Abstract
Current evidence favors the theory that, when the globular motor domain of myosin attaches to actin, the light chain binding domain or “lever arm” rotates, and thereby generates movement of actin filaments. Myosin is uniquely designed for such a role in that a long α-helix (≈9 nm) extending from the C terminus of the catalytic core is stabilized by two calmodulin-like molecules, the regulatory light chain (RLC) and the essential light chain (ELC). Here, we introduce a single-point mutation into the skeletal myosin RLC, which results in a large (≈50%) reduction in actin filament velocity (Vactin) without any loss in actin-activated MgATPase activity. Single-molecule analysis of myosin by optical trapping showed a comparable 2-fold reduction in unitary displacement or step size (d), without a significant change in the duration of the strongly attached state (τon) after the power stroke. Assuming that Vactin ≈ d/τon, we can account for the change in velocity primarily by a change in the step size of the lever arm without incurring any change in the kinetic properties of the mutant myosin. These results suggest that a principal role for the many light chain isoforms in the myosin II class may be to modulate the flexural rigidity of the light chain binding domain to maximize tension development and movement during muscle contraction.
Based on the atomic structure of the myosin head region [subfragment-1 (S1)], the “swinging lever-arm” model was proposed to explain motion generation by the myosin motor (1). The model assumed that small conformational changes originating in myosin's active site during ATP hydrolysis were amplified by a tilting motion of an ≈9-nm long α-helical neck or “lever,” the rigidity of which was maintained through its interactions with the regulatory (RLC) and essential (ELC) light chains. Subsequent structural and mechanical studies provided support for this model: specifically, atomic resolution structures of S1 (2, 3) and electron cryomicroscopy of S1-decorated actin filaments (4, 5) under varying nucleotide conditions were consistent with the neck swinging through as much as 70° to generate 5–10 nm of displacement at the end of the lever. The magnitude of this movement was in agreement with single myosin molecule displacement (i.e., step size) measurements made in the laser trap (reviewed in refs. 6 and 7). More recent tests of the lever model used genetically engineered myosins with necks of varying length. These mutants produced displacements in the laser trap that were linearly correlated with neck length, i.e., the most simple prediction of the lever-arm model (8–10). Nevertheless, challenges to this now widely accepted model still exist (7).
With the myosin neck serving such a key role in the mechanical properties of the motor, understanding the impact of the interactions between light chains and the underlying α-helical heavy chain on motor performance is clearly important. However, the function of the light chains and, in particular, their many isoforms, has always been poorly understood in striated muscles. Unlike smooth muscle myosin, and certain invertebrate muscle myosins, where light chain phosphorylation and calcium binding regulate myosin's activity, removal of the light chains from skeletal muscle myosin seems to have relatively little effect on enzymatic activity (11). Based on skinned fiber studies (12) and more recent in vitro motility assays (11, 13), it became apparent that light chain removal can have substantial effects on myosin force generation and velocity of movement. In the absence of light chains, the stability of the α-helical region is severely reduced, resulting in a shorter neck (14) and/or a less rigid lever arm, both of which could contribute to reduced force and motion.
To probe the functional significance of light chain–heavy chain interactions without incurring any significant changes in the global structure of the myosin molecule, we have introduced a single-point mutation (F102L) into the chicken skeletal myosin RLC and assessed the effect of this mutation on myosin's enzymatic activity and actin filament velocity (Vactin) in the motility assay. Unexpectedly, myosin with mutant RLC exhibited a large reduction in Vactin, independent of any change in steady-state ATPase activity. At the molecular level, Vactin ≈ d/τon, and thus the slower Vactin could result from the mutant RLC affecting the step size (d) and/or the kinetics of the actomyosin interaction by prolonging the period that myosin remains attached to actin after the powerstroke (τon). Based on our laser-trap studies, the presence of the mutant RLC does in fact decrease the step size by >50% without any apparent effect on myosin's kinetics. These data have been interpreted as evidence that the mutation to the RLC has altered its interaction with the heavy chain so that a significant compliance has been introduced into the lever. The profound mechanical effect that arises from this point mutation suggests that perturbations to the light chain-heavy chain interaction may also be the determining factor for human familial hypertrophic cardiomyopathy where point mutations have been identified in the ELC, RLC, and the underlying heavy chain region that binds the light chains (15).
Methods
Protein Preparation. Chicken pectoralis myosin was purified as described in Margossian and Lowey (16) and stored at –20°C in 50% glycerol, 0.6 M KCl, 25 mM sodium phosphate (pH 7.0), 1 mM DTT, 1 mM EDTA, and 1 mM NaN3. Native light chains were prepared by denaturing myosin, followed by fractionation using ion-exchange chromatography (11). Actin was prepared from chicken pectoralis powder (17) and stored as F-actin in 5 mM KCl, 5 mM imadazole (pH 7.5), 2 mM MgCl2, 3 mM NaN3, 0.5 mM DTT, and 1 μg/ml leupeptin at 4°C.
The expression vector pT7-7 containing WT (F102) or mutant (F102L) chicken skeletal regulatory light chain (RLC) cDNA has been described (18).
Single-headed myosin was prepared by brief papain digestion of myosin and isolated by hydrophobic interaction chromatography as described in Tyska et al. (19).
Purification of Expressed Protein. One-liter cultures of BL21(DE3) cells containing the pT7-7 (RLC) construct were grown 16–20 h at 37°C in enriched buffered media (2% Bacto-tryptone/1% yeast extract/0.5% NaCl/0.2% glycerol/50 mM potassium phosphate, pH 7.2/50 μg/ml carbenicillin). Cells were pelleted at 8,000 × g and washed once with 25 mM Tris (pH 8.0) and 1 mM EDTA. After resuspension in 200 ml of sonication buffer (25 mM Tris, pH 8.0/50 mM glucose/10 mM EDTA/1 μg/ml leupeptin), the cells were sonicated briefly on ice to break the cell membranes. Lysed cells were centrifuged to pellet the inclusion bodies, which were washed twice with 0.3% Triton X-100 in sonication buffer and once in buffer without Triton X-100. The pellet was solubilized in 6 M guanidine-HCl buffered with PBS (150 mM NaCl/10 mM sodium phosphate, pH 7.2/10 mM DTT/3mMNaN3) for 1 hr with stirring at 22°C. After clarification, the supernatant was dialyzed overnight at 4°C against PBS containing 1 μg/ml leupeptin. After centrifugation to remove denatured proteins, the supernatant was dialyzed for 3–4 h against buffer A (10 mM NaCl/20 mM imadazole, pH 7.5/5 mM DTT/1 mM EDTA/1 mM NaN3) and further purified on a 1.5 × 20-cm DEAE-Sephacel column equilibrated with buffer Aat4°C. The light chain was eluted with a 300 ml of gradient from 10–400 mM NaCl. Fractions containing pure RLC (determined by SDS/PAGE) were pooled, dialyzed overnight at 4°C against 5 mM sodium phosphate (pH 7.0), 0.2 mM EDTA, 0.5 mM DTT, and 3 mM NaN3, and freeze-dried in the presence of an equal weight of sucrose. Lyophilized RLC was stored at –20°C with desiccant.
Light Chain Exchange. Lyophilized LCs at 4–5mg/ml were dissolved in 6 M guanidine-HCl and 10 mM DTT as described in ref. 20. After dialysis overnight at 4°C against 0.6 M NaCl, 20 mM sodium phosphate (pH 7.5), 3 mM NaN3, and 1 mM DTT, the clarified light chains (0.2–2 mg/ml) were incubated with myosin (0.5 mg/ml) in 0.6 M NaCl, 20 mM sodium phosphate (pH 7.5), 3 mM NaN3, and 10 mM DTT, 5 mM EDTA, 1 mM EGTA, and 5 mM NaATP for 30 min at 40°C. After addition of magnesium chloride (20 mM), myosin was precipitated by an overnight dialysis against 40 mM NaCl, 5 mM sodium phosphate (pH 6.5), 5 mM MgCl2, 3 mM NaN3, and 1 mM DTT. The myosin pellet was washed once with dialysis buffer and resuspended in a minimal volume of 0.6 M NaCl, 25 mM sodium phosphate (pH 7.0), 1 mM MgCl2, 3 mM NaN3, and 1 mM DTT. When required, residual traces of LCs were removed from the myosin by pelleting with an equimolar concentration of actin or by gel filtration chromatography.
Light Chain Stripping and Reconstitution. Following the procedure in ref. 21, clarified myosin (6 mg/ml) in 4.7 M NH4Cl, 50 mM sodium phosphate (pH 7.5), 0.1 mM EGTA, 1 mM MgATP, and 8 mM DTT was loaded onto a 1.6 × 30-cm Superose 6 gel filtration column (Amersham Pharmacia Biosciences) equilibrated in 4.5 M NH4Cl, 50 mM sodium phosphate (pH 7.5), 2 mM EDTA, 3 mM NaN3, and 0.5 mM NaATP at 22°C. The protein was eluted directly into tubes containing 20 mM DTT, 2 mM ATP, 2- to 5-fold molar excess total light chains (prepared as described above) and incubated on ice. Fractions were pooled, and the ammonium chloride was removed by dialysis for 3–4 h against of 0.4 M NaCl, 5 mM MgCl2, 25 mM sodium phosphate (pH 7.5), 3 mM NaN3, and 1 mM DTT. Myosin was dialyzed overnight against 5 mM MgCl2, 5 mM sodium phosphate (pH 6.2), 3 mM NaN3, 0.5 mM MgATP, and 1 mM DTT, and pelleted to remove residual traces of added LCs. Pellets were superficially washed once with dialysis buffer and then resuspended in a minimal volume of 0.6 M NaCl, 25 mM sodium phosphate (pH 7.0), 3 mM NaN3, and 1 mM DTT.
Actin-Activated ATPase. The actin-activated ATPase activities of all myosin species were determined at 25°C in 50 mM KCl, 1 mM MgCl2, 2 mM MgATP, 10 mM imidazole (pH 7.5), 1 mM DTT, and 1 mM NaN3 (details in ref. 22).
In Vitro Motility Assay. To remove myosin that bound to actin in an ATP independent manner, samples were centrifuged with a 2-fold molar excess of actin in the presence of MgATP. Actin filament velocities were determined for each myosin species in an in vitro motility assay at 30°C in 25 mM KCl, 25 mM imidazole (pH 7.5), 4 mM MgCl2, 1 mM EGTA, 10 mM DTT, and 20 μg/ml (≈2-fold molar excess) light chains (ELC and the appropriate RLC) (details in ref. 22). Mean filament velocity was calculated from 7–15 consecutive video snapshots (0.1–0.3 s) of the digitized filament as described in ref. 23.
Optical Trap Experiments. Details of the optical-trap instrumentation and experimental procedures have been described (24). In brief, flow-cells were perfused with 1 μg/ml monomeric myosin in 0.3 M KCl, 0.5 mg/ml BSA, assay buffer (25 mM KCl/25 mM imidazole, pH 7.5/4 mM MgCl2/1 mM EGTA/10 mM DTT), 0.2 μg/ml (≈2-fold molar excess) lights chains (ELC and the appropriate RLC), and finally with phalloidin-rhodamine-labeled F-actin in assay buffer with oxygen scavengers, 1-μm polystyrene beads coated with N-ethylmaleimide-treated myosin, and 10 μMMgATP at pH 7.4. Experiments were initiated by capturing a myosin-coated polystyrene bead in each optical trap and securing an actin filament between the beads. The filament was pretensioned to 4 pN and brought into proximity of the myosin-coated surface to record the unitary events.
Mean-Variance Analysis. Estimates of displacement (d) and event durations (τon) were obtained from single-molecule measurements by mean-variance (MV) analysis (24). MV analysis involves a transform of the raw-time-series data into a 3D histogram, which is then fit to obtain estimates of unitary parameters. A MV histogram is created by passing a time window of variable width (20 ms for most of the data here) over the raw displacement data and plotting the mean and variance of each point. The histogram's third dimension is the number of points with a given mean and variance. Baseline is recognized as the population of points that have a high variance, and events appear as a population with lower variance. This variance reduction is caused by an increase in the effective stiffness as myosin attaches to actin and allows the event and baseline populations to be separated in two dimensions with higher resolution.
To estimate τon, the total counts of the event population are recorded at different window widths. The volume (count) at a given window width is representative of the number of events with a duration equal to or greater than that width. Accordingly, it is possible to estimate the average event duration by fitting volume vs. window width with the expression V = n × τon × e–W/τon. Here, V is the volume of the population, n is the number of events, τon is the average event duration, and W is the window width.
Results
Choice of RLC Mutation. The first clone for the RLC was isolated from a cDNA library prepared from chicken pectoralis muscle (25). The nucleotide sequence agreed with the protein sequence at all positions except for residue 102 (numbering starts with alanine), which coded for leucine (L) as opposed to phenylalanine (F) in the protein sequence (26). At the time, this difference was of little concern because a different chicken strain was used for the protein study. When the recombinant RLC was exchanged into chicken skeletal myosin, there was no detectable change in actin-activated ATPase activity; thus, this clone became the source for many mutagenesis studies in which cysteine residues were introduced at various locations for chemical modification by spectroscopic probes (18, 20). However, with the introduction of the in vitro motility assay, it was observed that myosin containing the recombinant light chain had a noticeably slower rate of actin filament movement (Vactin) than native myosin. Because the only known difference between the recombinant and the tissue RLC was the leucine at position 102, by mutating L102 to F we were able to restore Vactin to its original native value.
The F102L Mutation Slows in Vitro Motility. Examination of the RLC sequence for cardiac, skeletal, smooth muscle, and nonmuscle myosins from a number of species shows that the phenylalanine residue lies in a highly conserved region of the sequence (Fig. 1a). In the 3D structure, this residue is in the C-terminal lobe of RLC (1, 27), which serves to stabilize the long α-helix of the heavy chain (Fig. 1b). When we exchanged the expressed regulatory light chain containing the leucine mutation (hence referred to as F102L) into myosin, we found that it was difficult to replace >50% of the endogenous RLC, even with a 50-fold molar excess of RLC (Fig. 2a, lane 2). In contrast, exchange of F102 (WT) into myosin at 10-fold molar excess produced the 90% exchange expected for a light chain with the same binding affinity as the endogenous RLC (Fig. 2a, lane 3). When we compared the in vitro motility of myosin containing F102L with myosin containing F102, or to native myosin, the actin filament velocity was reduced by ≈30% in the mutant (Fig. 2b and Table 1).
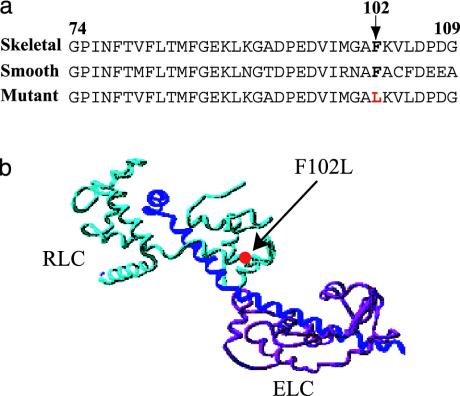
Fig. 1.
Location of the chicken skeletal RLC mutation. (a) Amino acid sequence alignment of chicken skeletal, smooth muscle, and mutant skeletal RLCs. A conserved phenylalanine (F102) and F102L mutation (red) are shown in bold. (b) The F102L RLC mutation (red circle) mapped to the E-helix in the ribbon structure of the scallop myosin regulatory domain (27). ELC, magenta; RLC, light blue; heavy chain, dark blue.
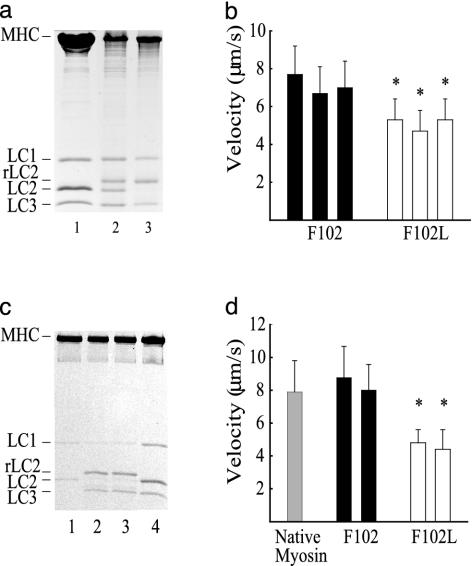
Fig. 2.
Characterization of chicken pectoralis muscle myosin containing WT (F102) and mutant (F102L) RLCs. (a and b) Exchange of recombinant RLC (rLC2) into myosin. (a) Twelve percent SDS/PAGE of control myosin (lane 1), F102L (lane 2), and F102 (lane 3). (b) Corresponding actin filament velocities of myosin with F102 (filled bars) and F102L (open bars). Average velocity for myosin with WT (F102) (7.1 ± 1.3 μm/s, mean ± SD) and F102L RLCs (5.1 ± 1.1 μm/s). Values represent three independent preparations. (c and d) Myosin heavy chain reconstituted with light chains. (c) Light chain-deficient myosin (lane 1), myosin reconstituted with F102L and LC3 (lane 2) and F102 and LC3 (lane 3), and control chicken skeletal myosin (lane 4). (d) Velocity of native chicken skeletal myosin (gray bar) and myosin reconstituted with F102 (filled bar) or F102L (open bar). Average velocity for myosin with F102L (4.6 ± 0.8 μm/s, mean ± SD) and F102 (8.4 ± 1.9 μm/s). Values represent two independent preparations. *, Statistically significant from myosin with F102 RLC at the P < 0.001 level by Student's t test.
Table 1. Summary of enzymatic and mechanical properties of myosin.
| Myosin | Vmax,* s-1 | Km,* μM | Vactin,† μm/s | d, nm | τon, ms |
|---|---|---|---|---|---|
| Native | 5.3 | 1.6 | 10.6 ± 1.7 (48) | 9.7 ± 2.4 (9) | 34.5 ± 8.1 (9) |
| Exchanged | |||||
| F102 | — | — | 7.1 ± 1.3 (82) | 8.1 ± 2.4 (12) | 30.1 ± 13.1 (15) |
| F102L | — | — | 5.1 ± 1.1** (83) | 6.1 ± 2.8** (27) | 29.5 ± 14.2 (27) |
| Reconstituted | |||||
| F102 | 5.4 | 1.4 | 8.4 ± 1.8 (77) | 9.6 ± 2.7 (19) | 25.9 ± 13.1 (11) |
| F102L | 5.6 | 1.2 | 4.6 ± 1.0** (82) | 3.8 ± 2.4** (8) | 31.6 ± 12.1 (8) |
| Single-headed | |||||
| F102 | 4.5 | 13.4 | 8.6 ± 1.5 (108) | — | — |
| F102L | 4.3 | 14.2 | 4.1 ± 0.8** (136) | — | — |
Data are presented as mean ± SD. Data in parentheses are the number of actin filaments measured for Vactin or the number of MV histograms contributing to the mean d and τon. Optical trap data were collected at 10 μM MgATP with 2- to 5-fold molar excess light chains. Except for the single-headed myosin, the in vitro motility assays were performed on the same preparations used for the unitary mechanical measurements.
Statistically significant vs. WT at the P < 0.001 level by Student's t test.
The ATPase parameters (Vmax and Km) are representative activity data for the different myosin preparations.
In addition to the actin filament velocities shown here, earlier data for exchanged and reconstituted double-headed myosin gave essentially the same results.
In an effort to increase the incorporation of mutant RLC into myosin, we first removed the endogenous RLC from myosin by gel filtration in a high concentration of ammonium chloride (4.5 M NH4Cl). We have shown (11, 21) that this procedure allows us to separate the myosin heavy chain from the dissociated light chains and isolate relatively pure heavy chain in the void volume (Fig. 2c, lane 1). Immediate addition of F102L (or F102) expressed RLC, together with essential light chain (LC3), to the heavy chain results in a myosin with a stoichiometric compliment of light chains (Fig. 2c, lanes 2 and 3). The actin-activated MgATPase activity of the reconstituted myosin was identical to that of native myosin, irrespective of the type of RLC (Fig. 3a and Table 1). The complete retention of enzymatic activity shows that exposure to concentrated salt solutions had no adverse, denaturing effect on the protein. The actin filament velocity for myosin containing mainly F102L (4.6 ± 0.8 μm/s) decreased by almost 50% compared with the velocity shown by WT (F102) myosin (8.4 ± 1.9 μm/s) (Fig. 2d). Thus, the ability of myosin to translocate actin can be correlated with the amount of mutant RLC incorporated into the myosin.
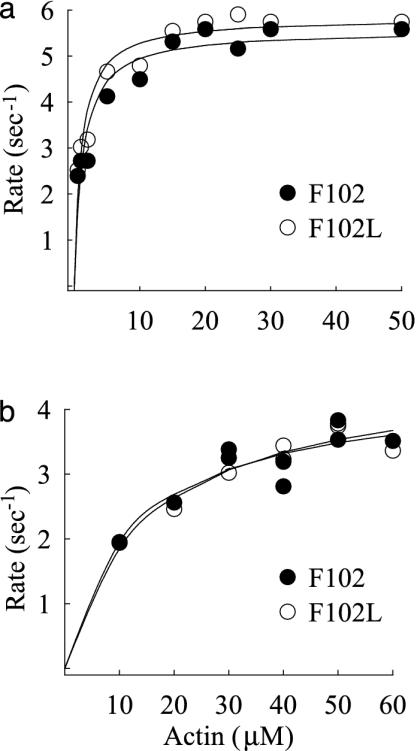
Fig. 3.
Actin-activated MgATPase activity for double-headed and single-headed myosins. (a) Myosin reconstituted with total light chains containing either F102 (filled circles) or F102L (open circles). Data were fit with a Michaelis–Menten equation to determine Vmax (≈5.6 s–1) and Km (1.2 μM). (b) Single-headed myosin with F102 or F102L, Vmax (≈ 4.4 s–1) and Km (≈13 μM). These values are all well within the range reported for filamentous native myosin at 25°C; single-headed subfragments typically have a higher Km than a double-headed species.
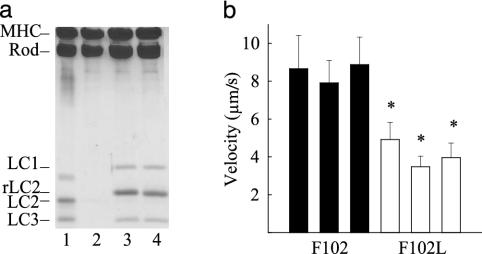
Head–Head Interactions Are Not a Factor in Slowing Motility. Whenever a change in the enzymatic and/or mechanical properties of myosin is observed, the question always arises as to whether an interaction between the heads is responsible. In the case of a thick-filament-regulated myosin such as smooth muscle myosin, the inhibition of activity in the dephosphorylated state has been attributed to one head interacting with the neighboring head in such a way as to render both heads inactive (28). Striated muscle myosins are thin-filament-regulated proteins, but, nevertheless, evidence for some degree of head–head interaction does exist (29, 19). To address the possibility that a mutant RLC might alter the functional relationship between the two heads, given its proximity to the head-rod junction, we prepared a single-headed myosin by brief proteolysis of myosin with papain. After separation of the single-headed species from rod and two-headed myosin by hydrophobic interaction chromatography (19), the light chains were removed from the heavy chain by chromatography in ammonium chloride as described above for myosin (Fig. 4a, lanes 1 and 2). Readdition of the expressed RLC (F102 or F102L) and ELC (LC1 and LC3) (Fig. 4a, lanes 3 and 4) resulted in enzymatically active preparations of mutant and WT single-headed myosins (Fig. 3b). The actin filament velocity of the single-headed species was remarkably similar to that displayed by the parent double-headed molecules, namely, the mutation in RLC reduced the velocity approximately 2-fold (8.6 μm/s to 4.1 μm/s) (Fig. 4b). These results suggest that the effect of the RLC point mutation is on the individual heads and does not involve any significant head–head interaction.
Fig. 4.
Characterization of single-headed chicken myosin reconstituted with F102 and F102L. (a) Twelve percent SDS/PAGE of single-headed (SH) myosin (lane 1), light chain-deficient SH-myosin (lane 2), and SH-myosin reconstituted with F102L (lane 3) and F102 (lane 4). Both ELC isoforms (LC1 and LC3) were included with the light chains. Papain digestion used to prepare single-headed myosin cleaves LC1 at the N terminus so that it migrates faster in the gel (lane 1). Note the two bands for MHC and rod, consistent with a single-headed species. (b) Average actin filament velocities for three different preparations of single-headed myosin with F102L (4.16 ± 0.75 μm/s, mean ± SD) compared with single-headed myosin with F102 (8.56 ± 1.5 μm/s). *, The difference between myosin with the F102 vs. F102L RLC was significant at the P < 0.001 level by Student's t test.
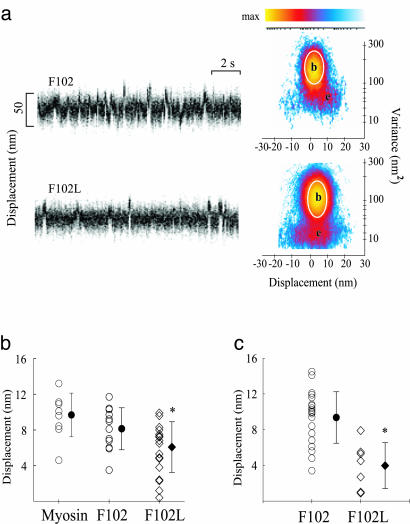
A Reduction in Step Size Accounts for Slower Movement. The slower velocity of the mutant myosin can be caused by a smaller step size or unitary displacement (d) of the actin filament as the myosin undergoes its power stroke, or by an increase in the time (τon) that myosin remains strongly bound to actin after the powerstroke, assuming Vactin ≈ d/τon. To determine the mechanism underlying the change in velocity, we measured d and τon directly in the optical trap. A segment of raw displacement data for myosin containing F102 or F102L is shown in Fig. 5a together with their respective MV histograms derived from the entire data records. In the MV histograms (Fig. 5a), the displacement event populations (e) are distinguished from the baseline population (b) when myosin is detached from actin, by a reduction in the displacement variance and a shift in the mean displacement position. The results from numerous MV histograms, based on the analysis of multiple data records from different myosin molecules, are summarized as scatter plots (Fig. 5 b and c), with each point representing the mean step size from one MV histogram. Myosin exchanged with WT F102 gives an average step size (8.1 nm) that is not significantly different from native myosin (9.7 nm) whereas myosin replaced with 50% F102L gives a significantly smaller step size (6.1 nm) (Fig. 5b and Table 1). The spread of data points is also appreciably larger for the mutant myosin, with an increased population of smaller displacements. When the myosin was reconstituted with 100% F102L regulatory light chain, the majority of displacements were small, with a mean value of 3.4 nm (Fig. 5c).
Fig. 5.
Single-molecule laser trap data from chicken skeletal myosin with F102 and F102L. (a) Representative data trace for F102 and F102L RLC-exchanged myosin. Myosin-binding events are characterized by a change in mean position associated with a reduction in variance. Shown are raw time traces (Left) and MV histograms (Right) representing myosin with F102 and F102L RLCs. MV histograms were calculated with a 20-ms time window by transforming the entire record (≅30–60 s) from which representative traces were taken. Note that the event population amplitudes, e, shown in the F102L histograms, are roughly half those of the F102 MV histogram. (b) Scatter plots representing the distributions of d produced by myosin, F102 RLC-exchanged myosin, and 60% F102L RLC-exchanged myosin. Mean values (filled symbols) and standard deviations are represented for each distribution. Values were obtained from three F102 (8.1 ± 2.4 nm) and F102L RLC-exchanged (6.1 ± 2.8 nm) myosin preparations. Each entry represents a fit from a single MV histogram, which may be comprised of 50–100 unitary events depending on the record duration and event density. (c) Scatter plots representing the distributions of d produced by myosin-reconstituted with total light chains. Mean values (filled symbols) ± SD are represented for each distribution. Values were obtained for two myosin preparations reconstituted with F102 (9.6 ± 2.7 nm) or F102L RLCs (3.8 ± 2.4 nm). *, The difference between myosin with the F102 vs. F102L RLC was significant at the P < 0.001 level by Student's t test.
Additional analysis of the MV histograms (see Methods) provided an estimate of the average time spent in the attached state, τon. There was no apparent effect of the mutant RLC on τon, with a value of ≈30 ms (see Table 1) being appropriate for the 10 μM ATP used in these experiments.
Discussion
Rotation of the myosin light chain binding domain or neck is generally accepted as the mechanism by which a single myosin head generates motion. In myosin II, the neck consists of two IQ-sequences, one of which binds the ELC and the other the RLC. In contrast, unconventional myosins (e.g., myosin V) can have up to six IQ-sequences that bind either tissue-specific light chains and/or calmodulin. Therefore, the light chains are essential in maintaining the structural rigidity of the neck required for its lever action. Here, we present evidence, from a single-molecule analysis of chicken skeletal myosin containing a point mutation in the RLC, that perturbation of the light chain–heavy chain interaction may increase the compliance of the neck sufficiently to prevent the efficient transmission of motion.
We have previously shown that RLC removal leads to a marked reduction in Vactin (>60%) without much change in ATPase activity or average isometric force (11, 13). A limitation of these experiments was that myosin deficient in RLC partially aggregates (30, 11), due to exposure of a strongly hydrophobic sequence (WPWM) on the C-terminal hook of the S1 heavy chain, which in the native state is shielded by interactions with the N-terminal lobe of the RLC (1, 27). The discovery that a single-point mutation in the RLC could produce almost the same reduction in sliding velocity, but without the confounding effects of aggregation, offered a unique opportunity to explore the consequences of altering the RLC-heavy chain interaction. Even though the mutant RLC (F102L) binds with lower affinity to the myosin heavy chain than native or WT (F102) RLC, as indicated by exchange experiments, once bound, there is no indication of any change in structure to the myosin neck. Electron micrographs of metal-shadowed preparations of single myosin molecules containing exclusively F102L were identical in appearance to native myosin (20), with no evidence of aggregation, or the more rounded, collapsed heads seen in light chain-deficient myosin (14). Thus, the effect of the mutant RLC on myosin's motion-generating capacity cannot be attributed to an overt structural change to the neck, but rather to a more subtle change that significantly affects myosin's functional properties.
A Mutant RLC Reveals an Inherently Compliant Lever. Under most conditions, myosin's actomyosin ATPase activity is correlated with its velocity of motion, whether in fibers (31) or in the motility assay (6). The fact that a single-point mutation in the RLC can slow the velocity to such a large extent, without any measurable effect on steady-state ATPase activity suggests that this apparent relationship has been uncoupled. An alteration to ADP release, and thus the rate of myosin detachment from actin, could explain the mutant RLC's effect on Vactin, but the step duration, τon, was unchanged at the 10-μM subsaturating ATP concentration used to enhance the detection of displacement events for a fast myosin. At this ATP concentration, τon is limited by ATP binding, and thus any potential change in the ADP release rate is difficult to determine. However, with the decrease in step size for the mutant RLC being sufficient to fully account for the reduction in actin filament velocity (i.e., Vactin ≈ d/τon), and with no effect of the mutation on ATPase activity, it seems that the reduction in step size is the principal determinant of changes to Vactin. These data are the first example of a point mutation in the lever arm of myosin II having a large effect on the power stroke, other than the more drastic structural changes brought about by the addition or deletion of IQ-motifs (8, 9).
Is it possible that the mutation to the RLC weakens its interaction with the underlying α-helix so that the inherent elastic properties of the α-helix are mechanically expressed? Howard and Spudich (32) proposed that the lever arm itself could be a major source of myosin's elasticity, the existence of which was first proposed by A. F. Huxley in 1957 (33). By modeling the neck region as a cantilevered beam that could bend under load, they estimated that flexural rigidity in this region could contribute an effective beam stiffness of ≈2 pN/nm, a value consistent with cross-bridge stiffness derived from fiber studies. If the normal interaction of the RLC with the α-helical heavy chain is altered by the mutation such that the effective stiffness of the lever is reduced, there would have to be a sizeable bending of the entire lever as the myosin generates motion against the force of the laser trap to account for a 50% reduction in step size. Given that the F102L mutation exists in the C terminus of the RLC, we propose instead that only a small defined segment of the α-helix is destabilized near the RLC–ELC junction, which then becomes a compliant element. As the lever rotates against the load of the trap, the ELC–IQ segment could undergo its full angular rotation, but the compliance associated with the articulation at the RLC-ELC junction would prevent the RLC–IQ segment from completing the swing. There is some evidence in support of such a model from the structural and biochemical studies on scallop myosin, a calcium-regulated protein (27). The crystal structure of the light chain-binding domain revealed that, although the Ca2+-binding site was located on the ELC, it required specific interactions with both the RLC and the heavy chain to chelate Ca2+ and activate enzymatic activity. The removal of Ca2+, and thereby the disruption of this ternary complex, results in a complete loss of activity. In a related fashion, one can speculate that a loss of specific interactions between the RLC, ELC, and the underlying α-helical heavy chain in chicken skeletal myosin might perturb the interface between the light chains sufficiently to introduce a compliant element into the lever arm.
Relevance of Single-Molecule Data to Muscle Fiber Mechanics. Can the molecular mechanics account for any of the mechanical properties observed in skeletal muscle fibers? Fortunately, there already exist a series of studies on rabbit psoas muscle fibers (34) in which about half of the endogenous RLC was replaced with an avian mutant RLC containing a point mutation in the N-terminal divalent cation-binding site (D47A). Interestingly, this mutant RLC (D47A) was constructed from the same clone as our recombinant RLC (25), and therefore inadvertently contained the F102L mutation in addition to the D47A mutation specifically introduced to study Ca2+ regulation (35). The mutant RLC(D47A), which was incapable of binding Ca2+ (and presumably Mg2+), reduced the maximum tension and stiffness of the skinned rabbit fibers by ≈60%, compared with control fibers (34). It was concluded that the decrease in tension could be due to a decrease in the number of cross-bridges generating force at any given time, and that the absence of a divalent-cation-binding site affected the kinetics of cross-bridge attachment and detachment. Alternatively, the D47A mutation might alter the structure of individual cross-bridges so that they could no longer develop normal force (34). On the basis of their data alone, the authors were unable to distinguish between these two mechanisms.
We suggest, on the basis of the molecular studies described here, that a less rigid myosin light chain-binding region most likely contributes to the decrease in stiffness and tension in fibers exchanged with mutant (D47A) RLC. Even though our light chain did not contain the D47A mutation, preliminary evidence showed that partial exchange of RLC (F102L) into muscle fibers caused a significant decrease in the unloaded shortening velocity as measured by the slack test (G. M. Diffee and R. L. Moss, personal communication). We predict that the additional mutation D47A would reduce the structural integrity of the lever arm even further because the absence of Mg2 is known to reduce the binding of RLC to the heavy chain. Thus, the combined effect of the two mutations in RLC may be sufficient to compromise the force-generating capacity of the individual cross-bridges, and thereby reduce the isometric tension observed in skinned fibers containing D47A exchange (34).
Alterations to light chain–heavy chain interactions are most likely a primary cause for human familial hypertrophic cardiomyopathy (FHC) in patients where point mutations to the RLC, ELC, and light chain-binding domain itself have been discovered (15). In fact, one such mutation was identified in the Ca2+/Mg2+-binding site of human cardiac RLC (36). The cardiac mutation N47K lies adjacent to the D47A mutation engineered in the chicken RLC. Both mutations abolish Ca2+ binding in RLC (37, 35). It is therefore possible that the mechanical performance of cardiac (slow skeletal) myosin will be affected in a similar manner to what has been described here for chicken skeletal myosin. This finding provides a plausible mechanism for how this and similar mutations might trigger FHC through their alteration in the myocardium's capacity to generate power. In conclusion, the results presented here suggest that light chain–heavy chain interactions are critical for optimal muscle performance, and thus specific light chain isoforms may be matched to their respective myosin II heavy chains to ensure a rigid lever for maximum motion generation.
Acknowledgments
We thank L. D. Saraswat (ArQule, Woburn, MA) for the cDNA clones of the regulatory light chain and the members of the Trybus and Warshaw laboratories for their assistance with protein preparations and the optical trap technology. This study was supported by National Institutes of Health Grants AR47906 and HL59408 (to S.L. and D.M.W.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: S1, myosin subfragment-1; RLC, regulatory light chain; ELC, essential light chain; LC1 and LC3, light chain isoforms of ELC; MV, mean-variance.
References
- 1.Rayment, I., Rypniewski, W. R., Schmidt-Base, K., Smith, R., Tomchick, D. R., Benning, M. M., Winkelmann, D. A., Wesenberg, G. & Holden, H. M. (1993) Science 261, 50–58. [DOI] [PubMed] [Google Scholar]
- 2.Fisher, A. J., Smith, C. A., Thoden, J. B. Smith, R., Sutoh, K., Holden, H. M. & Rayment, I. (1995) Biochemistry 34, 8960–8972. [DOI] [PubMed] [Google Scholar]
- 3.Dominguez, R., Freyzon, Y., Trybus, K. M. & Cohen, C. (1998) Cell 94, 559–571. [DOI] [PubMed] [Google Scholar]
- 4.Rayment, I., Holden, H. M., Whittaker, M., Yohn, C. B., Lorenz, M., Holmes, K. C. & Milligan, R. A. (1993) Science 261, 58–65. [DOI] [PubMed] [Google Scholar]
- 5.Whittaker, M., Wilson-Kubalek, E. M., Smith, J. E., Faust, L., Milligan, R. A. & Sweeney, H. L. (1995) Nature 378, 748–751. [DOI] [PubMed] [Google Scholar]
- 6.Tyska, M. J. & Warshaw, D. M. (2002) Cell Motil. Cytoskel. 51, 1–15. [DOI] [PubMed] [Google Scholar]
- 7.Yanagida, T., Kitamura, K., Tanaka, H., Iwane, A. H. & Esaki, S. (2000) Curr. Opin. Cell Biol. 12, 20–25. [DOI] [PubMed] [Google Scholar]
- 8.Warshaw, D. M., Guilford, W. H., Freyzon, Y., Krementsova, E., Palmiter, K. A., Tyska, M. J., Baker, J. E. & Trybus, K. M. (2000) J. Biol. Chem. 275, 37167–37172. [DOI] [PubMed] [Google Scholar]
- 9.Ruff, C., Furch, M., Brenner, B., Manstein, D. J. & Meyhofer, E. (2001) Nat. Struct. Biol. 8, 226–229. [DOI] [PubMed] [Google Scholar]
- 10.Moore, J. R., Kremetsova, E. B., Trybus, K. M. & Warshaw, D. M. (2004) J. Muscle Res. Cell Motil. 25, 29–35. [DOI] [PubMed] [Google Scholar]
- 11.Lowey, S., Waller, G. S. & Trybus, K. M. (1993) Nature 365, 454–456. [DOI] [PubMed] [Google Scholar]
- 12.Moss, R. L., Giulian, G. G. & Greaser, M. L. (1982) J. Biol. Chem. 257, 8588–8591. [PubMed] [Google Scholar]
- 13.VanBuren, P., Waller, G. S., Harris, D. E., Trybus, K. M., Warshaw, D. M. & Lowey, S. (1994) Proc. Natl. Acad. Sci. USA 91, 12403–12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flicker, P. F., Wallimann, T. & Vibert, P. (1983) J. Mol. Biol. 169, 723–741. [DOI] [PubMed] [Google Scholar]
- 15.Seidman, J. G. & Seidman, C. (2001) Cell 104, 557–567. [DOI] [PubMed] [Google Scholar]
- 16.Margossian, S. S. & Lowey, S. (1982) Methods Enzymol. 85, 55–71. [DOI] [PubMed] [Google Scholar]
- 17.Pardee, J. D. & Spudich, J. A. (1982) Methods Enzymol. 85, 164–181. [DOI] [PubMed] [Google Scholar]
- 18.Saraswat, L. D. & Lowey, S. (1991) J. Biol. Chem. 266, 19777–19785. [PubMed] [Google Scholar]
- 19.Tyska, M. J., Dupuis, D. E., Guilford, W. H., Patlak, J. B., Waller, G. S., Trybus, K. M., Warshaw, D. M. & Lowey, S. (1999) Proc. Natl. Acad. Sci. USA 96, 4402–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saraswat, L. D., Pastra-Landis, S. C. & Lowey, S. (1992) J. Biol. Chem. 267, 21112–21118. [PubMed] [Google Scholar]
- 21.Waller, G. S., Ouyang, G., Swafford, J., Vibert, P. & Lowey, S. (1995) J. Biol. Chem. 270, 15348–15352. [DOI] [PubMed] [Google Scholar]
- 22.Trybus, K. M. (2000) Methods 22, 327–335. [DOI] [PubMed] [Google Scholar]
- 23.Work, S. S. & Warshaw, D. M. (1992) Anal. Biochem. 202, 275–285. [DOI] [PubMed] [Google Scholar]
- 24.Guilford, W. H., Dupuis, D. E., Kennedy, G., Wu, J., Patlak, J. B. & Warshaw, D. M. (1997) Biophys. J. 72, 1006–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinach, F. C. & Fischman, D. A. (1985) J. Mol. Biol. 181, 411–422. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda, G., Suzuyama, Y., Maita, T. & Umegane, T. (1977) FEBS Lett. 84, 53–56. [DOI] [PubMed] [Google Scholar]
- 27.Xie, X., Harrison, D. H., Schlichting, I., Sweet, R. M., Kalabokis, V. N., Szent-Gyorgyi, A. G. & Cohen, C. (1994) Nature 68, 306–312. [DOI] [PubMed] [Google Scholar]
- 28.Wendt, T., Taylor, D., Trybus, K. M. & Taylor, K. (2001) Proc. Natl. Acad. Sci. USA 98, 4361–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conibear, P. B. & Geeves, M. A. (1998) Biophys. J. 75, 926–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pastra-Landis, S. C. & Lowey, S. (1986) J. Biol. Chem. 261, 14811–14816. [PubMed] [Google Scholar]
- 31.Barany, M. (1967) J. Gen. Physiol. 50, 197–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard, J. & Spudich, J. A. (1996) Proc. Natl. Acad. Sci. USA 93, 4462–4464. [PubMed] [Google Scholar]
- 33.Huxley, A. F. (1957) Prog. Biophys. 7, 255–318. [PubMed] [Google Scholar]
- 34.Diffee, G. M., Patel, J. R., Reinach, F. C., Greaser, M. L. & Moss R. L. (1996) Biophys. J. 71, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinach, F. C., Nagai, K. & Kendrick-Jones, J. (1986) Nature 322, 80–83. [DOI] [PubMed] [Google Scholar]
- 36.Andersen, P. S., Havndrup, O., Bundgaard, H., Moolman-Smook, J. C., Larsen, L. A., Mogensen, J., Brink, P. A., Borglum, A. D., Corfield, V. A., Kjeldsen, K., et al. (2001) J. Med. Genet. 38, E43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szczesna-Cordary, D., Guzman, G., Ng, S.-S. & Zhao, J. (2004) J. Biol. Chem. 279, 3535–3542. [DOI] [PubMed] [Google Scholar]