenomic analyses of families for heat shock transcription factors, heat shock protein 70, ABC transporters, crRLK1L kinase, and aquaporins in rice, and our results have suggested that individual members in each family showed functional diversity or redundancy, based on the integrated intensive transcriptome data to the context of phylogenetic tree (Jin et al., 2013; Nguyen et al., 2013; Nguyen Q. N. et al., 2014; Nguyen V. N. T. et al., 2014).
Like those earlier studies, we conducted such an analysis of the rice MLO family along with a comparative phylogenetic analysis. This included 12 rice MLOs, 15 Arabidopsis MLOs, and two barley MLOs. Data related to anatomical meta-expression, as well as responses to abiotic and biotic stresses, were examined within the context of a phylogenetic tree. Our objective was to estimate the functional redundancy or dominance among MLO family members and to gain extensive functional information about those genes. Another of our earlier investigations focused on light-responsive genes (Jung et al., 2008), in which we analyzed the expression patterns and defective phenotypes of light responsive family genes under various light conditions. We demonstrated there the benefit of selecting the predominant gene because defective phenotypes are revealed in loss-of-function mutants. Until now, function of a MLO gene (OsMLO12) in rice was only reported associated with pollen germination process (Yi et al., 2014). Based on the extensive expression data, we now propose that rice MLO members play broad roles in response to diverse environmental challenges, including pathogen infection, and cold and heat stresses, as well as the light response and developmental processes associated with the root, leaf, and pollen. Detailed data analysis and discussion will be presented.
Materials and methods
Multiple alignment and phylogenetic analysis
To perform our phylogenetic analysis of the MLO family, we used the protein sequences of 12 MLO genes identified in a previous global analysis of the rice MLO family (Liu and Zhu, 2008). We also developed a tree with rice, barley, and Arabidopsis MLO proteins, based on previous global analyses of this gene family (Jorgensen, 1992; Devoto et al., 2003). The protein sequences for our phylogenomic analysis were downloaded from TAIR, GreenPhyl and the Rice Genome Annotation Project Website (Rhee et al., 2003; Ohyanagi et al., 2006; Rouard et al., 2011). After multiple-alignment of those sequences with ClustalX (Higgins et al., 1996), we generated a phylogenetic tree using the Neighbor-Joining method, as incorporated in the MEGA5 tool kit for phylogenetic analysis (Tamura et al., 2011).
Analysis of microarray data and heatmap development
Microarray data including Affymetric and Agilent array data were downloaded from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and Genevestigator (https://genevestigator.com/gv/) (Zimmermann et al., 2004; Cao et al., 2012). We then uploaded the normalized data to the Multi Experiment Viewer (http://www.tm4.org/mev.html) and visualized the data via heatmaps. We used Genevestigator to compare levels of gene expression in several organs and to estimate the functional similarity among rice, barley, and Arabidopsis members.
Plant materials
Circadian rhythms and functional associations with rice gigantea (OsGI) were examined using wild-type (WT) plants and osgi mutants. Ten-day-old rice seedlings were grown on a Murashige Skoog medium under controlled conditions (28°C/25°C day/night, continuous light, and 78% relative humidity). They were then transferred to individual pots and placed in a growth chamber. Beginning at 30 d after germination, their leaves were sampled at 4-h intervals for 48 h. Light- and dark-dependent expression was analyzed using a heterozygous osdxr (rice 1-deoxy-D-xylulose 5-phosphate reductoisomerase) mutant line for which seeds were germinated as described above. Homozygous and WT plants were identified through genotyping, and leaves were collected from 10-day-old plants. Primers used for genotyping analysis are presented in Table S1.
Stress treatments and pathogen inoculation
Cold treatment consisted of transferring 10-day-old plants originally grown at 28°C to a 6°C ± 1°C lighted refrigerator. The control plants remained at 28°C. After 48 h, the chilled plants were returned to the 28°C chamber for recovery. Samples were collected at Hours 0, 24, and 48 of low-temperature treatment and after 24 and 48 h of recovery. To simulate heat stress, we exposed 10-day-old plants to 42°C ± 1°C for 0, 3, 6, and 12 h. The control plants remained at 28°C. Leaves of three plants were pooled for one biological replicate and each treatment had three repeats. To assess their genetic response to pathogen infection, we sprayed Magnaporthe oryzae conidia suspension with 0.005% Tween on to 3-week-old plants of Dongjin rice and then placed them in containers for 24 h to maintain 80% humidity under darkness before transferring them to a chamber under a 14-h photoperiod. Control and mock samples were collected by spraying water without and with 0.005% Tween, respectively.
RNA extraction and quantitative PCR
Leaf samples were frozen in liquid nitrogen and homogenized with a TissueLyser II (Qiagen, Hilden, Germany). Total RNA was extracted using RNAiso Plus according to the manufacturer's protocol (Takara Bio, Kyoto, Japan). The qPCR was performed by Qiagen Rotor-Gene Q real-time PCR cycler using follow thermal cycling procedure: 95°C for 10 s, 60°C for 30 s, and 72°C for 1 min. For evaluating tissue-specific expression patterns by real-time PCR, we used rice ubiquitin 5 (OsUbi5, LOC_Os01g22490) (Jain et al., 2006) as an endogenous control to normalize variance in the quality of RNA and the amount of cDNA. LATE ELONGATED HYPOCOTYL (OsLHY) served as the positive control when examining diurnal rhythms. The effects of cold and heat stress were monitored using OsNAC6 (LOC_Os01g66120) (Ohnishi et al., 2005) and OsHSP1 (LOC_Os04g01740) (Moon et al., 2014), respectively, as positive controls. Successful interaction between rice and M.oryzae was convinced by positive control OsPR10a (LOC_Os12g36880) (Lee et al., 2009). All of the primers for these real-time analyses are shown in Table S1.
Co-expression analysis
Twenty-five co-expressed genes of 6 individual MLO genes (OsMLO1, 2, 3, 4, 8, and 9) with top-score Pearson Correlation Coefficient (PCC) value were collected, based on the co-expression tool installed in Genevestigator (Zimmermann et al., 2004). Co-expressed genes might provide useful functional clues for associated MLO genes. Especially, functionally characterized genes collected from The Overview of functionally characterized Genes in Rice online (OGRO) database (Yamamoto et al., 2012) are more effective for this purpose.
Results
Integration of anatomical expression patterns with a phylogenetic tree of the rice MLO family
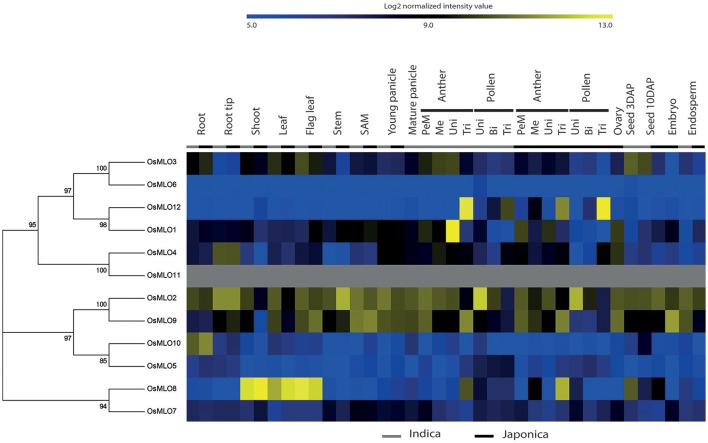
Using protein sequences for 12 MLO genes in rice, we generated a phylogenetic tree that incorporated subgroup information and anatomical meta-expression profiles based on 983 Affymetrix array data (Cao et al., 2012) (Figure 1). Starting at the top of the tree, OsMLO3 displayed a medium level of expression in almost all tissues, including roots, shoots, leaves, and flag leaves at the vegetative stage, and anthers and seeds at the reproductive stage. It was most closely linked with OsMLO6. However, very low expression by the latter in whole tissues suggested that OsMLO3 had a more dominant role. Uniquely, OsMLO12 showed strong anther-preferred expression in trinucleate and mature pollen. This profile was consistent with that reported by Yi et al. (2014). Expression of OsMLO1 was high in anthers at the uninucleate stage, especially in indica rice, although not at a level similar to that found in uninucleate pollen (Figure S1). Thus, OsMLO1 might be more active in the anthers than in the pollen grains themselves. The Affymetrix data revealed that OsMLO4 was preferentially expressed in the root tips but at only a low level in other tissues. In contrast, OsMLO2 was ubiquitously expressed. One of its paralogs, OsMLO9, showed dominant expression patterns in reproductive organs, and transcripts were particularly abundant in anthers at the uninucleate pollen stage while OsMLO2 was not detected there (Figure S1). Although OsMLO5 and OsMLO10, as well as OsMLO7 and OsMLO8, are thought to have resulted from segmental duplication, each pairing appears to have retained differential expression patterns from an anatomical perspective. While expression of OsMLO5 was extremely low in the roots, that of OsMLO10 was much higher than for OsMLO5. Moreover, when compared with OsMLO7, OsMLO8 was more predominantly expressed in the shoots, leaves, and anthers at the trinucleate pollen stage (Figure 1). All of these findings suggested that functional specificity and dominance is much greater for OsMLO8 and OsMLO10 than for their homologs OsMLO7 and OsMLO5, respectively. Therefore, with the exception of OsMLO11, for which no probe was printed on the Affymetrix chip, we were able to use meta-expression data to estimate the anatomical functions of all MLO genes examined here.
Figure 1.
Meta-analysis of OsMLO gene expression patterns using 983 Affymetrix anatomical array data. Blue, low level of log2 intensity; yellow, high level. Gray bar, indica samples; black bar, japonica samples. OsMLO11 has no probe on rice Affymetrix chip. SAM, shoot apical meristem; PMe, pre-meiotic; Me, meiotic; Uni, uninucleate stage; Bi, binucleate stage; Tri, trinucleate stage; DAP, days after pollination.
Confirmation of anatomical expression patterns for rice MLO family genes by real-time
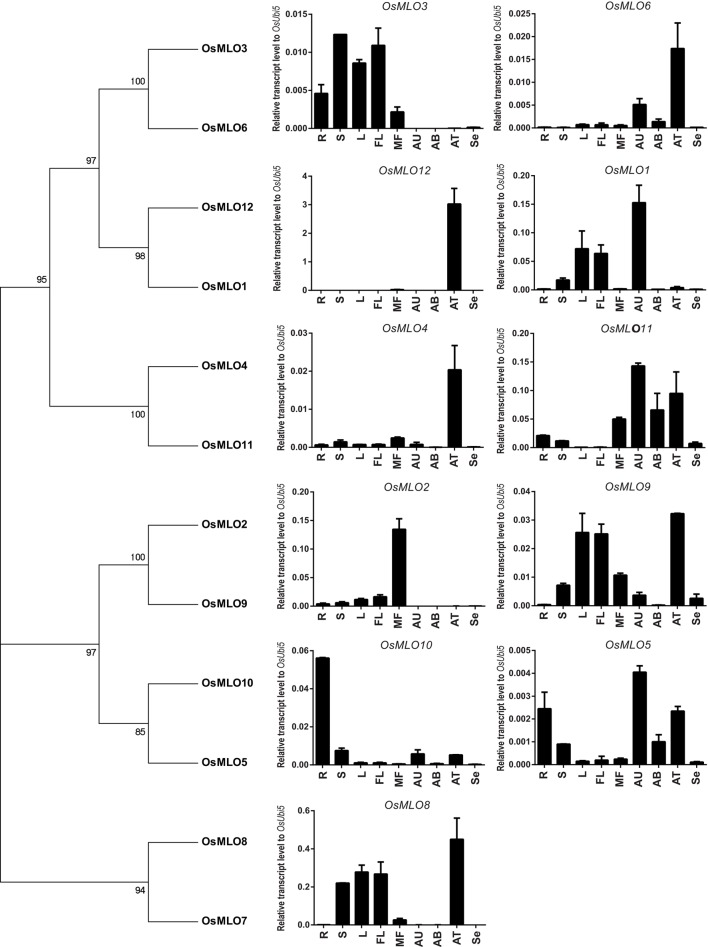
To confirm the results from our meta-analysis of anatomical expression profiles, we performed real-time PCR with 12 rice MLO genes that are expressed in the roots, shoots, leaves, young panicles, mature flowers, and seeds (at 6 d post-pollination), as well as in anthers sampled at the uni-, bi-, and trinucleate stages (Figure 2). Our findings here closely matched those obtained from meta-analyses of tissue-specific expression profiles. However, we failed to detect any expression of OsMLO7, probably because its transcript level was extremely low. We determined that OsMLO11 was highly expressed in mature flowers and in uni-, bi-, and trinucleate anthers. These results demonstrated the high reliability of meta-expression profiles based on a large collection of transcriptome data.
Figure 2.
Real-time expression profiles for 12 OsMLO genes. Anatomical samples were prepared from root (R), shoot (S), mature leaf (L), young panicle (3cm-YP), mature flower (MF), anther [uni- (AU), bi- (AB), and tricellular (AT)], and seed at 6 days after pollination (Se). Rice ubiquitin (OsUbi5) was served as internal control.
Phylogenetic and comparative expression analyses of MLO proteins in rice, barley, and Arabidopsis
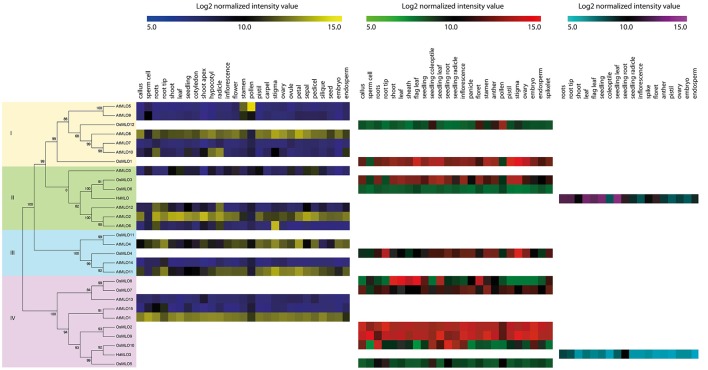
Although the roles of rice MLO family members have long been studied, only one of those genes has known biological functions. It is known that OsMLO12 regulates pollen hydration (Yi et al., 2014). However, we believed that the expression patterns of other genes in that family might be good indicators for predicting their biological functions. We also expected that a comparative transcriptome analysis of MLO family members from rice and Arabidopsis would provide more accurate evidence for functional conservancy among orthologous gene groups in those two species. Therefore, we combined phylogenetic and meta-expression data from Genevestigator, which uses a similar platform to present expression patterns within an anatomical context.
Our phylogenetic tree covered 29 protein sequences that were aligned from rice (12 MLOs), Arabidopsis (15), and barley (2). This established four clades, i.e., I through IV (Figure 3). Dividing the number of clade for MLO family has yet to be unanimous (Feechan et al., 2008; Pessina et al., 2014; Kusch et al., 2016), we separated 29 MLOs into four clades base on primary evolution and relative known function gene with small numbers of MLO genes. This tree implied that the evolutionary processes for MLO genes from monocots and dicots were rather independent. For example, the 29 members included only two orthologous pairings—AtMLO4/OsMLO11 in Clade III and OsMLO10/HvMLO3 in Clade IV.
Figure 3.
Comparisons of expression patterns among OsMLO genes. Log2 intensity values (colored by species) ranged from 5 to 15: Arabidopsis thaliana (blue to yellow), Oryza sativa (green to red), and Hordeum vulgare (cerulean to violet). OsMLO11 expression pattern was not available in Genevestigator data. Clade number is indicated on left side of phylogenetic tree.
For Clade I, the anatomy tool installed in Genevestigator indicated similar expression between AtMLO5 and OsMLO12 (Figure 2). Both AtMLO8 and OsMLO1 were ubiquitously expressed except in the pollen (Figure 2). AtMLO10 was predicted to function in the roots, hypocotyls, radicles, and endosperm.
In Clade II, expression of OsMLO3 in almost all organs/tissues was the most similar to the patterns found for AtMLO2 than AtMLO6, and AtMLO12 expression. Elliott et al. (2002) have demonstrated that introducing OsMLO3 into the barley MLO null mutant genotype mlo-5 leads to 50% susceptibility restoration. Therefore, it appears that OsMLO3 in rice might have a biological function in plant defenses. However, it is unclear whether the presence of a single copy of that gene is sufficient to confer pathogen resistance. Because OsMLO6 was expressed at low levels in most tissues/organs while AtMLO6 and AtMLO12 were highly expressed in roots and radicles, we suspect that the latter two genes have roles in root development that are conserved with AtMLO2.
For Clade III, the expression patterns of AtMLO4, AtMLO11, and OsMLO4 indicated that they have similar functions in the development of root tips and stigma, suggesting that the roles of MLO genes within that clade are conserved. Although data for OsMLO11 were not available using the Affymetrix array platform, we were able to determine its spatio-temporal expression patterns using the Agilent array platform. In doing so, we found that this gene was highly expressed in the roots, inflorescences, pistils, palea/lemma, and ovaries (Figure S2). Therefore, expression was similar between OsMLO11 and OsMLO4 at the reproductive stage. While AtMLO4 displayed similar expression to AtMLO11 in every tissues/organ, AtMLO14 expression is disparity. However, whereas knockouts of AtMLO4 and AtMLO11 caused an abnormal root phenotype, the paralog AtMLO14 was not involved in root development. This might explain their different patterns of expression even though AtMLO11 and AtMLO14 were the closest members in the evolutionary tree (Figure 3). Nevertheless, we expect that the osmlo4 mutant would display the same behavior upon tactile stimulation as that shown by mutants of AtMLO4 or AtMLO11.
In Clade IV, OsMLO8 was preferentially expressed in the leaves and shoots. However, we were unable to find similar expression patterns for Arabidopsis genes in this clade. Instead, AtMLO1, OsMLO2, and OsMLO9 were ubiquitously expressed in most of the anatomical samples, suggesting that these MLO family members have housekeeping functions.
Diurnal regulation associated with the MLO family
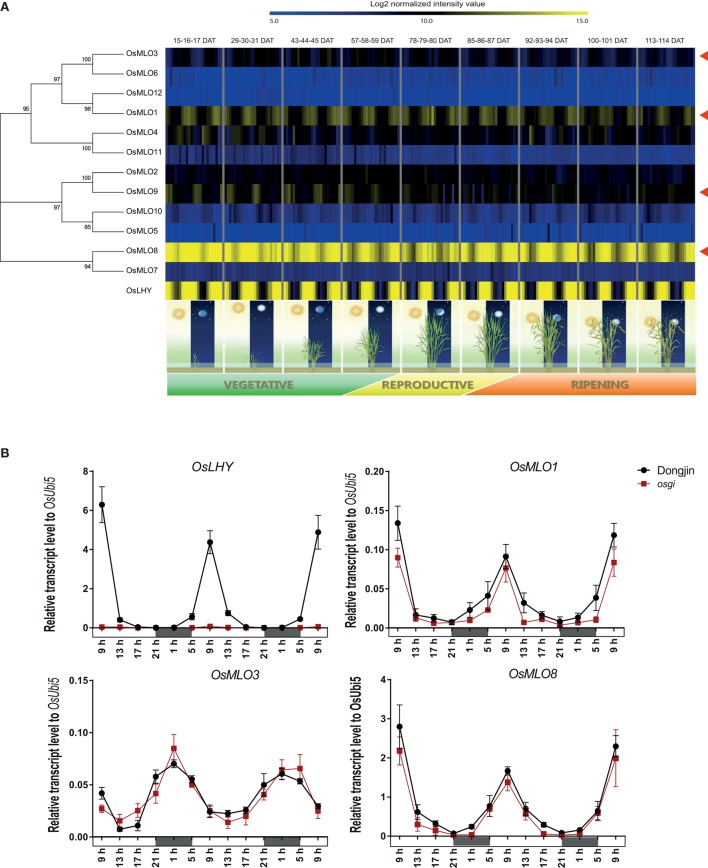
Diurnal rhythm is synchronized with the day/night cycle and is regulated by two mechanisms: light and the circadian clock. To elucidate the diurnal rhythm of our MLO genes, we analyzed their expression patterns using publicly available Agilent 44k array data obtained for diurnal and circadian gene expression in the leaf over the entire life span of field-grown rice plants (Sato et al., 2013). Among the 12 MLO genes, leaf preferentially expressed OsMLO8 showed high expression that was obviously diurnal, while OsMLO1, OsMLO3, and OsMLO9 were also diurnally expressed, albeit at a more moderate level, in mature leaves (Figure 4A). Real-time PCR analyses of genes from 4-week-old leaves indicated that OsMLO1, OsMLO3, and OsMLO8 exhibited cyclic expression every 24 h (Figure 4B).
Figure 4.
Diurnal expression patterns of OsMLO genes (red arrows indicate diurnal rhythm) in mature leaves, using available Agilent 44k array data over entire plant life span (A), or evaluated at 13 time points over 48-h period in “Dongjin” rice and osgi mutant (B). As a standard marker gene for diurnal rhythm, expression of OsLHY was peaked during daytime. OsUbi5 was served as internal control. DAT, days after transplanting. Continuous white and black bar indicates day and night time, respectively.
To obtain insight into the mechanism that regulates the diurnal rhythm of MLO genes, we evaluated a rice mutant. In osgi, diurnal expression of marker gene OsLHY was dramatically down-regulated across all time points whereas the rhythm of expression for OsMLO1, OsMLO3, and OsMLO8 was similar to that detected in WT plants (Figure 4B). These findings indicated that the genes are not related to OsGI or the circadian mechanism.
Light response associated with the MLO family
To determine whether the MLO genes are involved in light-/dark-response mechanisms, we evaluated their expression using Agilent 60K microarray data for a mutant of the light-responsive osdxr, which belongs to the methylerythritol 4-phosphate pathway (Jung et al., 2008) (Figure 5A). Real-time PCR analysis confirmed that OsMLO1 expression was decreased while that of OsMLO3 was much lower in the mutant than in the WT (Figure 5B). Leaf-specific expression of OsMLO8 was suppressed completely in osdxr, suggesting a close connection between OsMLO8 and the light-response pathway. Our results implied that the MLO genes are diurnally expressed and that OsMLO3 is dark-inducible while OsMLO1 and OsMLO8 are light-inducible (Figure 5).
Figure 5.
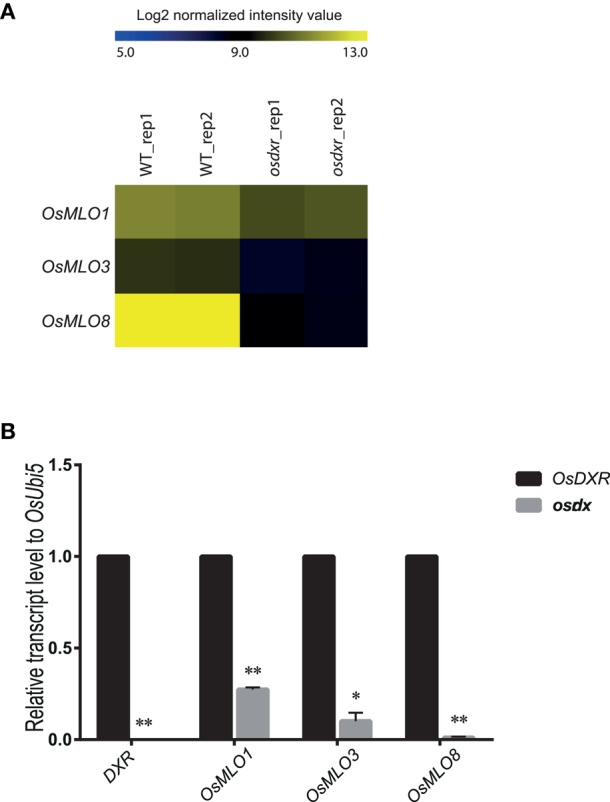
Heatmap for light- or dark-inducible expression of OsMLO genes in wild-type (WT/DXR) rice and osdxr mutant. (A) Analysis of 3 genes using Agilent 60K microarray data for osdxr mutant. (B) Validation of expression of OsMLO1, OsMLO3, and OsMLO8 in WT (DXR) and osdxr mutant. Transcripts of OsDxr was absent in knockout plant. **p < 0.01; *0.01 < p < 0.05.
The MLO proteins interact with calmodulin as the second messenger to transfer a signal forward to downstream pathways (Kim et al., 2002a,b; Stein and Somerville, 2002). To investigate whether the diurnal response-dependent OsMLOs function with calmodulin proteins, we examined the differential expression of calmodulin (CaM) and CaM-like (CML), searching for genes with log2-fold changes >1.8 and p-values <0.05 in the osdxr mutant (Table S2). Subsequent analysis of the microarray data presented four CML genes—OsCML1, OsCML16, OsCML24, and OsCML28—with decreased expression in that mutant (Figure 6A). Real-time PCR analysis also confirmed that all four were significantly down-regulated in osdxr (Figure 6B). Therefore, we propose that these OsCML genes would be primary targets for studying functional relationships among diurnal response-dependent OsMLOs.
Figure 6.
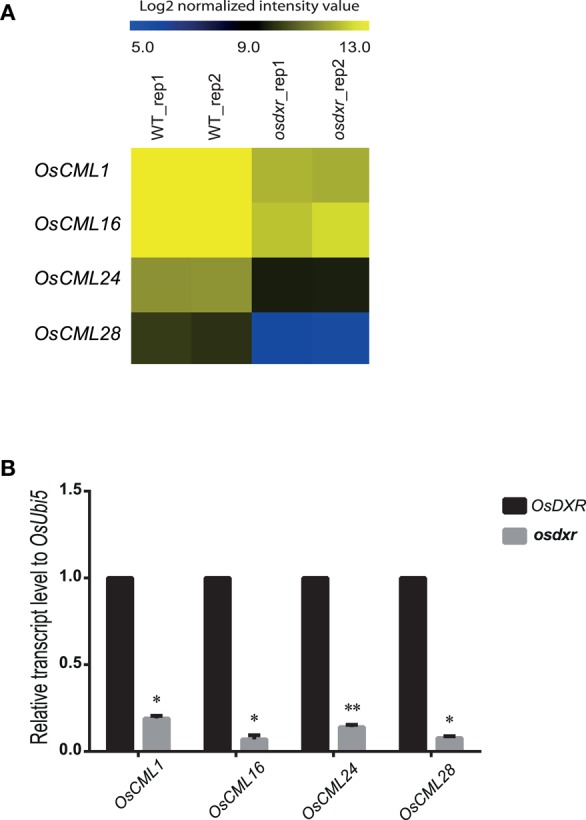
Heatmap for expression patterns of OsCML genes in wild-type (WT/DXR) rice and osdxr mutant. (A) Expression analysis of 4 OsCML genes using Agilent 60K microarray data for WT vs. osdxr knockout line. (B) Downregulation of OsCML genes in osdxr mutant demonstrated by real-time PCR analysis. **p < 0.01; *0.01 < p < 0.05.
Responses of the MLO family to high- and low-temperature stresses
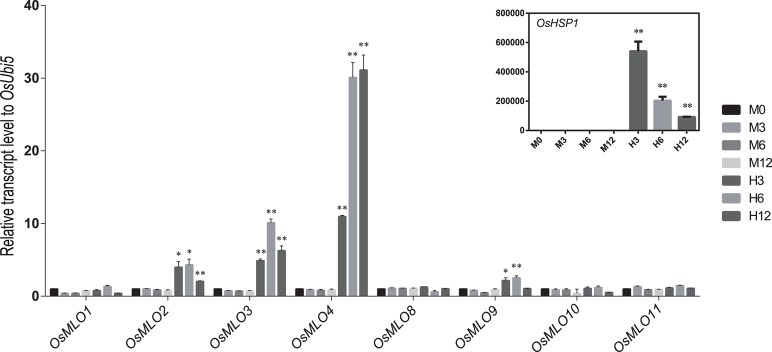
Although the barley MLO genes are affected by abiotic stresses such as leaf-wounding and herbicides (Piffanelli et al., 2002), little is known about how genes in that family are influenced by other sources of stress. We used the log2 fold-change data in response to abiotic stresses (Figure S3) to investigate the meta-expression patterns of 12 rice MLO genes. Differential expression was monitored via real-time PCR with samples under either heat- or cold-stress conditions. As expected, OsMLO2, OsMLO3, OsMLO4, and OsMLO9 were rapidly up-regulated during the first 3 h of heat treatment (Figure 7). Their expression began to decline after 12 h of treatment but was still higher than that measured from the WT control. Among those four genes, OsMLO4 was the most responsive to high temperature, with expression being >30-fold higher than that in control plants after 6 h and remaining at that level for 12 h of treatment. Because OsMLO5, OsMLO6, and OsMLO7 were expressed only at low levels in the leaves, they were eliminated from further analysis of temperature sensitivity.
Figure 7.
Expression profiles of OsMLO genes under heat stress evaluated by real-time PCR analysis. OsHsp1 served as a positive marker for heat-stress response. OsUbi5 was used as internal control. M, control treatment; H, high-temperature treatment. Numbers after M and H indicate time points (hours) after stress treatment. **p < 0.01; *0.01 < p < 0.05.
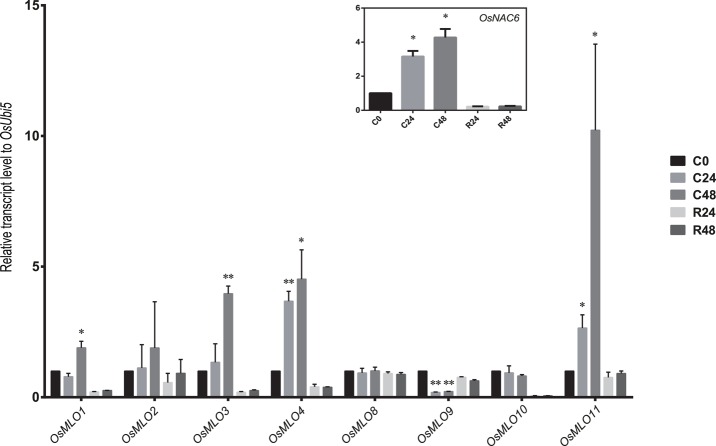
In response to cold stress, OsMLO1 and OsMLO3 were up-regulated at 48 h after treatment was applied. However, their level of expression declined to that measured from the control when the chilled plants were allowed to recover at 28°C (Figure 8). Both OsMLO4 and OsMLO11 were quickly induced by low temperatures, with expression increasing after just 24 h of treatment and transcripts being retained at higher levels until Hour 48. By contrast, OsMLO9 was down-regulated by 24 h of chilling. This response was the opposite of its upregulation by heat stress, implying that the gene has separate functions in determining the plant response to temperature extremes. All of these findings provided evidence that the expression of these rice MLO genes is influenced by cold and/or heat stress.
Figure 8.
Expression profiles of OsMLO genes under cold stress evaluated by real-time PCR. OsNAC6 served as a positive marker for cold-stress response. OsUbi5 was used as internal control. C, cold treatment; R, recovery. Numbers after C and R indicate time points (hours) after stress treatment. **p < 0.01; *0.01 < p < 0.05.
Interactions between rice MLO genes and Magnaporthe oryzae
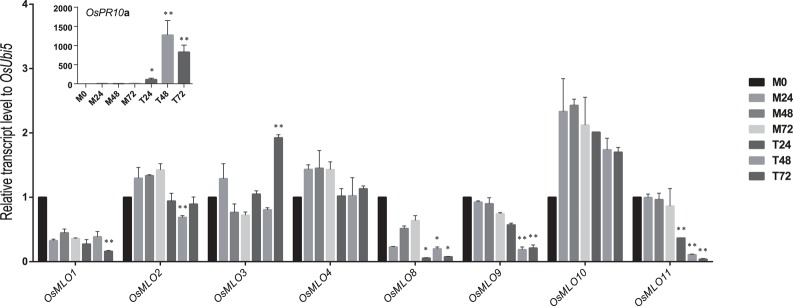
“Dongjin” rice plants were inoculated with M. oryzae PO6-6 to monitor the response of MLO genes to pathogen infection. We had expected that OsMLO3 would show differential expression similar to that demonstrated by its homologous members in Arabidopsis and barley. In fact, OsMLO3 was significantly induced at 72 h after infection (Figure 9). Expression of two light-inducible genes—OsMLO1 and OsMLO8—was relatively decreased after 72 h and 24 h, respectively, two closely linked members—OsMLO2 and OsMLO9—were slightly down-regulated at 48 h, and OsMLO11 were significantly reduced expression at 48 h after treatment. These data indicated that rice MLO family members have possible roles in the pathogen response.
Figure 9.
Real-time analysis of OsMLO genes in Dongjin rice variety inoculated with M. oryzae PO6-6. Samples from mock treatment (M), and fungal inoculation treatment (T) were examined after inoculation for 24, 48, and 72 h. Numbers after C and R indicate time points (hours) after stress or MOCK treatment. Rice PR10a served as a positive marker for infection. OsUbi5 was used as internal control. **p < 0.01; *0.01 < p < 0.05.
Co-expression analysis suggests potential downstream genes of MLO signaling
Twenty five co-expressed genes of 6 individual MLO genes (OsMLO1, 2, 3, 4, 8, and 9) with the highest PCC value were selected by co-expression analysis tool installed in Genvestigator with 2532 anatomical samples (Table S3). To identify the possible function in which each MLO gene is involved, we searched rice genes with known functions from the OGRO database out of the co-expressed genes. Unfortunately, progress of functional genomics in rice is still restricted and explains functions for 2 or 3% of genes in rice genome (Chandran et al., 2016). Thus, we only found two known genes from co-expressed genes to OsMLO3 or OsMLO8. A known gene, OsHsfA4d/Spl7, one of co-expressed genes to OsMLO3, encodes heat shock transcription factor (Yamanouchi et al., 2002), which activates heat shock genes under high temperature stress. spl7 mutant causes scattered brown spot on leaf and dead cells increase since exposing under heat stress or UV solar radiation. spl7 also accumulated higher amounts of H2O2 in response to rice blast fungal elicitor (Kojo et al., 2006). Although the co-expression score (PCC value) of OsMLO3 and Spl7 is 0.6847, this linkage suggests one of downstream responsive genes of OsMLO3 associated with heat stress, cell death and H2O2 accumulation. In case of fructose-1,6-bisphosphatase (OscFBP1/MOC2), the co-expression score with OsMLO8 is 0.9314. OscFBP1 converts triose-phosphates to sucrose in the day (Daie, 1993). Loss function of OscFBP1 exhibits monoculm phenotype and reduced photosynthetic rate (Lee et al., 2008; Koumoto et al., 2013). Since leaf- preferred OsMLO8 gene showed the peak of expression level under daylight, its co-expression with OscFBP1 suggests the important role in photosynthesis.
Discussion
Although members of the MLO family generally function in defense responses, some of those genes are also involved in developmental processes, such as pollen tube elongation or root formation (Chen et al., 2009; Kessler et al., 2010; Bidzinski et al., 2014). The functional implication of each MLO member can be ascertained by meta-expression data based on a large collection of transcriptome data. Expression patterns for genes associated with the development of pollen tubes or roots are closely linked with their corresponding tissues/organs, thereby making them useful indicators when examining currently uncharacterized MLO genes. Phylogenomic analysis combined to whole-genome transcriptome provides diverse information about the function of MLOs. The further studies can be followed to address the biological processes of each MLO gene by using protein-protein interaction or co-expression analysis (Jin et al., 2013; Nguyen V. N. T. et al., 2014). In particular, their patterns of expression in response to cold or heat stress can give researchers novel directions for functional studies of this gene family. Those tools also allow us to estimate functional redundancy among closely linked family members and can inspire more effective strategies for identifying defective phenotypes in loss-of-function studies. For example, although AtMLO2, AtMLO6, and AtMLO12 are redundant, their roles are not equal in conferring a defense response against fungi. Likewise, single knockout mutants of AtMLO4 and AtMLO11 show a similar root-wave phenotype, but this phenotype is not enhanced in a double mutant. Among MLO genes in rice, OsMLO3 and OsMLO6 are supposed to be originally tandem duplication (Liu and Zhu, 2008). We also determined that OsMLO3 and OsMLO6 are closely related from our phylogenomic data, but expression of OsMLO3 is stronger than that of OsMLO6, meaning that OsMLO3 might have a predominant role between them. Additionally, two pairs of OsMLO10/OsMLO5 and OsMLO8/OsMLO7 are believed to be the result of segmental duplication, but the stronger and unique level of expression of OsMLO10 and OsMLO8 suggests their predominant roles relative to their closest members. These results suggest that functional redundancy in rice MLO family might occur to maintain robustness of few members which might play the important roles in rice during evolution.
One advantage of a phylogenomic analysis is that it informs researchers about which experimental conditions are most suitable when facilitating studies of genes of interest to identify defective phenotypes. Here, we identified MLO genes associated with specific tissue/organ types, including OsMLO12, which is uniquely expressed in pollen at the trinucleate stage (Yi et al., 2014); OsMLO10, which is preferentially expressed in the roots; and OsMLO8, which is preferentially expressed in shoots and in anthers at the trinucleate stage. Because ubiquitous expression of the MLO family members may not be beneficial at all stages of plant development, it is critical that we make careful selections of those genes that have conditional or tissue-preferential regulation when we are developing more desirable rice cultivars. Our combination of microarray data and real-time results demonstrated that OsMLO1, OsMLO2, OsMLO3, OsMLO4, OsMLO9, and OsMLO11 are involved in the response to heat and/or chilling in rice. This suggests that MLO proteins have roles in plant adaptations to temperature stresses. Under those stressful conditions, accumulations of reactive oxygen species (ROS) act as a signal for plant defense response (Knight and Knight, 2001), even that phenomenon can put plants at risk. Hence, exposure to extreme temperatures is a stress that quickly induces H2O2 accumulations and damages plants (O'Kane et al., 1996; Rizhsky et al., 2002; Vacca et al., 2004). Besides, other researchers have reported a negative relationship between MLO protein activity and H2O2 accumulations under biotic stress (Opalski et al., 2005; Kim and Hwang, 2012). Thus, we suggest that the absence of MLO proteins in rice might cause a reduction of H2O2 that enhances tolerance against those environmental challenges. Nevertheless, the biochemical functions of MLO proteins remain to be elusive.
Even though the functions of MLO proteins may have diverged evolutionarily between species, the rice MLO family appears to have a role in defenses against fungal attacks. Piffanelli et al. (2002) initially showed that the induction of hydrogen peroxide (H2O2) accumulations upon infection by a fungal pathogen was stronger in a barley mlo mutant than in the WT. And in this study, upregulation of OsMLO3 at 72 h after pathogen infection might induce the production and accumulation of H2O2, leading to cell death in rice leaves. However, it is unclear why OsMLO1, OsMLO2, OsMLO8, OsMLO9, and OsMLO11 are down-regulated by such infections.
The role of MLO genes has never been mentioned as light responsive genes. In this study, we identified that OsMLO1, OsMLO3, and OsMLO8 in particular showed diurnal behavior during the light/dark cycle. This result suggests that changing day/night cycle effects on the expression of those MLO genes.
Altogether, MLO genes in rice are supposed to share their roles into different processes of development and defense. The seven transmembrane motif-containing MLO proteins perhaps play the role as the perception of various stimuli from environment. Since plants are exposed to variety of stress, increasing intracellular Ca2+ level results in change the level of H2O2 through binding to CaM/CML, ubiquitous calcium-binding protein (McCormack et al., 2005; Reddy et al., 2011; Lee et al., 2012). Moreover, CaM was discovered to be important for defense response in plant by the interaction with MLO (Kim et al., 2002b). This finding suggested the potential communication between plant abiotic responses and immunity via signaling from MLO to Ca2+—dependent CaM/CML, which stimulates the production of H2O2. Additional, co-expression gene of OsMLO3 also convinced the relationship between heat stress and cell death since spl7 mutant increased death cell under heat stress and retained more H2O2 than wildtype. Although no direct evidence of an association between the light pathway and ROS signaling has been described previously, photoperiod is critically related to oxidative signaling. In Arabidopsis, the catalase2 mutant causes oxidative signaling to be activated during short days but not long days, specifically under photorespiratory conditions (Queval et al., 2007). A link between day length and H2O2 accumulations has also been demonstrated with transcriptome data showing that many H2O2-responsive genes are day length-dependent (Queval et al., 2012). As we have suggested here, a MLO mediating relationship among abiotic stress, biotic stress, photoperiod, and H2O2 might exist in rice.
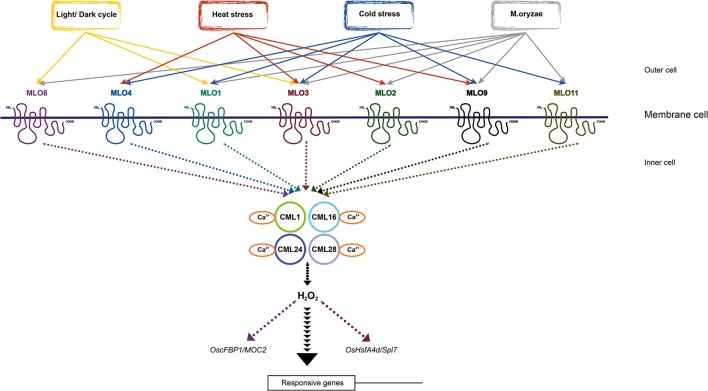
In summary, we have gathered evidence that the MLO family in rice is involved in a light signaling pathway associated with the diurnal rhythm. The results from this work also underscore the roles that these genes have in both abiotic- and biotic-stress responses, as shown in Table 1 and the model depicted in Figure 10. The featured expression patterns were also summarized in Table 1. Environment aspects might stimulate the changing of H2O2 level by means of the interaction MLO and CaM/CML-Ca2+. The production of H2O2 might play as a messenger and stimulate the expression of the responsive genes to adapt to the environmental change. However, further researches will be required to obtain a deeper understanding of the rice MLO genes and their functional implications.
Table 1.
Summary of MLO family members in rice with highlighted meta-expression patterns.
 |
Preferred tissue(s) revealed by qRT-PCR | Diurnal rhythm | Heat stress | Cold stress | Magnaporthe oryzae infection |
|---|---|---|---|---|---|
| OsMLO1 | Leaf, uninucleate anther | Light-inducible |
 a a
|
 b b
|
|
| OsMLO2 | Mature flower |
 a a
|
 b b
|
||
| OsMLO3 | Shoot, leaf | Dark-inducible |
 a a
|
 a a
|
 a a
|
| OsMLO4 | Trinucleate anther |
 a a
|
 a a
|
||
| OsMLO5 | Root, uninucleate anther | ||||
| OsMLO6 | Trinucleate anther | ||||
| OsMLO7 | – | ||||
| OsMLO8 | Leaf, trinucleate anther | Light-inducible |
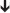 b b
|
||
| OsMLO9 | Leaf, trinucleate anther |
 a a
|
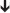 b b
|
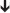 b b
|
|
| OsMLO10 | Root | ||||
| OsMLO11 | Uninucleate anther |
 a a
|
|||
| OsMLO12 | Trinucleate anther |
Up-regulated expression relative to untreated control.
Down-regulated expression relative to untreated control.
Figure 10.
Simplified model of OsMLO mediating stress-response pathways. Model includes 7 OsMLO genes that respond to different experimental conditions, plus OsCML genes in downstream signaling pathway of OsMLO family genes and potential responsive genes in signaling pathways. OsHsfA4d/Spl7 and OscFBP1/MOC2 were presented as downstream factors of OsMLO3 and OsMLO8, respectively. Solid arrows show relationships analyzed in this study; dashed arrows indicate unclear relationships. Individual MLO pathway was indicated by different color.
Author contributions
VN and KV performed the experiments; VN, KV, and KJ analyzed the data; and VN, KV, HP, JJ, and KJ wrote the paper. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was conducted with the supports of “Integrating Research Program (Project No. 20150645 to KJ)” by Kyung Hee University and of Next-Generation BioGreen 21 Program (PJ01100401 to KJ) by Rural Development Administration, Republic of Korea.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01413
Details of microarray data for OsMLO genes from anthers and pollen of indica and japonica rice cultivars. ACP, archespore; PMe, pre-meiotic; Me, meiotic; U, uninucleate stage; B, binucleate stage; T, trinucleate stage; FL, anther at flowering time; MP, mature pollen; GP, geminated pollen.
Agilent 44K data of OsMLO genes, including expression patterns in 13 tissues/organs. Log2 intensities range from 5 (blue) to 15 (yellow).
Expression patterns based on Affymetrix arrays. Phylogenomic analysis of 12 OsMLO genes with integration of abiotic stress meta-data obtained in response to drought, salt, cold, heat, or submergence. Red boxes, genes up-regulated by stress treatment relative to control; green boxes, down-regulated by stress treatment; gray boxes, data not considered in microarray analysis.
Primer sequences used for real-time/RT PCR and genotyping analyses.
Details of microarray data for OsCaM and OsCML genes examined in this study with wild-type rice and osdxr mutant.
Summary of co-expression genes with OsMLO1, 2, 3, 4, 8, and 9 genes in 2532 anatomical samples using co-expression analysis tool in Genevestigator.
References
- Bidzinski P., Noir S., Shahi S., Reinstädler A., Gratkowska D. M., Panstruga R. (2014). Physiological characterization and genetic modifiers of aberrant root thigmomorphogenesis in mutants of Arabidopsis thaliana MILDEW LOCUS O genes. Plant Cell Environ. 37, 2738–2753. 10.1111/pce.12353 [DOI] [PubMed] [Google Scholar]
- Büschges R., Hollricher K., Panstruga R., Simons G., Wolter M., Frijters A., et al. (1997). The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88, 695–705. 10.1016/S0092-8674(00)81912-1 [DOI] [PubMed] [Google Scholar]
- Cao P., Jung K.-H., Choi D., Hwang D., Zhu J., Ronald P. C. (2012). The rice oligonucleotide array database: an atlas of rice gene expression. Rice 5:17. 10.1186/1939-8433-5-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran A. K. N., Lee G.-S., Yoo Y.-H., Yoon U.-H., Ahn B.-O., Yun D.-W., et al. (2016). Functional classification of rice flanking sequence tagged genes using MapMan terms and global understanding on metabolic and regulatory pathways affected by dxr mutant having defects in light response. Rice 9:17. 10.1186/s12284-016-0089-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Noir S., Kwaaitaal M., Hartmann H. A., Wu M.-J., Mudgil Y., et al. (2009). Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in Arabidopsis root thigmomorphogenesis. Plant Cell 21, 1972–1991. 10.1105/tpc.108.062653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consonni C., Humphry M. E., Hartmann H. A., Livaja M., Durner J., Westphal L., et al. (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38, 716–720. 10.1038/ng1806 [DOI] [PubMed] [Google Scholar]
- Daie J. (1993). Cytosolic fructose-1,6-bisphosphatase: a key enzyme in the sucrose biosynthetic pathway. Photosynth. Res. 38, 5–14. 10.1007/BF00015056 [DOI] [PubMed] [Google Scholar]
- Devoto A., Hartmann H. A., Piffanelli P., Elliott C., Simmons C., Taramino G., et al. (2003). Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J. Mol. Evol. 56, 77–88. 10.1007/s00239-002-2382-5 [DOI] [PubMed] [Google Scholar]
- Devoto A., Piffanelli P., Nilsson I., Wallin E., Panstruga R., Von Heijne G., et al. (1999). Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J. Biol. Chem. 274, 34993–35004. 10.1074/jbc.274.49.34993 [DOI] [PubMed] [Google Scholar]
- Elliott C., Zhou F., Spielmeyer W., Panstruga R., Schulze-Lefert P. (2002). Functional conservation of wheat and rice Mlo orthologs in defense modulation to the powdery mildew fungus. Mol. Plant. Microbe. Interact. 15, 1069–1077. 10.1094/MPMI.2002.15.10.1069 [DOI] [PubMed] [Google Scholar]
- Feechan A., Jermakow A. M., Torregrosa L., Panstruga R., Dry I. B. (2008). Identification of grapevine MLO gene candidates involved in susceptibility to powdery mildew. Funct. Plant Biol. 35, 1255–1266. 10.1071/FP08173 [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Thompson J. D., Gibson T. J. (1996). Using CLUSTAL for multiple sequence alignments, in Computer Methods for Macromolecular Sequence Analysis, ed Doolittle R. F. (San Diego, CA: Academic Press; ), 383–402. 10.1016/S0076-6879(96)66024-8 [DOI] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Tyagi A. K., Khurana J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345, 646–651. 10.1016/j.bbrc.2006.04.140 [DOI] [PubMed] [Google Scholar]
- Jin G. H., Gho H. J., Jung K. H. (2013). A systematic view of rice heat shock transcription factor family using phylogenomic analysis. J. Plant Physiol. 170, 321–329. 10.1016/j.jplph.2012.09.008 [DOI] [PubMed] [Google Scholar]
- Jorgensen J. H. (1992). Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 66, 141–152. 10.1007/BF00023919 [DOI] [Google Scholar]
- Jung K.-H., Lee J., Dardick C., Seo Y.-S., Cao P., Canlas P., et al. (2008). Identification and functional analysis of light-responsive unique genes and gene family members in rice. PLoS Genet. 4:e1000164. 10.1371/journal.pgen.1000164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S. A., Shimosato-Asano H., Keinath N. F., Wuest S. E., Ingram G., Panstruga R., et al. (2010). Conserved molecular components for pollen tube reception and fungal invasion. Science 330, 968–971. 10.1126/science.1195211 [DOI] [PubMed] [Google Scholar]
- Kim D. S., Hwang B. K. (2012). The pepper MLO gene, CaMLO2, is involved in the susceptibility cell-death response and bacterial and oomycete proliferation. Plant J. 72, 843–855. 10.1111/tpj.12003 [DOI] [PubMed] [Google Scholar]
- Kim M. C., Lee S. H., Kim J. K., Chun H. J., Choi M. S., Chung W. S., et al. (2002a). Mlo, a modulator of plant defense and cell death, is a novel calmodulin-binding protein. Isolation and characterization of a rice Mlo homologue. J. Biol. Chem. 277, 19304–19314. 10.1074/jbc.M108478200 [DOI] [PubMed] [Google Scholar]
- Kim M. C., Panstruga R., Elliott C., Müller J., Devoto A., Yoon H. W., et al. (2002b). Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416, 447–451. 10.1038/416447a [DOI] [PubMed] [Google Scholar]
- Knight H., Knight M. R. (2001). Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci. 6, 262–267. 10.1016/S1360-1385(01)01946-X [DOI] [PubMed] [Google Scholar]
- Kojo K., Yaeno T., Kusumi K., Matsumura H., Fujisawa S., Terauchi R., et al. (2006). Regulatory mechanisms of ROI generation are affected by rice spl mutations. Plant Cell Physiol. 47, 1035–1044. 10.1093/pcp/pcj074 [DOI] [PubMed] [Google Scholar]
- Koumoto T., Shimada H., Kusano H., She K. C., Iwamoto M., Takano M. (2013). Rice monoculm mutation moc2, which inhibits outgrowth of the second tillers, is ascribed to lack of a fructose-1,6-bisphosphatase. Plant Biotechnol. 30, 47–56. 10.5511/plantbiotechnology.12.1210a [DOI] [Google Scholar]
- Kusch S., Pesch L., Panstruga R. (2016). Comprehensive phylogenetic analysis sheds light on the diversity and origin of the MLO family of integral membrane proteins. Genome Biol. Evol. 8, 878–895. 10.1093/gbe/evw036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Yun H. S., Kwon C. (2012). Molecular communications between plant heat shock responses and disease resistance. Mol. Cells 34, 109–116. 10.1007/s10059-012-0121-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. K., Jeon J. S., Börnke F., Voll L., Cho J. I., Goh C. H., et al. (2008). Loss of cytosolic fructose-1,6-bisphosphatase limits photosynthetic sucrose synthesis and causes severe growth retardations in rice (Oryza sativa). Plant Cell Environ. 31, 1851–1863. 10.1111/j.1365-3040.2008.01890.x [DOI] [PubMed] [Google Scholar]
- Lee S. K., Song M. Y., Seo Y. S., Kim H. K., Ko S., Cao P. J., et al. (2009). Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics 181, 1627–1638. 10.1534/genetics.108.099226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zhu H. (2008). Molecular evolution of the MLO gene family in Oryza sativa and their functional divergence. Gene 409, 1–10. 10.1016/j.gene.2007.10.031 [DOI] [PubMed] [Google Scholar]
- McCormack E., Tsai Y. C., Braam J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389. 10.1016/j.tplants.2005.07.001 [DOI] [PubMed] [Google Scholar]
- Moon J. C., Ham D. J., Hwang S. G., Park Y. C., Lee C., Jang C. S. (2014). Molecular characterization of a heat inducible rice gene, OsHSP1, and implications for rice thermotolerance. Genes Genomics 36, 151–161. 10.1007/s13258-013-0152-y [DOI] [Google Scholar]
- Nguyen M. X., Moon S., Jung K. H. (2013). Genome-wide expression analysis of rice aquaporin genes and development of a functional gene network mediated by aquaporin expression in roots. Planta 238, 669–681. 10.1007/s00425-013-1918-9 [DOI] [PubMed] [Google Scholar]
- Nguyen Q.-N., Lee Y.-S., Cho L.-H., Jeong H.-J., An G., Jung K.-H. (2014). Genome-wide identification and analysis of Catharanthus roseus RLK1-like kinases in rice. Planta 241, 603–613. 10.1007/s00425-014-2203-2 [DOI] [PubMed] [Google Scholar]
- Nguyen V. N. T., Moon S., Jung K.-H. (2014). Genome-wide expression analysis of rice ABC transporter family across spatio-temporal samples and in response to abiotic stresses. J. Plant Physiol. 171, 1276–1288. 10.1016/j.jplph.2014.05.006 [DOI] [PubMed] [Google Scholar]
- O'Kane D., Gill V., Boyd P., Burdon R. (1996). Chilling, oxidative stress and antioxidant responses in Arabidopsis thaliana callus. Planta 198, 371–377. 10.1007/BF00620053 [DOI] [PubMed] [Google Scholar]
- Ohnishi T., Sugahara S., Yamada T., Kikuchi K., Yoshiba Y., Hirano H. -Y., et al. (2005). OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet. Syst. 80, 135–139. 10.1266/ggs.80.135 [DOI] [PubMed] [Google Scholar]
- Ohyanagi H., Tanaka T., Sakai H., Shigemoto Y., Yamaguchi K., Habara T., et al. (2006). The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 34, D741–D744. 10.1093/nar/gkj094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalski K. S., Schultheiss H., Kogel K. H., Hückelhoven R. (2005). The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. Plant J. 41, 291–303. 10.1111/j.1365-313X.2004.02292.x [DOI] [PubMed] [Google Scholar]
- Pessina S., Pavan S., Catalano D., Gallotta A., Visser R. G. F., Bai Y., et al. (2014). Characterization of the MLO gene family in Rosaceae and gene expression analysis in Malus domestica. BMC Genomics 15:618. 10.1186/1471-2164-15-618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli P., Zhou F., Casais C., Orme J., Jarosch B., Schaffrath U., et al. (2002). The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 129, 1076–1085. 10.1104/pp.010954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G., Issakidis-Bourguet E., Hoeberichts F. A., Vandorpe M., Gakière B., Vanacker H., et al. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52, 640–657. 10.1111/j.1365-313X.2007.03263.x [DOI] [PubMed] [Google Scholar]
- Queval G., Neukermans J., Vanderauwera S., van Breusegem F., Noctor G. (2012). Day length is a key regulator of transcriptomic responses to both CO2 and H2O2 in Arabidopsis. Plant Cell Environ. 35, 374–387. 10.1111/j.1365-3040.2011.02368.x [DOI] [PubMed] [Google Scholar]
- Reddy A. S. N., Ali G. S., Celesnik H., Day I. S. (2011). Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23, 2010–2032. 10.1105/tpc.111.084988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S. Y., Beavis W., Berardini T. Z., Chen G., Dixon D., Doyle A., et al. (2003). The Arabidopsis Information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 31, 224–228. 10.1093/nar/gkg076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L., Liang H., Mittler R. (2002). The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 130, 1143–1151. 10.1104/pp.006858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouard M., Guignon V., Aluome C., Laporte M. A., Droc G., Walde C., et al. (2011). GreenPhylDB v2.0: comparative and functional genomics in plants. Nucleic Acids Res. 39, 1095–1102. 10.1093/nar/gkq811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Takehisa H., Kamatsuki K., Minami H., Namiki N., Ikawa H., et al. (2013). RiceXPro Version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 41, 1206–1213. 10.1093/nar/gks1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M., Somerville S. C. (2002). MLO, a novel modulator of plant defenses and cell death, binds calmodulin. Trends Plant Sci. 7, 379–380. 10.1016/S1360-1385(02)02322-1 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca R. A., de Pinto M. C., Valenti D., Passarella S., Marra E., De Gara L. (2004). Production of reactive oxygen species, alteration of cytosolic ascorbate peroxidase, and impairment of mitochondrial metabolism are early events in heat shock-induced programmed cell death in tobacco Bright-Yellow 2 cells. Plant Physiol. 134, 1100–1112. 10.1104/pp.103.035956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E., Yonemaru J., Yamamoto T., Yano M. (2012). OGRO: The overview of functionally characterized genes in rice online database. Rice 5:26. 10.1186/1939-8433-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi U., Yano M., Lin H., Ashikari M., Yamada K. (2002). A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc. Natl. Acad. Sci. U.S.A. 99, 7530–7535. 10.1073/pnas.112209199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J., An S., An G. (2014). OsMLO12, encoding seven transmembrane proteins, is involved with pollen hydration in rice. Plant Reprod. 27, 169–180. 10.1007/s00497-014-0249-8 [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632. 10.1104/pp.104.046367 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of microarray data for OsMLO genes from anthers and pollen of indica and japonica rice cultivars. ACP, archespore; PMe, pre-meiotic; Me, meiotic; U, uninucleate stage; B, binucleate stage; T, trinucleate stage; FL, anther at flowering time; MP, mature pollen; GP, geminated pollen.
Agilent 44K data of OsMLO genes, including expression patterns in 13 tissues/organs. Log2 intensities range from 5 (blue) to 15 (yellow).
Expression patterns based on Affymetrix arrays. Phylogenomic analysis of 12 OsMLO genes with integration of abiotic stress meta-data obtained in response to drought, salt, cold, heat, or submergence. Red boxes, genes up-regulated by stress treatment relative to control; green boxes, down-regulated by stress treatment; gray boxes, data not considered in microarray analysis.
Primer sequences used for real-time/RT PCR and genotyping analyses.
Details of microarray data for OsCaM and OsCML genes examined in this study with wild-type rice and osdxr mutant.
Summary of co-expression genes with OsMLO1, 2, 3, 4, 8, and 9 genes in 2532 anatomical samples using co-expression analysis tool in Genevestigator.