Abstract
The classification performance of Kraken was evaluated in terms of sensitivity and specificity when using short and long 16S rRNA sequences. A total of 440,738 sequences from bacteria with complete taxonomic classifications were downloaded from the high quality ribosomal RNA database SILVA. Amplicons produced (86,371 sequences; 1450 bp) by virtual PCR with primers covering the V1–V9 region of the 16S-rRNA gene were used as reference. Virtual PCŔs of internal fragments V3–V4, V4–V5 and V3–V5 were performed. A total of 81,523, 82,334 and 82,998 amplicons were obtained for regions V3–V4, V4–V5 and V3–V5 respectively. Differences in depth of taxonomic classification were detected among the internal fragments. For instance, sensitivity and specificity of sequences classified up to subspecies level were higher when the largest internal fraction (V3–V5) was used (54.0 and 74.6% respectively), compared to V3–V4 (45.1 and 66.7%) and V4–V5 (41.8 and 64.6%) fragments. Similar pattern was detected for sequences classified up to more superficial taxonomic categories (i.e. family, order, class…). Results also demonstrate that internal fragments lost specificity and some could be misclassified at the deepest taxonomic levels (i.e. species or subspecies). It is concluded that the larger V3–V5 fragment could be considered for massive high throughput sequencing reducing the loss of sensitivity and sensibility.
Keywords: Biological sciences, Microbiology, Bioinformatics, Genetics
1. Introduction
Research aimed to study microbial communities using 16S rRNA gene sequences must be based on accurate taxonomy classifications. Next generation sequencing (NGS) technologies emerged at the end of the last decade as the most powerful and promising tool for the study and classification of prokaryotic communities thriving in any of the diverse natural environments. These technologies led to a decreased cost per megabase and as consequence, an increase in the number and diversity of sequenced genes and genomes (Goodwin et al., 2016). Several platforms such as Roche GS20, Roche 454, Applied Biosystems 3730xl and GS-FLX, Applied Biosystems Ion Torrent, Illumina Mi Seq and HiSeq, and others have been developed and improved during the last decade.
However, current NGS platforms work with high-throughput short-read sequences (300 bp max in each sense) and therefore consider only a fraction of the 16S rRNA. Universal primers targeting internal conserved regions have been designed to elucidate the bacterial profile of a given sample; however, no primer is truly universal and the coverage of these primers depends on the environment; furthermore, regions of the rRNA gene differ in taxonomic informativeness (Soergel et al., 2012). Thus, selected primers are required to be not only highly accurate, but to amplify those regions containing greater information than the others (Martínez-Porchas and Vargas-Albores, 2015). Therefore, current methods used for taxonomy classification have to be adapted to the capabilities of the NGS technologies; i.e. considering shorter sequences and using internal primers.
Novel algorithms have been created to study these short sequences including k-mers. Herein, Wood and Salzberg (2014) designed a novel computational program (Kraken) based on ultrafast-metagenomic sequence classification using exact alignments and its use is being extended rapidly (Flygare et al., 2016; Lindgreen et al., 2016; Lu et al., 2016; Susilawati et al., 2016; Valenzuela-González et al., 2016, Yang et al., 2016a; Yang et al., 2016b). This tool assigns taxonomic labels to noisy sets of DNA sequences and has demonstrated greater speed and sensitivity than state-of-the-art tools. For instance, Valenzuela-González et al. (2016), demonstrated that Kraken not only analyzes short DNA sequences, but has an acceptable classification performance analyzing long Sanger-sequenced samples compared to the Ribosomal Database Project classifier (RDP classifier).
Kraken performs alignments of short and large sequences, mapping each sequence to the lowest ancestor, forming subtrees and assigning specific weight to each node (equal to the number of sequences associated with the node’s taxon). Despite using exact alignments avoids the generation of chimeric subtrees, there is still a very small probability to have chimeras causing false classifications. Furthermore, Soergel et al. (2012) asserted that taxonomic classifications using short reads should be treated with skepticism. Current NGS technologies are aimed at the generation of longer read lengths (up to 5 kbp) (Steinbock and Radenovic, 2015) and thus to improve classification sensitivity; however, the use of short reads is still the most economically viable strategy and therefore it is of paramount relevance to evaluate how much information is sacrificed when using short sequences instead of the complete 16S rRNA gene. Considering the above information, the aim of this study was to evaluate the classification performance of Kraken in terms of sensitivity and specificity when using short and long 16S rRNA sequences generated in silico from the robust rRNA database SILVA release 123.
2. Materials and methods
2.1. Sequences
A total of 513,309 bacteria sequences were downloaded from the high quality ribosomal RNA database SILVA (version 123). From these, only 440,738 that had complete taxonomic classifications (from phylum to species) were used for virtual PCR. Virtual amplicons with identical sequence were grouped and considered as a single amplicon.
2.2. Virtual PCR
A homemade PHP script was used to simulate in silico PCR reactions, using primers for the amplification of the complete 16S rRNA gene (Table 1). Identical sequences were grouped and the in silico PCR reaction for the amplification of internal fragments was performed on these unique large sequences (unilarge) using the corresponding internal primers (Table 1). Primers isoforms were generated by substituting degenerate nucleotides with the corresponding bases. Each isoform was searched in the gene sequence; if not found, the length of the primer was reduced, eliminating one nucleotide of the 5′ end. This routine was repeated until a full match was achieved. Finally, if primer size was less than 12 nucleotides, a negative result or no reaction was considered. Primer sequences (forward and reverse) were then excluded from resulting amplicon.
Table 1.
Primers used for the amplification of the complete 16S rRNA gene (V1–V9) and the internal fractions (V3–V4, V4–V5 and V3–V5).
| Name | Sequences | References |
|---|---|---|
| Large (V1–V9) |
Fw: AGAGTTTGATYMTGGCTCAG Rv: GTCRTAACAAGGTAACC |
(Baker et al., 2003; Edwards et al., 1989) |
| V3–V5 | Fw: CCTACGGGNGGCNGCA Rv: CCGNCNATTNNTTTNAGTTT |
(Baker et al., 2003; Perreault et al., 2007) |
| V4–V5 | Fw: GCCAGCAGCCGCGGTAA Rv: CCGNCNATTNNTTTNAGTTT |
(Liu et al., 2007; Reysenbach et al., 1992) |
| V3–V4 | Fw: CCTACGGGNGGCWGCAG Rv: GACTACHVGGGTATCTAATCC |
(Claesson et al., 2010; Muyzer et al., 1993) |
The selection of primers for the virtual amplification of internal segments (Table 1) was based on previous successful results reported; for instance, primers amplifying V3–V4 region are commonly used by NGS platforms such as MiSeq Illumina (Klindworth et al., 2013); primers for V4–V5 were reported by Soergel et al. (2012) as one of the best set of primers after the evaluation of thousands of combinations; whereas primers for V3–V5 are commonly used together with DGGE technology (Perreault et al., 2007; Rettedal et al., 2010).
2.3. Taxonomic classification
The resulting amplicons (without primer sequences) were formatted as FASTq file. The former large fragments covering the complete 16S rRNA gene were considered as Sanger-sequenced in both directions, whereas the internal fragments (V3–V4, V4–V5 and V3–V5) were considered as sequenced in a MiSeq (Illumina, USA) next generation sequencing platform (2 × 300 cycles). All sequence sets were analyzed by the Kraken classifier (Wood and Salzberg, 2014) installed at Illumina BaseSpace app (MiniKraken 20141208) (https://basespace.illumina.com/apps).
2.4. Classification performance
Classification performance using the different internal fragments (V3–V4, V4–V5 and V3–V5) was estimated considering the classification output registered by the unilarge sequences as reference. The specificity and sensitivity of the taxonomic classification obtained by using the different fragments were estimated according to the method described by Baldi et al. (2000) and adjusted by Diaz et al. (2009), Krause and Whitaker (2015). Specificity and sensitivity were calculated for the classification output at the levels of genus, species and subspecies, considering the sensitivity and specificity obtained in these categories. Herein, the sensitivity (Sni) for a taxonomic class I was defined as the percentage of fragments from class I correctly classified and it was computed as follows:
The specificity (Spi) was considered as the proportion of fragments correctly assigned to a particular lineage, other studies refer as Specificity, using the following equation:
Let the i-th taxonomic class of taxonomic rank r be denoted as class i. Further, let Zi be the total number of sequences from class i, assigned by Kraken to Large sequences. The true positives (TPi) the number of sequences correctly assigned to class i, the false positives (FPi) the number of sequences from any class j ≠ i that is wrongly assigned to i.
3. Results
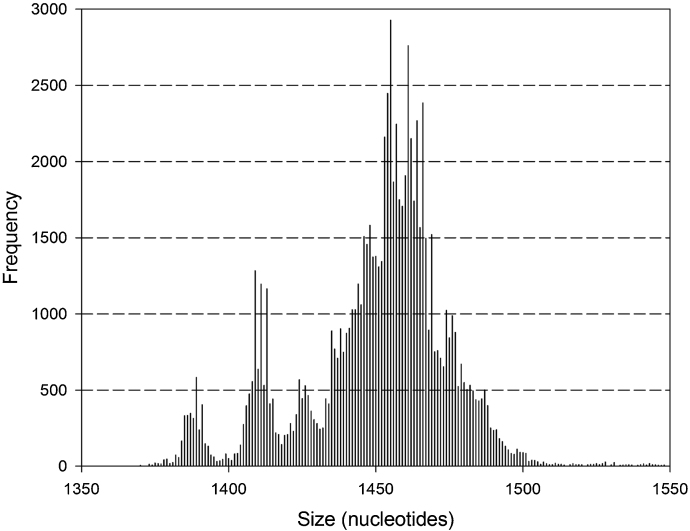
Silva data base (release 123) contains 440,738 non-redundant sequences with taxonomic description from phylum to species or strain. However, not all of them were virtually amplified using the primer set for the complete gene. A total of 102,101 amplicons (23%) were obtained; from these, 86,371 resulted to be unique large fragments with an average size of 1450.3 bp. Considering sequences within the size range of mean ±2 standard deviations (S.D.) resulted in a homogeneous group that accounted for 99.1% of the sequences ranging from 1378 to 1522 bp (1450 ± 1.9 bp; Fig. 1) and covering variable regions V1 to V9. Thereafter virtual PCR was performed on this group of sequences (85,594) by using primers for the amplification of segments containing hypervariable regions V3–V4, V4–V5 and V3–V5 (Table 1).
Fig. 1.
Distribution size of amplicons obtained after virtual PCR of complete 16S rRNA gene (V1–V9). Amplicons with extreme sizes outside the range of mean ± 2 standard deviations were excluded.
A total of 81,523 amplicons (95.2%) were obtained with primers for V3–V4 region, whereas 82,998 (96.9%) and 82,334 (96.1%) were detected when using primers for regions V3–V5 and V4–V5 respectively. Regarding amplicon sizes produced by V3–V4, V4–V5 and V3–V5 primers with a confidence range of 99%, these ranged from 393 to 440, 363 to 383 and 515 to 563 bp, respectively.
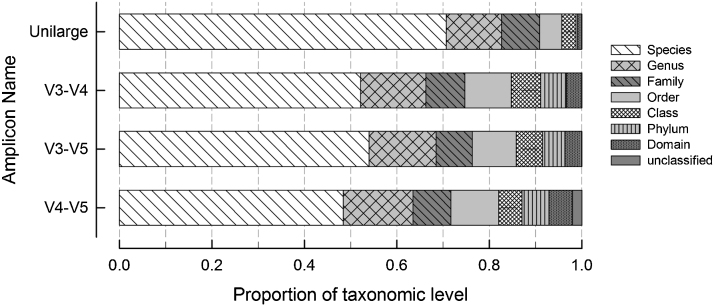
Significant differences were obtained in the depth of taxonomic classification using the different regions; for instance, ≥70% of the sequences of the unilarge amplicons (V1–V9) were assigned up to species level, whereas ≤53% of the sequences obtained after the amplification of the internal fragments (V3–V4, V4–V5 or V3–V5) were assigned to species (Fig. 2); however, some discrepancies (detailed below) were detected regarding the final classification label assigned to particular sets of internal sequences.
Fig. 2.
Cumulate proportion of taxonomic levels assigned to each amplicon type (complete gene or internal fractions) after being submitted to Kraken classifier. Unilarge is the set of sequences depurated by elimination of redundant amplicons and included in the 99% confidence range.
Differences were detected regarding the classification output when using the different internal fractions. For instance, 45.4% of the internal sequences obtained for the V3–V4 region received the same classification than their perfect matches of the unilarge V1–V9 fragments (Table 2); whereas 41.1% and 52.39% of the sequences obtained for V4–V5 and V3–V5 regions were similarly classified than their perfect matches of the unilarge fragments (Table 2). Most of these differences were associated only to a deeper classification level for the unilarge sequences; however, in few other cases, internal sequences were assigned to different taxonomic groups.
Table 2.
Proportion of sequences of the internal fragments (V3–V4, V4–V5, V3–V5) that received exactly the same classification than their unilarge (complete 16S) perfect matches. Proportion of sequences with a different classification result are also showed.
| V3–V4 | V3–V5 | V4–V5 | |
|---|---|---|---|
| Equal to large | 36,973 (45.3%) | 43,485 (52.4%) | 33,794 (41.0%) |
| Different to large | 44,170 (54.2%) | 39,243 (47.3%) | 47,542 (57.8%) |
| No reaction | 380 (0.5%) | 270 (0.3%) | 998 (1.2%) |
| Total | 81,523 | 82,998 | 82,334 |
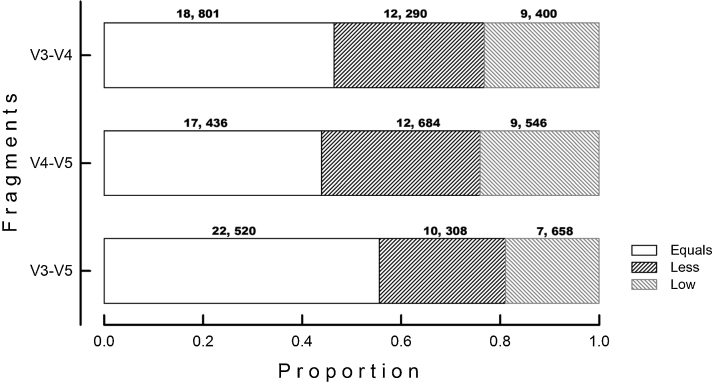
For instance, 41,685 unilarge sequences (V1–V9) resulted to be classified up to Subspecies level; however, when used as internal V3–V5 fragments, only 22,520 (55.6%) sequences received the same classification than the unilarge fragments (subspecies), whereas 10,308 (25.5%) were similarly classified but to less deep taxonomic levels (species, or genus, or family, and so on) (Fig. 3). The rest of the sequences received a different (and erroneous) classification output; for instance, 7,669 (18.9%) sequences obtained after the amplification of the V3–V5 region were labeled as a different organism (other species, or genus, or family, and so on); 11 of these sequences were inclusively classified up to Subspecies, but to a different specimen.
Fig. 3.
Classification output obtained with the internal fractions (V3–V4, V4–V5, V3–V5) respect to the complete 16S rRNA gene sequences (V1–V9). Proportion of sequences with same classification results using either the complete sequence or the internal fragment sequence are labeled as “Equal”; whereas sequences of internal fractions with similar classification but to more superficial level are represented as “Less”, and sequences with different taxonomic classification compared to the respective complete sequence (V1–V9) are named as “Low”.
Regarding V3–V4 fragments, only 18,801 (46.4%) sequences received the same classification than the unilarge fragments (subspecies), whereas 12,290 (30.3%) were similarly classified but to more superficial levels (species, or genus, or family, and so on) (Fig. 3). The rest of the sequences received a different (and erroneous) classification output; for instance, 9,400 (23.2%) sequences obtained after the amplification of the V3–V4 region were labeled as a different organism (other species, or genus, or family, and so on) of these sequences were inclusively classified up to Subspecies, but to a different specimen.
For V4–V5 fragments, 39,677 sequences resulted to be perfect matches of V1–V9 fragments. However, when used as internal V4–V5 fragments, only 17,436 (43.9%) sequences received the same classification than the unilarge fragments (subspecies), whereas 12,684 (32%) were similarly classified but to more superficial levels (species, or genus, or family, and so on) (Fig. 3). The rest of the sequences received a different (and erroneous) classification output; for instance, 9,546 (24.1%) sequences obtained after the amplification of the V4–V5 region were labeled as a different organism (other species, or genus, or family, and so on); 11 of these sequences were inclusively classified up to subspecies, but to a different specimen.The same pattern was observed when the unilarge sequences were classified to lower taxonomic levels (species, genus, or family) as maximum result; a fraction of the internal fragments received exactly the same classification than the unilarge fragments, whereas the rest were similarly classified but to more superficial levels and others received a completely different classification (different species, or genus, or family).
Regarding sensitivity and specificity analyses, both indicators exhibited higher values for the three taxonomic levels (subspecies, species and genus) when the largest internal fraction (V3–V5) was used, compared to V3–V4 and V4–V5 fragments. Results also demonstrate that internal fragments lost specificity at the deepest taxonomic level (i.e. species and subspecies). Moreover, V3–V5 fragments registered the lowest proportion of false positive results, followed by V3–V4 and V4–V5 fragments respectively (Table 3).
Table 3.
Sensitivity and specificity obtained by the different internal 16S rRNA gene fractions, considering the complete sequence of the 16S rRNA gene. Results were calculated considering the number of large sequences classified to subspecies, species or genus as maximum result.
| Sequences | Index | Subspecies | Species | Genus |
|---|---|---|---|---|
| Unilarge | Total (Z) | 41,685 | 57,616 | 68,331 |
| V3–V5 | Sensitivity | 54.02% | 56.41% | 67.57% |
| Specificity | 74.62% | 86.34% | 89.54% | |
| True Positives | 22,520 | 32,501 | 46,173 | |
| False Positives | 7,658 | 5,140 | 5,395 | |
| V4–V5 | Sensitivity | 41.83% | 44.00% | 57.27% |
| Specificity | 64.62% | 78.86% | 83.94% | |
| True Positives | 17,436 | 25,351 | 39,135 | |
| False Positives | 9,546 | 6,795 | 7,485 | |
| V3–V4 | Sensitivity | 45.10% | 48.32% | 60.38% |
| Specificity | 66.67% | 80.13% | 85.11% | |
| True Positives | 18,801 | 27,842 | 41,255 | |
| False Positives | 9,400 | 6,902 | 7,216 |
Note: Let Z be the total number of sequences assigned by Kraken to the different taxonomic levels.
4. Discussion
The overcome of NGS technologies to study microbial communities has contributed to the study of microbial diversity form a deeper perspective (Shokralla et al., 2012; van Dijk et al., 2014); these technologies are able to consider those poorly represented bacteria or differentiating bacteria with very similar sequences but varying on at least a single nucleotide. However, results demonstrated that this deeper insight is not necessarily accompanied with greater sensitivity and/or specificity. In contrast, both features have to be sacrificed when using current NGS technologies based on short sequences.
The better classification specificity achieved when using the unilarge fragments (V1–V9) compared to any of the internal fragments (V3–V4, V4–V5 or V3–V5) was an expected result, considering that several of the shorter sequences provided information corresponding to the most superficial taxonomic levels, but lacking some elements to be classified to more specific levels (species or subspecies). However, the loss of specificity can be considered as incomplete information, but that can be analyzed to some extent. For instance, only 54.5% of the largest internal fragments (V3–V5) received the same classification label than their respective unilarge matches, suggesting a loss of information or elements that provide identity to these sequences.
Shorter NGS-type sequences (V3–V4, V4–V5) similar to those used in the current NGS platforms exhibited poorer results. To this respect, other reports have documented that most of the information is usually contained in the V3–V6 region for the phylogenetic analysis of most bacterial phyla, while V2 and V8 are the least reliable regions (Yang et al., 2016a; Yang et al., 2016b). However, sequence divergence is not distributed evenly along the 16S rRNA gene, leading to interpretation problems such as over-representation and limited coverage; this discord may affect analyses of diversity, relative abundance and species richness. Herein, Kim et al. (2011) evaluated different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes and concluded that no partial sequence region could estimate OTU richness or define OTUs as reliably as nearly full-length genes. Moreover, Guo et al. (2013) asserted that despite the advantages of NGS technologies to achieve a deeper understanding on bacterial diversity for complex environmental samples, these also introduce blurring due to the relatively low taxonomic capability of short reads. Therefore, despite these short sequences may cover an information-rich fraction of the 16S rRNA gene, the classification outputs have to be interpreted as partial results, probably leading to partial conclusions.
On the other hand the lack of sensitivity when using short sequences is a worrying outcome, as this means that a considerable proportion of the results could be erroneous, and these kinds of errors can affect the overcome conclusions of any research process. The decrease in specificity could be considered as loss of information, but the results could at least provide any idea of the bacterial populations thriving in any environment; however, the loss of sensitivity implies erroneous results with the consequent misinterpretations. Furthermore, the considerable proportion of sequences reported as erroneously classified (false positives) could have a significant impact on the correct analysis of any dataset. This could be also a greater problem when using other hypervariable regions with less information richness.
Higher specificity and sensitivity values have been reported in other studies analyzing short sequences (∼200 bp); however, they used very limited sample sizes (Ounit et al., 2015; Wood and Salzberg, 2014), while in this study the sample size was closest to the universe of sequences recorded in databases. In spite of the low sensitivity and sensitivity achieved with short sequences, these results cannot be associated to a poor performance of the taxonomic classifier used in this study. For instance, the Ribosomal Database Project-Classifier (RDP), has been compared to other classification methods and has been reported (Lan et al., 2012) as an adequate program for the analysis of short 16S rRNA gene sequences; however, Valenzuela-González et al. (2016) demonstrated that deeper classification can be achieved using Kraken compared to RDP, for either short or long sequences.
The recent comparative study performed by Lindgreen et al. (2016) concluded that none of the most advanced classifiers can be considered as the best for a complete sequence analysis. In terms of sensitivity and specificity at genus level all methods (with their respective algorithms) showed adequate performances (CLARK, Genometra, GOTTCHA, Kraken, LMAT, MG-RAST, OneCodex); however CLARK and Kraken resulted to be the best tools in terms of prediction of relative abundances of bacterial phyla (Lindgreen et al., 2016).
Finally, the larger V3–V5 internal fragment (amplicon size range: 515–563 bp) which has been commonly used to study bacterial diversity through DGGE, could be considered as candidate for massive throughput sequencing.
Declarations
Author contribution statement
Marcel Martínez-Porchas: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Enrique Villalpando-Canchola: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Francisco Vargas-Albores: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Competing interest statement
The authors declare no conflict of interest.
Funding statement
This work was supported by the National Council for Science and Technology (CONACyT), Mexico, grant 84398 (to FVA).
Additional information
No additional information is available for this paper.
References
- Baker G.C., Smith J.J., Cowan D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Baldi P., Brunak S., Chauvin Y., Andersen C.A.F., Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- Claesson M.J., Wang Q., O'Sullivan O., Greene-Diniz R., Cole J.R., Ross R.P., O'Toole P.W. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz N., Krause L., Goesmann A., Niehaus K., Nattkemper T. TACOA—taxonomic classification of environmental genomic fragments using a kernelized nearest neighbor approach. BMC Bioinformatics. 2009;10:56. doi: 10.1186/1471-2105-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards U., Rogall T., Blöcker H., Emde M., Böttger E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flygare S., Simmon K., Miller C., Qiao Y., Kennedy B., Di Sera T., Graf E.H., Tardif K.D., Kapusta A., Rynearson S., Stockmann C., Queen K., Tong S., Voelkerding K.V., Blaschke A., Byington C.L., Jain S., Pavia A., Ampofo K., Eilbeck K., Marth G., Yandell M., Schlaberg R. Taxonomer: an interactive metagenomics analysis portal for universal pathogen detection and host mRNA expression profiling. Genome Biol. 2016;17:111. doi: 10.1186/s13059-016-0969-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Ju F., Cai L., Zhang T. Taxonomic precision of different hypervariable regions of 16s rRNA gene and annotation methods for functional bacterial groups in biological wastewater treatment. PLoS One. 2013;8:e76185. doi: 10.1371/journal.pone.0076185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Morrison M., Yu Z. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J. Microbiol. Methods. 2011;84:81–87. doi: 10.1016/j.mimet.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glöckner F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause D.J., Whitaker R.J. Inferring speciation processes from patterns of natural variation in microbial genomes. Syst. Biol. 2015;64:926–935. doi: 10.1093/sysbio/syv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y., Wang Q., Cole J., Rosen G. Using the RDP classifier to predict taxonomic novelty and reduce the search space for finding novel organisms. PLoS One. 2012;7:e32491. doi: 10.1371/journal.pone.0032491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgreen S., Adair K.L., Gardner P. An evaluation of the accuracy and speed of metagenome analysis tools. Sci. Rep. 2016;6:19233. doi: 10.1038/srep19233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Lozupone C., Hamady M., Bushman F.D., Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Breitwieser F.P., Thielen P., Salzberg S.L. Bracken: estimating species abundance in metagenomics data. Biorxiv. 2016:051813. [Google Scholar]
- Martínez-Porchas M., Vargas-Albores F. Microbial metagenomics in aquaculture: a potential tool for a deeper insight into the activity. Rev. Aquacult. 2015:1–15. [Google Scholar]
- Muyzer G., de Waal E.C., Uitterlinden A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ounit R., Wanamaker S., Close T.J., Lonardi S. CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics. 2015;16:236. doi: 10.1186/s12864-015-1419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault N.N., Andersen D.T., Pollard W.H., Greer C.W., Whyte L.G. Characterization of the Prokaryotic diversity in cold saline perennial springs of the canadian high arctic. Appl. Environ. Microbiol. 2007;73:1532–1543. doi: 10.1128/AEM.01729-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettedal E.A., Clay S., Brozel V.S. GC-clamp primer batches yield 16S rRNA gene amplicon pools with variable GC clamps, affecting denaturing gradient gel electrophoresis profiles. FEMS Microbiol. Lett. 2010;312:55–62. doi: 10.1111/j.1574-6968.2010.02097.x. [DOI] [PubMed] [Google Scholar]
- Reysenbach A.L., Giver L.J., Wickham G.S., Pace N.R. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokralla S., Spall J.L., Gibson J.F., Hajibabaei M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 2012;21:1794–1805. doi: 10.1111/j.1365-294X.2012.05538.x. [DOI] [PubMed] [Google Scholar]
- Soergel D.A.W., Dey N., Knight R., Brenner S.E. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J. 2012;6:1440–1444. doi: 10.1038/ismej.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbock L.J., Radenovic A. The emergence of nanopores in next-generation sequencing. Nanotechnology. 2015;26:074003. doi: 10.1088/0957-4484/26/7/074003. [DOI] [PubMed] [Google Scholar]
- Susilawati T.N., Jex A.R., Cantacessi C., Pearson M., Navarro S., Susianto A., Loukas A.C., McBride W.J.H. Deep sequencing approach for investigating infectious agents causing fever. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:1137–1149. doi: 10.1007/s10096-016-2644-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-González F., Martínez-Porchas M., Villalpando-Canchola E., Vargas-Albores F. Studying long 16S rDNA sequences with ultrafast-metagenomic sequence classification using exact alignments (Kraken) J. Microbiol. Methods. 2016;122:38–42. doi: 10.1016/j.mimet.2016.01.011. [DOI] [PubMed] [Google Scholar]
- van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Wood D., Salzberg S. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Wang Y., Qian P.Y. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinf. 2016;17:135. doi: 10.1186/s12859-016-0992-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Noyes N.R., Doster E., Martin J.N., Linke L.M., Magnuson R.J., Yang H., Geornaras I., Woerner D.R., Jones K.L., Ruiz J., Boucher C., Morley P.S., Belk K.E. Use of metagenomic shotgun sequencing technology to detect foodborne pathogens within the microbiome of the beef production chain. Appl. Environ. Microbiol. 2016;82:2433–2443. doi: 10.1128/AEM.00078-16. [DOI] [PMC free article] [PubMed] [Google Scholar]