Abstract
In Bacillus subtilis and other Gram-positive bacteria, pyrimidine-mediated regulation of the pyrG gene, which encodes CTP synthetase, occurs through an attenuation mechanism involving an intrinsic transcription terminator in the pyrG leader region. Low intracellular levels of CTP prevent termination at the attenuator by a mechanism that requires the nontemplate strand sequence GGGC at the pyrG transcription initiation site (first G =+1) and the leader transcript sequence GCUCCC located at the 5′ end of the terminator RNA hairpin. In this study, we demonstrate that reiterative transcription adds G residues (up to at least 10) to the 5′ end of pyrG transcripts when B. subtilis cells are starved for pyrimidines but not when cells are grown with excess cytidine. Regulated repetitive addition of G residues, as well as pyrimidine-mediated pyrG regulation, requires the sequence GGGC or GGGT at the start of pyrG transcription. Mutational insertion of four extra G residues at the 5′ end of the pyrG transcript (i.e., 5′-GGGGGGGC) results in constitutive pyrG expression. We propose that the incorporation of extra G residues by reiterative transcription at the wild-type promoter occurs when normal transcription elongation is stalled at position +4 by low levels of the incoming substrate, CTP, during pyrimidine limitation. The poly(G) extensions on the 5′ ends of pyrG transcripts act to prevent transcription attenuation by base pairing with the sequence CUCCCUUUC located in the 5′ strand of the terminator hairpin. This control mechanism is likely to operate in other Gram-positive bacteria containing similar pyrG leader sequences.
The bacterial pyrG gene encodes the enzyme CTP synthetase, which aminates UTP to form CTP as the final step of the pyrimidine nucleotide biosynthetic pathway. Recent studies indicate that pyrG expression in the Gram-positive bacteria Bacillus subtilis and Lactococcus lactis is regulated by a CTP-sensitive transcription attenuation control mechanism (1–3). In this mechanism, intrinsic transcription termination is controlled at an attenuator immediately preceding the pyrG structural gene, but the means by which the attenuator is regulated by CTP was not determined (2, 3).
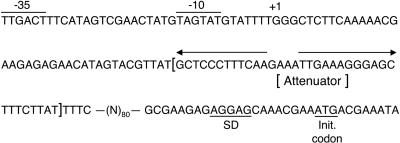
A clue to the mechanism of pyrG regulation came from a comparison of pyrG leader sequences of several Gram-positive bacteria (2). This comparison showed that the lengths and sequences of the leader regions are not conserved except for three short segments that specify GGGC at the 5′ end of the pyrG transcript and two complementary sequences, GCUCCC and GGGAGC, which form the base of the stem of the terminator hairpin (Fig. 1). Systematic mutagenesis of the leader region revealed that the conserved sequences specifying GGGC (or a mutant version GGGU) and the hairpin sequence GCUCCC were critical cis-acting elements required for pyrimidine-mediated regulation of pyrG expression in B. subtilis (3). Based on these results, it was proposed that transcription termination at the pyrG attenuator was controlled by an RNA-binding protein (3). However, a search for the gene encoding this protein was unsuccessful, suggesting an alternative control mechanism.
Fig. 1.
Nucleotide sequence of the pyrG promoter-regulatory region of B. subtilis. The sequence of the nontemplate strand is shown. The –10 and –35 regions of the promoter are overlined and labeled. The transcription start site is labeled +1. The pyrG attenuator sequence is enclosed by brackets, and its dyad symmetry is indicated by arrows. The pyrG Shine–Dalgarno sequence (SD) and initiation codon are underlined and labeled. Sequence of an 80-base spacer region was omitted.
In this study, we provide evidence that control of pyrG expression in B. subtilis occurs by a previously uncharacterized attenuation control mechanism. We show that pyrimidine limitation, apparently by inducing a pause before the insertion of the C residue at +4 in the pyrG transcript, facilitates reiterative transcription (4) after the nontemplate strand sequence GGG of the pyrG initially transcribed region. This reaction adds extra G residues to the 5′ end of the transcript, which permits extensive base pairing between the poly(G) end of the transcript and a tract of nine pyrimidine residues that forms most of the upstream half of terminator hairpin. The formation of this secondary structure precludes formation of the terminator hairpin and allows read-through transcription. Thus, this mechanism provides for high levels of pyrG expression when the cell needs more CTP. We discuss the likelihood that analogous mechanisms control pyrG expression in other Gram-positive bacteria.
Materials and Methods
General Microbiological Materials and Methods. Bacterial strains, growth media, and culture conditions have been described, as were the methods used for transformation of bacteria (2). The pyrimidine auxotrophic strain B. subtilis HC11 (pyrB::Spcr) (5) and its derivatives were grown in minimal medium supplemented with either excess cytidine (200 μg of cytidine per ml) or limiting pyrimidines (100 μg of orotate per ml). Growth on excess cytidine provides ample intracellular levels of CTP and UTP, whereas growth on 100 μg of orotate per ml is suboptimal and results in low intracellular levels of CTP and UTP. The assay for β-galactosidase activity in strains bearing pyrG′-lacZ fusions was as described (2).
DNA and RNA Isolation, Manipulation, and Analysis. Plasmid DNA from Escherichia coli was isolated as described by Sambrook et al. (6) and purified by using the plasmid minikit from Qiagen (Valencia, CA). PCR was performed in a Perkin–Elmer Cetus DNA thermal cycler using TaqDNA polymerase (Invitrogen) under conditions recommended by the supplier. DNA sequencing was performed at the University of Illinois Genetic Engineering Facility. Total RNA was extracted from B. subtilis by using the RNeasy minikit from Qiagen. Cells were harvested in exponential phase. For cells grown in minimal medium with cytidine, 50 ml of culture was harvested at OD600 ≈ 0.6. For cells grown in minimal medium with orotate, 100 ml of culture was harvested at OD600 ≈ 0.3.
Plasmid and Strain Constructions. Plasmid pMS46U, in which four G residues (nontemplate stand sequence) were inserted at position +1 of the pyrG promoter region, was constructed by using two oligodeoxynucleotides, pyrG-46 and pyrG-U. The sequences of the pyrG-46 and pyrG-U oligos were 5′-CGGAATTCCTTCAAAACGATCTTGACTTTCATAGTCGAACTATGTAGTATGTATTTTGGGGGGGCTCTTCAAAAACGAAGAGAGAAC and 5′-CGGGATCCGGTGAAAATAAGAAAGCTCCCTTTCAATTTCTTGAAAGGGAGCATAACGTACTATGTTCTCTCTTCGTTTTTGAA, respectively. In each sequence, a 20-nt region complementary to the other oligo is underlined, the inserted G residues (in pyrG-46) are in italic type, and an embedded (EcoRI or BamHI) restriction site is in bold type. The two oligos (100 pmol of each) were annealed, and the overhanging DNA ends were filled in by using the Klenow (exo–) fragment of E. coli DNA polymerase I as described (7). The double-stranded DNA products were purified with the MinElute reaction clean-up kit from Qiagen, digested with EcoRI and BamHI, and ligated to EcoRI + BamHI-digested plasmid pDH32 (8), yielding plasmid pMS46U. This plasmid was integrated into the chromosomal amyE gene of B. subtilis strain HC11 as described (2) to generate strain QM425.
Plasmids containing the wild-type or single-base-replacement mutant versions of the B. subtilis pyrG promoter region were constructed by PCR amplification of nucleotides –49 to +81 of the desired promoter regions. The DNA templates used for PCR were previously described plasmids containing wild-type and mutant pyrG′-lacZ fusions (3). The PCR primers used were pyrG-44 (5′-CGGGCTGCAGCTTCAAAACGATCTTGACT; the PstI site is underlined) and pyrG-H (2). The PCR products were digested with PstI and BamHI and inserted between the PstI and BamHI sites of the high-copy-number plasmid pEB112 (9). These plasmids were transformed into B. subtilis HC11 with selection for neomycin (7 μg/ml) resistance, and the transformants were used for primer extension mapping of wild-type and mutant pyrG transcripts. The derivative of plasmid pEB112 containing the wild-type pyrG promoter region was designated plasmid pEBWT.
Primer Extension Mapping. Primer extension mapping of pyrG transcripts was performed as described by Saxild et al. (10). Primer C (2) (60 pmol), which is complementary to nucleotides 15–34 in the pyrG transcript, was labeled with 0.22 μM [γ-33P]ATP [10 μCi (1 Ci = 37 GBq), total radioactivity] by using T4 polynucleotide kinase (Invitrogen) as suggested by the supplier. The labeled primer was purified by gel filtration using a Sephadex G-50 column (Amersham Pharmacia). One picomole of labeled primer C, 2 μg of total cellular RNA, and 10 units of avian myeloblastosis virus reverse transcriptase (Promega) were included in the reaction mixtures, which were incubated at 37°C for 1 h. To produce the sequence ladder used to identify primer extension products, dideoxy sequencing of the pyrG promoter region was performed by using 5′-33P-labeled primer C, plasmid pJH4133 (11) as template, and the T7 Sequenase v2.0 DNA sequencing kit (United States Biochemical). Primer extension and sequencing reaction products were electrophoretically separated on a denaturing 10% polyacrylamide gel and visualized with a Molecular Dynamics PhosphorImager.
Sequencing 5′ Ends of pyrG Transcripts. The 5′ ends of pyrG transcripts were sequenced by using a modified version of the ligation-anchored PCR procedure described by Troutt et al. (12). In this method, cDNA copies of the 5′ ends of pyrG transcripts were generated by primer extension essentially as described above. The cellular RNA used as a source of template was isolated from B. subtilis strain HC11 or HC11/pEBWT grown under conditions of pyrimidine limitation. The amount of cellular RNA added to the primer extension reaction mixture was 2 and 10 μg for strains HC11 and HC11/pEBWT, respectively. After primer extension and analysis of the products by polyacrylamide gel electrophoresis, the cDNAs corresponding to 5′-extended pyrG transcripts were isolated. T4 RNA ligase (10 units, New England Biolabs) was then used to ligate the cDNAs to an anchor oligodeoxynucleotide (10 pmol) with the sequence 5′-GCCAAAGACAGTGCGGGGGATCCGCC. The anchor oligo contained a 5′ phosphate and a 3′ dideoxy-C residue, with the latter designed to prevent ligation to this end of the molecule. The products of the ligation reaction were used as templates for PCR using a primer pair consisting of a primer specific for the anchor oligo (5′-CGCGGATCCCCCGCACTGTCTTTGGC; the BamHI site is underlined) and another specific for sequence downstream of the pyrG promoter region (5′-GCGAATTCCGTACTATGTTCTCTCTTCG; the EcoRI site is underlined). The PCR products were analyzed by agarose gel electrophoresis. DNA within the expected size range were purified by using the QIAXII kit (Qiagen), digested with EcoRI and BamHI, and inserted into EcoRI + BamHI-digested plasmid pZero2.1 (Invitrogen). Recombinant plasmids were transformed into E. coli, and several transformants were used to prepare plasmid DNA. Plasmids containing EcoRI–BamHI fragments between 65 and 100 bp were identified, and these fragments were sequenced.
Results
Reiterative Transcription Adds G Residues to the 5′ Ends of pyrG Transcripts During Pyrimidine Limitation. Previously, we demonstrated by primer extension mapping that only one major pyrG transcript was synthesized in B. subtilis HC11 (pyrB::Spcr) cells grown with excess cytidine (2). This transcript was initiated at a G residue located 8 bases downstream of the –10 region of the pyrG promoter (Fig. 1). In contrast, primer extension mapping of pyrG transcripts synthesized in cells limited for pyrimidines produced a ladder of primer extension products, which indicated the addition of up to 10 extra nucleotides to the 5′ ends of pyrG transcripts (2). (A repeat of this experiment is shown below.) Significantly, the initially transcribed region of the pyrG promoter begins with three G residues in the nontemplate strand (Fig. 1). Previous studies indicate that such homopolymeric tracts can support reiterative transcription that will add extra G residues to the 5′ ends of nascent transcripts (4, 13–15). Such reiterative transcription is facilitated by a reduced rate of normal transcription elongation after the polymerization of the first three G residues (16), which may occur by slow insertion of a C residue at position +4in B. subtilis cells starved for pyrimidines. Thus, it seemed likely that the multiple pyrG transcripts indicated by primer extension mapping were the products of reiterative transcription and contained up to 10 extra G residues at their 5′ ends.
To determine the sequence of the 5′ ends of the apparently extended pyrG transcripts, strain B. subtilis HC11 (pyrB::Spcr) was grown under conditions of pyrimidine limitation. RNA was isolated from these cells and used in a modified version of the ligation-anchored PCR procedure (12) to amplify DNA corresponding to 5′ ends of pyrG transcripts. The PCR products were cloned, and three independent clones were sequenced. The results indicated that the sequences of the corresponding pyrG transcripts were as specified by the pyrG DNA except that they contained 5, 7, and 7 extra 5′ G residues, and thus these transcripts possessed 5′ poly(G) tracts of 8, 10, and 10 residues, respectively. This experiment was repeated by using cellular RNA from pyrimidine-limited cells of B. subtilis HC11/pEBWT, which carries multiple copies of the wild-type pyrG promoter region. One clone was sequenced, and this sequence contained 8 extra G residues, indicating a 5′ poly(G) tract of 11 G residues. These results establish that, under conditions of pyrimidine limitation, reiterative transcription occurs after the first three residues of the initially transcribed region of the pyrG promoter and produces transcripts containing poly(G) tracts up to at least 11 residues in length.
Effects of Regulatory Mutations in the pyrG Initially Transcribed Region on Reiterative Transcription. We previously constructed single base-pair substitution mutations in the initially transcribed region of the B. subtilis pyrG promoter. These mutations were incorporated into pyrG′-lacZ fusions, which were integrated individually into the B. subtilis chromosome. The effects of the mutations on pyrimidine-mediated regulation of pyrG expression were measured (3). Our results showed that a G-to-A substitution (nontemplate strand sequence) at position +1, +2, or +3 reduced pyrG expression to extremely low levels and prevented derepression of pyrG expression by pyrimidine limitation (Table 1). To assess the effects of the same mutations on reiterative transcription at the pyrG promoter, the transcripts initiated at the mutant promoters were analyzed by primer extension mapping. For comparison, transcripts initiated at the wild-type pyrG promoter were also analyzed. To improve sensitivity, cellular RNA was isolated from derivatives of B. subtilis strain HC-11(pyrB::Spcr) carrying either the wild-type or a mutant pyrG promoter region on a recombinant version of the high-copy-number plasmid pEB112. Strains were grown with either excess cytidine or limiting pyrimidines. These growth conditions result in ample or low intracellular levels of pyrimidine nucleoside triphosphates, respectively.
Table 1. Effects of mutations in the pyrG initially transcribed region on regulation of pyrG′-lacZ expression.
| β-Galactosidase activity*
|
|||
|---|---|---|---|
| Strain (pyrG′-lacZ genotype) | + Cytidine | + Orotate | Fold regulation |
| QM401 (wild type) | 40 ± 4 | 600 ± 34 | 15 |
| QM416 (G + 1 to A) | <1 | 1.4 ± 0.6 | |
| QM404 (G + 2 to A) | 5 ± 0.4 | 1.3 ± 0.2 | 0.3 |
| QM405 (G + 3 to A) | <1 | <1 | |
| QM412 (C + 4 to A) | 100 ± 36 | 200 ± 51 | 2 |
| QM413 (C + 4 to T) | 14 ± 2 | 550 ± 92 | 39 |
| QM425 (G4-insertion) | 960 ± 135 | 1990 ± 54 | 2 |
All data, except for strain QM425, are reprinted with permission from ref. 3.
Expressed in Miller units; mean of six determinations.
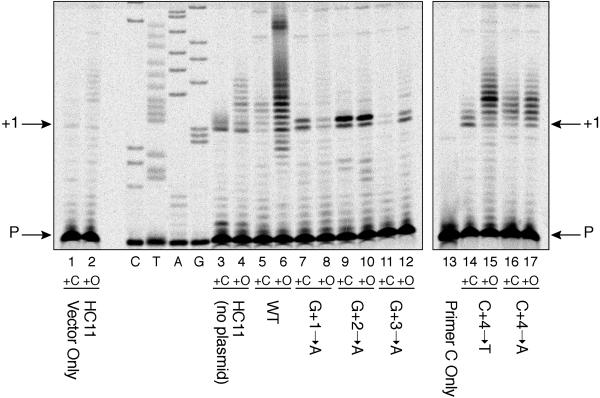
The results show that extensive reiterative transcription occurred at the wild-type promoter under conditions of pyrimidine limitation (+O) but not when cells were grown with excess cytidine (+C) (Fig. 2). This pattern was observed with the chromosomal promoter in the HC11 transformant carrying the cloning vector pEB112 (lanes 1 and 2), with the chromosomal promoter of strain HC11/no plasmid (lanes 3 and 4), and with the promoter on the multicopy plasmid (lanes 5 and 6). One difference observed between the plasmid and chromosomal wild-type promoters was that, in cells grown with excess cytidine, there was more (although clearly restricted) reiterative transcription at the promoter carried on a multicopy plasmid. The reason for this basal level of reiterative transcription is not known. In contrast to the results with the wild-type promoter, reiterative transcription was not observed with the mutant promoters (Fig. 2, lanes 7–12). In cells grown with excess cytidine or limiting pyrimidines, two transcripts were synthesized: one seemed to be the “normal” pyrG transcript and the other was 1 nt longer. This longer transcript could indicate the efficient use of a second start site, seven bases downstream of the –10 region, at the mutant promoters. In any case, each G-to-A mutation simultaneously eliminated reiterative transcription and high levels of pyrG expression under conditions of pyrimidine limitation.
Fig. 2.
Primer extension mapping of pyrG transcripts of B. subtilis strain HC11 (pyrB::Spcr) and HC11 cells containing plasmid pEB112-borne wild-type and mutant pyrG 5′ promoter-leader regions. Cells were grown with excess cytidine (+C) or the poor pyrimidine source orotate (+O). Primer extension products are shown for strain HC11 bearing pEB112 only (lanes 1 and 2), HC11 without a plasmid (lanes 3 and 4), and HC11 bearing pEB112-derived plasmids containing pyrG promoter-leader sequences as follows: wild type (lanes 5 and 6), G+1-to-A mutation (lanes 7 and 8), G+2-to-A mutation (lanes 9 and 10), G+3-to-A mutation (lanes 11 and 12), C+4-to-T mutation (lanes 14 and 15), and C+4-to-A mutation (lanes 16 and 17). Lane 13 shows the position of radiolabeled primer C only (also labeled “P” and indicated by an arrow). C, T, A, and G indicate the lanes of a dideoxy sequencing ladder produced with a pyrG DNA template and radiolabeled primer C. Note that we labeled the sequencing ladder to correspond to nontemplate strand sequence. The position of bands corresponding to transcripts initiated at G+1 is indicated.
In our previous study (3), we also constructed and characterized mutations that replace the C residue at position +4 (Fig. 1) with either an A or a T residue. The effects of these mutations on pyrG expression were different. The C-to-T mutation had only moderate effects on the levels of pyrG expression and slightly increased pyrimidine-mediated regulation, whereas the C-to-A mutation caused constitutive pyrG expression (Table 1). [The residual 2-fold increase in expression in pyrimidine-starved cells seen with QM412 is probably a nonspecific effect of growth limitation that is unrelated to attenuation (2, 3).] The effects of these two mutations on reiterative transcription at the pyrG promoter were analyzed by primer extension mapping as described above. In the case of the C-to-T mutation, the results were essentially the same as those observed with the wild-type promoter. They indicated little reiterative transcription with excess cytidine and extensive reiterative transcription during pyrimidine limitation (Fig. 2, lanes 14 and 15). These results show a correlation between extensive reiterative transcription at the pyrG promoter and high levels of pyrG expression. In the case of the C-to-A mutation, the pattern of primer extension products indicated a significant level of reiterative transcription, which was only modestly affected by the pyrimidine source (Fig. 2, lanes 16 and 17). These results, coupled with the effects of the C-to-A mutation on pyrG expression and regulation (3), reinforce the direct correlation between reiterative transcription and pyrG expression.
It should be noted that in several independent primer extension mapping experiments such as the one shown in Fig. 2, the general patterns of bands were highly reproducible. However, we observed significant variation in the amount of primer extension product. Consequently, we avoided drawing conclusions based on levels of primer extension products.
Four Extra G Residues at the 5′ End of the pyrG Transcript Are Sufficient to Cause Constitutive pyrG Expression. To test directly whether the addition of extra G residues to the 5′ ends of pyrG transcripts was sufficient to cause constitutive pyrG expression, we constructed a pyrG′-lacZ fusion in which four G residues (nontemplate strand sequence) were inserted at position +1 of the pyrG promoter. This promoter region specifies pyrG transcripts containing a 5′ poly(G) tract of at least seven residues even under conditions of cytidine excess. The mutant pyrG′-lacZ fusion was integrated into the chromosome of B. subtilis strain HC11. This strain, designated QM425, and an isogenic strain with a wild-type promoter in the pyrG′-lacZ fusion, designated QM401, were grown with excess cytidine and limiting pyrimidines. Exponential-phase cells were harvested, and β-galactosidase levels were measured (Table 1). In strain QM425 (mutant fusion) grown with excess cytidine and limiting pyrimidines, the β-galactosidase levels were 960 and 1,990 Miller units, respectively. In strain QM401 (wild-type fusion) grown with excess cytidine and limiting pyrimidines, the β-galactosidase levels were 40 and 600 Miller units, respectively. These results clearly demonstrate that the addition of at least four G residues to pyrG transcripts is sufficient to cause high levels of pyrG expression, which is only modestly reduced under conditions of cytidine excess.
Discussion
Reiterative transcription, which is also referred to as pseudotemplated transcription, transcriptional slippage, and RNA polymerase stuttering, is a reaction catalyzed by bacterial, phage, viral, and eukaryotic RNA polymerases (4, 14, 15, 17). In this reaction, nucleotides are repetitively added to the 3′ end of a nascent transcript caused by slippage between the transcript and the DNA or RNA template. Typically, slippage occurs between a homopolymeric sequence in the transcript and at least three complementary bases in the template (18, 19). In most cases, the mechanism involves one or more rounds of a one-base upstream shift of the transcript so that the same nucleotide in the template specifies multiple residues in the transcript (20, 21). Reiterative transcription can occur during transcription initiation or elongation, resulting in transcripts that can be immediately released from RNA polymerase (4, 22, 23) or extended into productive transcripts after a switch to nonreiterative nucleotide addition (17, 22). Reiterative transcription can involve the addition of any of the nucleotide substrates; however, these substrates are not equivalent. For example, during transcription initiation, repetitive U addition, but not A or G addition, generally causes the nascent transcripts to be aborted (14, 22). Recent studies indicate that reiterative transcription plays an important role in the expression and regulation of a number of bacterial and viral genes by a variety of mechanisms (4, 21, 22, 24).
In the present study, we demonstrated that reiterative transcription can occur after the first three bases (i.e., GGG, nontemplate strand sequence) of the initially transcribed region of the pyrG promoter and add as many as 10 extra G residues to the 5′ ends of pyrG transcripts in B. subtilis. This reaction is conditional, occurring at a high level only when cells are grown under conditions of pyrimidine limitation. Pyrimidine limitation causes low intracellular levels of CTP, which seems to be the negative effector of pyrimidine-mediated attenuation control of pyrG expression in B. subtilis and other Gram-positive bacteria (1, 2). Mutational analysis of the pyrG initially transcribed region demonstrated a direct correlation between extensive reiterative transcription and high levels of pyrG expression in B. subtilis. This analysis also revealed that the nucleotide at position +4, which is a C residue in the initially transcribed region of the wild-type pyrG promoter, is important for regulation. Changing the position +4 residue to an A nearly eliminated pyrimidine-mediated regulation. However, changing this residue to a T (specifying a U in the transcript) only slightly affected regulation. Under the conditions we used to measure pyrimidine-mediated regulation, the intracellular levels of both CTP and UTP vary in parallel. Our results indicate that pyrimidine-mediated regulation of pyrG expression requires that a pyrimidine nucleotide be specified by position +4 of the initially transcribed region. Presumably, low intracellular levels of this nucleotide result in a transcription pause that would facilitate reiterative transcription. In fact, in comparable situations, such a transcription pause has been shown to enhance reiterative transcription (16, 25, 26). Finally, we showed that a mutation that specifies four extra G residues at the 5′ end of the pyrG transcript (for a total of seven) causes constitutive pyrG expression. This result indicates that the extra G residues, even without pyrimidine limitation and presumably without reiterative transcription, are sufficient to allow transcription through the pyrG attenuator.
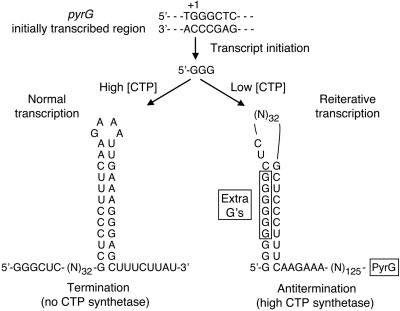
Based on these observations, we propose the following model for regulation of pyrG expression in B. subtilis. Transcription is initiated eight bases downstream of the –10 region of the pyrG promoter, the nascent transcript with the sequence GGG is synthesized, and RNA polymerase is positioned to insert the next nucleotide, a C residue according to the sequence of the pyrG initially transcribed region. In cells grown with ample pyrimidines and containing high levels of CTP, a C residue is readily incorporated at the fourth position in the transcript. This transcript is elongated normally until the pyrG attenuator is transcribed. Then the 3′ end of the transcript forms a terminator hairpin followed by a U-rich tract, which together cause termination of the pyrG transcript (Fig. 3). Thus, the pyrG gene is not expressed and CTP synthetase is not made when the cell does not need additional CTP. On the other hand, in cells grown under pyrimidine-limiting conditions and containing low levels of CTP, the addition ofaCresidue to the nascent GGG transcript is slow and transcription pauses. This pause provides time for the nascent transcript to slip upstream (relative to the DNA template) and allow an extra G residue to be added to the nascent transcript. This process can be repeated multiple times (e.g., up to 10 times) until eventually a C residue is inserted. The transcript is then elongated normally until the attenuator sequence that specifies the upstream segment of the terminator hairpin is transcribed. The sequence of this RNA segment in B. subtilis is 5′-GCUCCCUUUCAA, which includes a tract of nine pyrimidines. Because both C and U residues base pair with G residues (27), the run of pyrimidines will immediately base pair with the poly(G) tract at the 5′ end of the transcript. The stability of this base pairing depends on the number of G residues at the 5′ end of the transcript; seven or more G residues are apparently sufficient for the formation of a stable “antiterminator” secondary structure (Fig. 3). As RNA polymerase continues elongating the pyrG transcript, formation of the terminator hairpin is precluded (by formation of the antiterminator hairpin), and full-length pyrG transcripts are synthesized. These transcripts are translated to make CTP synthetase, which is needed by the cells starved for pyrimidines. Although the model describes pyrG expression at high and low intracellular levels of CTP, we presume that regulation occurs continuously over a wide range of CTP concentrations. These concentrations influence the extent of reiterative transcription, which in turn controls the level of read-through transcription at the pyrG attenuator.
Fig. 3.
Model for CTP-mediated regulation of pyrG expression in B. subtilis. The model shows the effects of CTP concentration on the fate of the pyrG transcript after the first three G residues have been incorporated into the nascent transcript. High CTP concentrations allow normal transcript elongation until intrinsic termination occurs in the pyrG leader region. Low CTP concentrations induce a transcription pause that allows reiterative transcription and the addition of extra G residues, which participate in the formation of an antiterminator hairpin. The extra G residues are boxed. The model shows the insertion of six extra G residues, but as many as 10 extra residues can be added.
No trans-acting regulatory protein is involved in this mechanism, nor does the regulatory effector (CTP) bind to the leader region of the transcript, as has been demonstrated recently for regulatory systems referred to as riboswitches (28, 29). Our model also provides an explanation for the previous observation that the pyrG attenuator functions as an unregulated terminator of transcription of the upstream rpoE gene (2). Presumably, the rpoE transcript does not contain the extra G residues, which are only added during initiation at the pyrG promoter, and is unable to form the antiterminator hairpin.
Although our model is consistent with all available data related to the regulation of pyrG expression, there are several aspects of the model and supporting data that require further clarification. For instance, the model assumes that the antiterminator hairpin forms quickly and sequesters RNA needed to form the terminator hairpin. This assumption seems reasonable, because essentially the same type of competition is known to occur during attenuation control of amino acid biosynthetic operons in enteric bacteria (30). Also, in the model, extra G residues at the 5′ end of the pyrG transcript are necessary to form the antiterminator hairpin, but the exact number of extra G residues needed to accomplish this task is not clear. Our data indicate that four or more extra G residues are sufficient and that reiterative transcription can add at least 10 extra G residues. However, the ability to form a stable antiterminator hairpin remains to be determined for poly(G) tracts of various length. Related to this issue is the possibility that the number of extra G residues added to the 5′ end of pyrG transcripts is limited, perhaps by the architecture of the transcription initiation complex. In fact, such a limitation has been reported for a T7 promoter, at which reiterative transcription occurs, but no more than 11 extra G residues are added (15). Finally, it is not clear why the C-to-A mutation at position +4 of the pyrG promoter enhances reiterative transcription. One possibility is that this mutation creates a transcription pause site at position +4 and pausing occurs in cells grown in both media examined in this study.
Our model is fundamentally different from a model recently proposed by Jørgensen et al. (1) for CTP-sensitive attenuation control of pyrG expression in L. lactis. In their model, low CTP concentrations cause RNA polymerase to pause at a stretch of C residues in the pyrG leader, thereby allowing an antiterminator (not the same as proposed in this study) to form and transcription to proceed. The exact location of the regulatory stretch of C residues was not identified, and the proposed antiterminator structure is likely to be unstable. We believe that the mechanism proposed by Jørgensen et al. is incorrect. The upstream half and loop of the terminator hairpin of the L. lactis pyrG leader region contains a run of nine pyrimidine residues, and the first four bases of the initially transcribed region of the L. lactis pyrG promoter are identical to the corresponding region of B. subtilis. Thus, it seems likely that attenuation control of pyrG expression in L. lactis occurs by the same mechanism as we propose for this regulation in B. subtilis.
It was recently reported that the sequences of the initially transcribed regions and attenuators in the pyrG promoter-leader regions of many Gram-positive bacteria were strongly conserved (1, 2). Specifically, the first four (nontemplate strand) bases of the initially transcribed region are always GGGC, and the upstream half of the attenuator contains a long run of pyrimidines. Therefore, we suspect that the mechanism that we have proposed for pyrG regulation in B. subtilis also regulates pyrG expression in other Gram-positive bacteria possessing the conserved sequences. This model for pyrG regulation adds to the rapidly growing list of remarkable mechanisms of gene regulation in which the leader transcript plays a central role in sensing the level of its operon's end-product and then appropriately adjusting the expression of this operon (4, 22, 28–32).
Acknowledgments
This research was supported by National Institutes of Health Grants GM47112 (to R.L.S.) and GM29466 (to C.L.T.).
References
- 1.Jørgensen, C. M., Hammer, K. & Martinussen, J. (2003) J. Bacteriol. 185, 6562–6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meng, Q. & Switzer, R. L. (2001) J. Bacteriol. 183, 5513–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng, Q. & Switzer, R. L. (2002) J. Bacteriol. 184, 6734–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qi, F. & Turnbough, C. L., Jr. (1995) J. Mol. Biol. 254, 552–565. [DOI] [PubMed] [Google Scholar]
- 5.Hu, P. & Switzer, R. L. (1995) Arch. Biochem. Biophys. 316, 260–266. [DOI] [PubMed] [Google Scholar]
- 6.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 7.Bonner, E. R., D'Elia, J. N., Billips, B. K. & Switzer, R. L. (2001) Nucleic Acids Res. 29, 4851–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grandoni, J. A., Fulmer, S. B., Brizzio, V., Zahler, S. A. & Calvo, J. M. (1993) J. Bacteriol. 175, 7581–7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leonhardt, H. & Alonso, J. C. (1988) J. Gen. Microbiol. 134, 605–609. [DOI] [PubMed] [Google Scholar]
- 10.Saxild, H. H., Jacobsen, J. H. & Nygaard, P. (1995) Microbiology 141, 2211–2218. [DOI] [PubMed] [Google Scholar]
- 11.Trach, K., Chapman, J. W., Piggot, P., LeCoq, D. & Hoch, J. A. (1988) J. Bacteriol. 170, 4194–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Troutt, A. B., McHeyzer-Williams, M. G., Pulendran, B. & Nossal, G. J. V. (1992) Proc. Natl. Acad. Sci. USA 89, 9823–9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nüsslein, C. & Schaller, H. (1975) Eur. J. Biochem. 56, 563–569. [DOI] [PubMed] [Google Scholar]
- 14.Jacques, J. P. & Kolakofsky, D. (1991) Genes Dev. 5, 707–713. [DOI] [PubMed] [Google Scholar]
- 15.Martin, C. T., Muller, D. K. & Coleman, J. E. (1988) Biochemistry 27, 3966–3974. [DOI] [PubMed] [Google Scholar]
- 16.Guajardo, R., Lopez, P., Dreyfus, M. & Sousa, R. (1998) J. Mol. Biol. 281, 777–792. [DOI] [PubMed] [Google Scholar]
- 17.Linton, M. F., Raabe, M., Pierotti, V. & Young, S. G. (1997) J. Biol. Chem. 272, 14127–14132. [DOI] [PubMed] [Google Scholar]
- 18.Xiong, X. F. & Reznikoff, W. S. (1993) J. Mol. Biol. 231, 569–580. [DOI] [PubMed] [Google Scholar]
- 19.Cheng, Y., Dylla, S. M. & Turnbough, C. L., Jr. (2001) J. Bacteriol. 183, 221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, H.-C. & Roberts, J. W. (1990) Biochemistry 29, 10702–10709. [DOI] [PubMed] [Google Scholar]
- 21.Hausmann, S., Garcin, D., Delenda, C. & Kolakofsky, D. (1999) J. Virol. 73, 5568–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, C., Heath, L. S. & Turnbough, C. L., Jr. (1994) Genes Dev. 8, 2904–2912. [DOI] [PubMed] [Google Scholar]
- 23.Barr, J. N. & Wertz, G. W. (2001) J. Virol. 75, 6901–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen, B., Wills, N. M., Nelson, C., Atkins, J. F. & Gesteland, R. F. (2000) Proc. Natl. Acad. Sci. USA 97, 1683–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vidal, S., Curran, J. & Kolakofsky, D. (1990) EMBO J. 9, 2017–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, D. J. (1996) J. Biol. Chem. 271, 11659–11667. [PubMed] [Google Scholar]
- 27.Jaeger, J. A., Turner, D. H. & Zuker, M. (1989) Proc. Natl. Acad. Sci. USA 86, 7706–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandal, M., Boese, B., Barrick, J. E., Winkler, W. C. & Breaker, R. R. (2003) Cell 113, 577–586. [DOI] [PubMed] [Google Scholar]
- 29.Grundy, F. J. & Henkin, T. M. (2004) Curr. Opin. Microbiol. 7, 126–131. [DOI] [PubMed] [Google Scholar]
- 30.Landick, R., Turnbough, C. L., Jr., & Yanofsky, C. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, eds. Neidhardt, F. C., Curtiss, R., III, Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (Am. Soc. Microbiol., Washington, DC), Vol. 1, pp. 1263–1286. [Google Scholar]
- 31.Wilson, H. R., Archer, C. D., Liu, J. & Turnbough, C. L., Jr. (1992) J. Bacteriol. 174, 514–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler, W. C., Nahvi, A., Roth, A., Collins, J. A. & Breaker, R. R. (2004) Nature 428, 281–286. [DOI] [PubMed] [Google Scholar]