Abstract
Outbreaks of pseudorabies (PR) have occurred in southern China since late 2011, resulting in significant economic impacts on the swine industry. To identify the cause of PR outbreaks, especially among vaccinated pigs, 11 pseudorabies virus (PRV) field strains were isolated from Guangdong province during 2013–2014. Their major viral genes (gE, TK, gI, PK, gD, 11K, and 28K) were analyzed in this study. Insertions or deletions were observed in gD, gE, gI and PK genes compared with other PRV isolates from all over the world. Furthermore, sequence alignment showed that insertions in gD and gE were unique molecular characteristics of the new prevalent PRV strains in China. Phylogenetic analysis showed that our isolates were clustered in an independent branch together with other strains isolated from China in recent years, and that they showed a closer genetic relationship with earlier isolates from Asia. Our results suggest that these isolates are novel PRV variants with unique molecular signatures.
Keywords: molecular characterization, phylogenetic analysis, pseudorabies virus
Introduction
Pseudorabies (PR), which is also known as Aujeszky's disease (AD), has resulted in significant impacts on the swine industry since it was first identified by Aujeszky in 1902. This disease is caused by the pseudorabies virus (PRV), a double-stranded DNA virus that belongs to the Herpesviridae family and the Alphaherpesvirinae subfamily. PRV infection is characterized by respiratory, reproductive, and neurological symptoms, varying according to the age of the pigs and the virulence of the strain. While efforts to eradicate PRV in the United States and Europe have shown great progress, pseudorabies remains an endemic problem in many countries [6,15].
Since late 2011, a few outbreaks of disease in pigs have caused large economic losses to the swine industry in China. It was been shown that the pathogenic PRV was one of the etiologic agents of the epidemic, and PRV variants were isolated from different region of China. New PRV isolates were also reported to have significantly increased virulence and significant differences in antigenicity compared to previous isolates [1,12,17,18,19].
Therefore, to elucidate the cause of PR outbreaks among Bartha-K61-vaccinated pigs, clinical samples from pigs with suspected PRV infections were collected in Guangdong province from 2013 to 2014 and 11 PRV strains were isolated in our laboratory. A number of major viral genes were selected for molecular characterization of the new prevalent PRV strains. Conventional live attenuated vaccine strains, such as Bartha and Norden, have a natural deletion of a large fragment in the unique short region of the genome, including the gE, gI, 11K, and 28K genes [7,13,14]. Moreover, deletion of a large fragment in the 28K gene was also found in vaccine strain BUK-TK900 [3]. Based on these findings, these deletions are the likely cause of attenuated virulence since gE, TK, gI and PK are known to contribute greatly to the virulence of PRV in pigs [4,5,8,17]. Furthermore, as a constituent of the viral envelope, glycoprotein gD is essential for PRV replication and induces neutralizing antibodies [16]. Thus, the gE, TK, gI, gD, PK, 11K and 28K genes of the isolated strains were selected for analysis in this study.
The goal of this study was to establish phylogenetic relationships between PRVs circulating in southern China from 2013 to 2014 with other strains based on molecular characterization, elucidate the prevalence of the virus, and generate useful guidelines for the epidemiological control of pseudorabies in China.
Materials and Methods
Virus strains
To investigate the cause of recent PR outbreaks, clinical samples were collected from different pig farms in Guangdong province from 2013 to 2014, where PRV Bartha-K61 vaccines were used to protect their pigs against pseudorabies. Overall, 11 PRV strains were isolated in our laboratory and designated GD-1-2013, GD-2-2013, GD-3-2013, GD-4-2013, GD-5-2014, GD-6-2014, GD-7-2014, GD-8-2014, GD-9-2014, GD-10-2014, and GD-11-2014 (Supplementary Table 1).
Viral genome extraction and PCR amplification
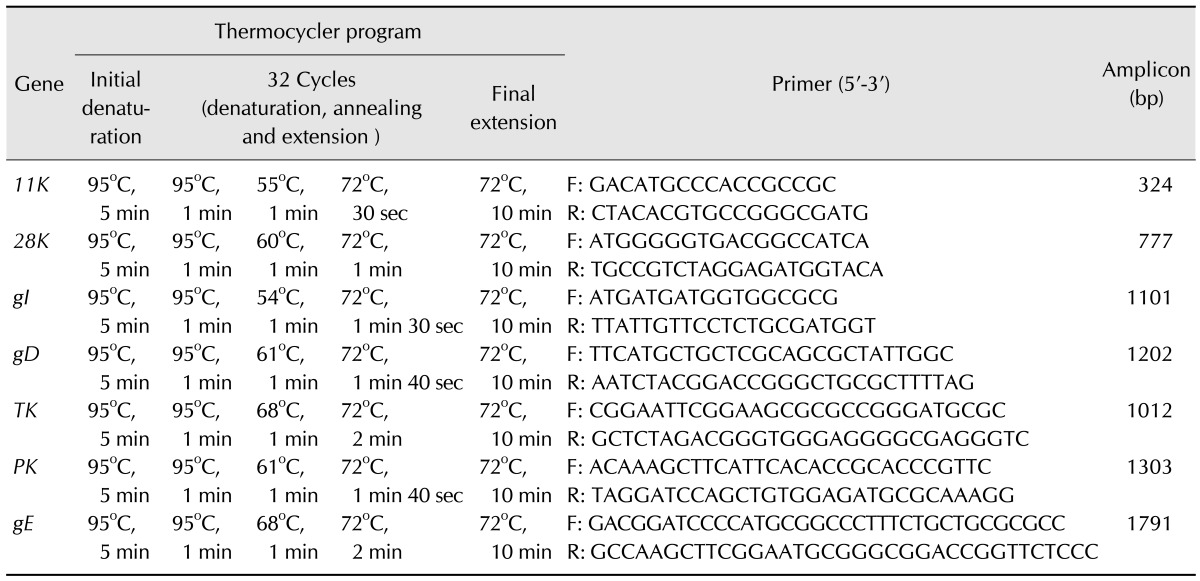
DNA extraction was performed using an AxyPrep Body Fluid Viral DNA/RNA Miniprep Kit (Axygen, China) according to the manufacturer's instructions. For sequencing, the major viral genes (gE, TK, gI, gD, PK, 11K and 28K) of these PRV strains were amplified by PCR in a total reaction volume of 50 µL containing 2.5 U of LA Taq DNA polymerase (Takara Bio, China), 25 µL of 2× GC buffer І (Takara Bio), 20 pmol of each primer (Table 1), 2 mM of each deoxynucleoside triphosphate (dNTP), and 2 µL of extracted DNA. The thermocycler programs used are shown in Table 1. The PCR products were visualized on 1.0% agarose gels stained with ethidium bromide after running for 30 min at 110 volts. The PCR products were sent to BGI Tech Solutions (China) for sequencing in both directions.
Table 1. Thermocycler programs and primers used in this study for amplification of pseudoravies virus viral genes (11K, 28K, gI, gD, TK, PK and gE).
Sequence analysis and alignment of nucleotide sequences
The sequencing results were edited and analyzed using the DNAstar software (ver. 7.1.0). Clustal X 2.0 was used to compare the nucleotide sequences of strains isolated in this study with those of other representative PRV strains (Supplementary Table 2). The nucleotide sequence homologies were further assessed using Clustal W (DNAstar software). Phylogenetic trees were constructed using the neighbor-joining method with the MEGA program (ver. 5.05) and bootstrap analyses were conducted using 1,000 replicates.
Results
Homology analysis
Homology analysis was performed to compare the nucleotide sequences of the viral genes of the 11 isolated strains with other strains available in GenBank. The homologies of the nucleotide sequences of gE, TK, gI, gD, PK, 11K and 28K among the PRV isolates under investigation were 99.8%–100.0%, 99.9%–100.0%, 100.0%, 99.9%–100.0%, 99.9%–100.0%, 100.0% and 100.0%, respectively. The gE, TK, gI, gD, PK, 11K and 28K genes of the strains under investigation shared 99.7–99.9%, 99.9–100.0%, 99.9%, 99.9–100.0%, 99.9%–100.0%, 99.3% and 100.0% nucleotide homology, respectively, with the TJ strain (China, 2012), indicating a very close phylogenetic relationship. The gE, TK, gI, gD, PK, 11K and 28K genes of the PRV isolates under investigation shared identities of 99.5%–99.7%, 99.7%–99.9%, 99.7%, 99.4%–99.5%, 98.9%–99.0%, 99.3%, and 99.7%, respectively, compared with the previously isolated Ea strain (China, 1992), and identities of 97.7%–98.1%, 99.5%–99.7%, 96.1%–96.3%, 98.9%–99.4%, 98.4%–99.6%, 98.0%, and 96.2%–97.4%, respectively, compared with the occidental strains Becker (USA, 1967) and Kaplan (Hungary, 1959).
Alignment of nucleotide sequences
Alignment of nucleotide sequences was conducted to identify differences between strains isolated in the Western and Eastern regions on a global scale. When compared with Kaplan and Becker, there were two insertions of six discontinuous nucleotides at positions 142–144 and 1,488–1,490 in the gE gene of the strains under investigation, which was similar to other strains isolated from China during 2012–2013 (panel A in Fig. 1). There was a 6 nt deletion at position 82–87 in the PK gene of the tested strains compared with Becker (panel B in Fig. 1). When compared with Kaplan and Becker, there was a 3 nt insertion at position 737–739 and a 3 nt deletion at position 518–520 in the gI gene of the tested strains (panel C in Fig. 1). A 6 nt insertion was identified in the gD gene of our isolated strains at nucleotide position 831–836 compared with Kaplan and Bartha (panel D in Fig. 1). The rest of the genes investigated in this study had no nucleotide insertions or deletions in common with the other PRV isolates.
Fig. 1. Alignment of gE (A), PK (B), gI (C) and gD (D) genes of pseudorabies virus at the nucleotide level. Black box indicates the region of insertion or deletion.
To provide a further understanding of the variations in Chinese PRV isolates, differences among the 11 isolates under investigation and previously isolated Chinese strains were analyzed. We found that both our isolates and Ea (China, 1992) had the same insertions or deletions in the gI and PK genes (panels B and C in Fig. 1). A 6 nt insertion was identified in the gD gene in all the tested strains, as well as in Fa (China, 1989), which was absent from PRV-FZ (China, 2007), LA (China, 2002) and Min-A (China, 1996) (panel D in Fig. 1). Furthermore, two insertions were identified in the gE gene of the tested strains when compared with Kaplan and Becker. The first insertion site was found in all 11 isolates as well as in LA (China, 2002), Ea (China, 1992), and GDSH (China, 2007), but was absent from SH (China, 1999). With regard to the second insertion site, our isolates shared the same characteristics with LA (China, 2002), but were different from Ea (China, 1992), GDSH (China, 2007) and SH (China, 1999) (panel A in Fig. 1).
In addition, compared with the other strains, the strains under investigation shared the same nucleotide insertions or deletions as the TJ strain.
Phylogenetic analysis
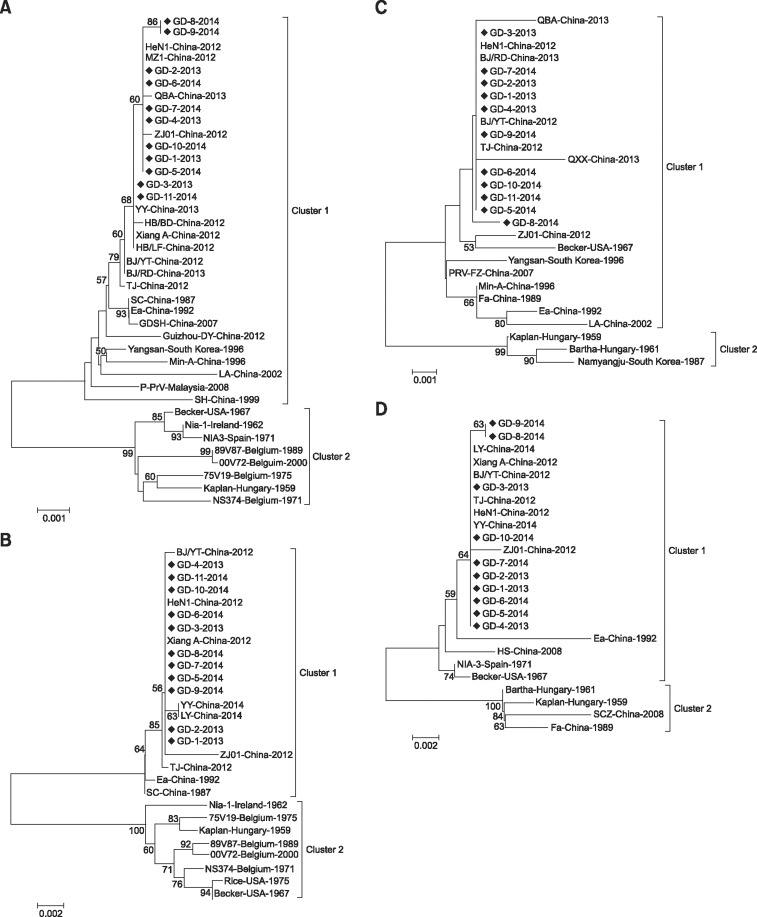
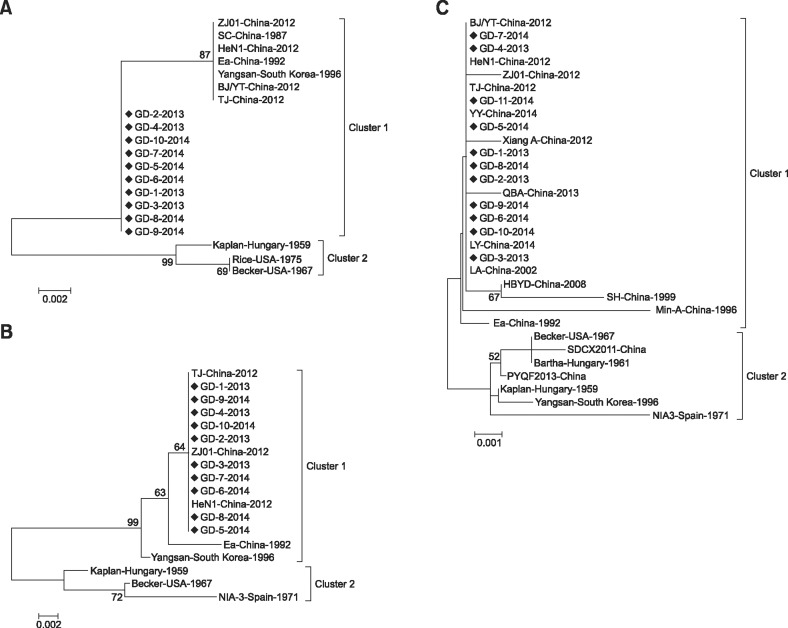
To determine the phylogenetic relationships between the PRV circulating in southern China from 2013 to 2014 and other strains, phylogenetic trees were constructed based on the major viral genes (gE, TK, gI, PK, gD, 11K and 28K). Phylogenetic analysis of multiple sequences generated trees with similar profiles for all genes except PK and gD, which were classified in two primary clusters. Cluster 1 comprised strains from eastern countries (i.e., China, South Korea and Malaysia) exclusively, while cluster 2 represented strains from western countries and a small number of other Asian strains. Based on the genes involved in this study, PRV strains isolated in China from 2012 to 2014 were clustered into one group and also formed a relatively independent branch. In addition, PRV strain TJ (China, 2012) exhibited a marked genetic relationship with the strains under investigation (Figs. 2 and 3).
Fig. 2. Phylogenetic trees for gE (A), gI (B), gD (C) and PK (D) genes. Only bootstrap values greater than 50 are shown. Strains marked with black diamonds were isolated in our laboratory during 2013–2014.
Fig. 3. Phylogenetic trees for 11K (A), 28K (B) and TK (C) genes. Only bootstrap values greater than 50 are shown. Strains marked with black diamonds were isolated in our laboratory during 2013–2014.
Phylogenetic analysis of the genes in this study revealed that our isolates were not only closely related to other strains isolated in China in recent years, but also had a relatively close genetic relationship with earlier isolates from Asia, including Ea (China, 1992), Min-A (China, 1996), P-PrV (Malaysia, 2008), and Yangsan (South Korea, 1996). Moreover, it was found that the 11 strains under investigation were relatively distant in genetic terms from isolates of western countries, including Becker (USA, 1967), Kaplan (Hungary, 1959) and NIA-3 (Spain, 1971) (Figs. 2 and 3).
Discussion
Since late 2011, outbreaks of Aujeszky's disease have occurred in many Bartha-K61-vaccinated pig farms in China, including those in Guangdong province, causing great economic losses in the swine industry. These outbreaks suggest that PRV strains have evolved new types of pathogenicity. Furthermore, all 11 strains investigated in this study were isolated from Bartha-K61-vaccinated pig herds, indicating that this vaccine fails to provide effective protection against the new prevalent PRV strains in China.
Based on these observations, we investigated the cause of this increasing virulence and changes in the immunogenicity of the new PRV isolates at the molecular level by analyzing the sequences of the major viral genes (gE, TK, gI, PK, gD, 11K and 28K). Nucleotide sequence alignments revealed insertions or deletions in the gD, gE, gI and PK genes of the newly isolated PRV strains compared with other PRV isolates from all over the world. When compared with previously isolated Chinese strains, a 6 nt insertion in gD and two insertions in gE were found to be unique molecular characteristics of the new prevalent PRV strains in China. These changes could be considered molecular markers of the variant PRVs prevailing in China, and it is speculated that these genes play a role in the variation of Chinese PRV strains.
PRV virions are coated with glycoproteins, which are crucial for viral pathogenesis. These molecules are involved in the initiation of viral replication and spreading, mediating important interactions between the virion and host cell; thus, the glycoproteins are good candidates for the rational design of vaccination strategies [10]. Although glycoprotein gE is not essential for viral replication, the absence of this protein reduces virulence [11]. Furthermore, glycoprotein gD, which is essential for viral replication, is required for viral penetration into the cell [16] and induces neutralizing antibodies [2]. Thus, it can be assumed that variations in the gD and gE genes are related to virulence alteration and immunogenic changes in PRV strains. However, the potential relationship between the insertions or deletions in viral genes identified in this study and the virulence of PRV, which is thought to be determined by multiple factors, remains to be clarified.
Phylogenetic analysis of the major viral genes in this study showed that the newly isolated strains are closely related to the strains isolated in China in recent years. These findings indicate that the isolates from the outbreaks of PR occurring in China from 2012 to 2014 share the same origin. However, the strains under investigation exhibit a relatively closer phylogenetic relationship with the Asian PRV strains than those from western countries, indicating that these newly isolated strains may not originate from virulent strains within China.
It should be noted that the strains under investigation share a significantly high level of homology with the PRV strain TJ (China, 2012), and also share the same nucleotide insertions or deletions as other strains. Furthermore, our strains also formed a tight cluster with TJ in all phylogenetic trees. When compared with the classical PRV SC strain, which was isolated in China in 1987 and classified as a virulent strain [20], the TJ strain is associated with higher mortality in pigs [9]. Together with the results of experimental infections of pigs with the GD-1-2013 strain (a mortality of 60% to 120-day-old pigs) (unpublished data), the high levels of identities between the isolates under investigation and the TJ strain indicate that these newly emerging strains are also highly virulent.
In summary, the results demonstrate that the newly emerging strains are PRV variants with unique molecular signatures. Further studies are required to explore the relevance of the differences within sequences to the virulence alteration and immunogenic characteristics of these PRV variants.
Acknowledgments
The authors thank Guangdong Wen's Food Co. Ltd. for assistance collecting samples, and Doctor Ling Zhu, Faculty of Veterinary and Agricultural Sciences, University of Melbourne, Parkville, for critically reviewing the manuscript.
Footnotes
Conflict of Interest: There is no conflict of interest.
Supplementary Material
Supplementary data is available at http://www.vetsci.org only.
PRV strains isolated from Guangdong province of southern China during 2013–2014
Reference strains used in this work
References
- 1.An TQ, Peng JM, Tian ZJ, Zhao HY, Li N, Liu YM, Chen JZ, Leng CL, Sun Y, Chang D, Tong GZ. Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg Infect Dis. 2013;19:1749–1755. doi: 10.3201/eid1911.130177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eloit M, Fargeaud D, L'Haridon R, Toma B. Identification of the pseudorabies virus glycoprotein gp50 as a major target of neutralizing antibodies. Arch Virol. 1988;99:45–56. doi: 10.1007/BF01311022. [DOI] [PubMed] [Google Scholar]
- 3.Ficińska J, Bieńkowska-Szewczyk K, Jacobs L, Płucienniczak G, Płucienniczak A, Szewczyk B. Characterization of changes in the short unique segment of pseudorabies virus BUK-TK900 (Suivac A) vaccine strain. Arch Virol. 2003;148:1593–1612. doi: 10.1007/s00705-003-0121-x. [DOI] [PubMed] [Google Scholar]
- 4.Kimman TG, de Wind N, Oei-Lie N, Pol JMA, Berns AJM, Gielkens ALJ. Contribution of single genes within the unique short region of Aujeszk's disease virus (suid herpesvirus type 1) to virulence, pathogenesis and immunogenicity. J Gen Virol. 1992;73:243–251. doi: 10.1099/0022-1317-73-2-243. [DOI] [PubMed] [Google Scholar]
- 5.Kit S, Kit M, Pirtle EC. Attenuated properties of thymidine kinase-negative deletion mutant of pseudorabies virus. Am J Vet Res. 1985;46:1359–1367. [PubMed] [Google Scholar]
- 6.Klupp BG, Hengartner CJ, Mettenleiter TC, Enquist LW. Complete, annotated sequence of the pseudorabies virus genome. J Virol. 2004;78:424–440. doi: 10.1128/JVI.78.1.424-440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lomniczi B, Blankenship ML, Ben-Porat T. Deletions in the genomes of pseudorabies virus vaccine strains and existence of four isomers of the genomes. J Virol. 1984;49:970–979. doi: 10.1128/jvi.49.3.970-979.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lü S, Guo G, Wei F, Wang W, Dong B, Bi Y, Miao L, Shen Z. Study on safety and immune efficiency of pseudorabies virus alive vaccine with gE−/gI−2;/TK− gene deleted. Prog Vet Med. 2014;35:1–5. [Google Scholar]
- 9.Luo Y, Li N, Cong X, Wang CH, Du M, Li L, Zhao B, Yuan J, Liu DD, Li S, Li Y, Sun Y, Qiu HJ. Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Vet Microbiol. 2014;174:107–115. doi: 10.1016/j.vetmic.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Mettenleiter TC. Immunobiology of pseudorabies (Aujeszky's disease) Vet Immunol Immunopathol. 1996;54:221–229. doi: 10.1016/s0165-2427(96)05695-4. [DOI] [PubMed] [Google Scholar]
- 11.Mulder WAM, Jacobs L, Priem J, Kok GL, Wagenaar F, Kimman TG, Pol JMA. Glycoprotein gE-negative pseudorabies virus has a reduced capability to infect second- and third-order neurons of the olfactory and trigeminal routes in the porcine central nervous system. J Gen Virol. 1994;75:3095–3106. doi: 10.1099/0022-1317-75-11-3095. [DOI] [PubMed] [Google Scholar]
- 12.Peng J, An T, Zhao H, Liu Y, Chen J, Leng C, Sun Y, Chang D, Tian Z, Tong G. Identification and antigenic variation of new epidemiology of pseudorabies virus from swine. Chin J Prev Vet Med. 2013;35:1–4. [Google Scholar]
- 13.Peng J, Chen J, Tian Z, Wang Y, Zhou Y, An T, Tong G. Identification of the deletion in the Us region of pseudorabies virus Bartha K61 strain. Chin Vet Sci. 2008;38:646–649. [Google Scholar]
- 14.Petrovskis EA, Timmins JG, Gierman TM, Post LE. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J Virol. 1986;60:1166–1169. doi: 10.1128/jvi.60.3.1166-1169.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pomeranz LE, Reynolds AE, Hengartner CJ. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev. 2005;69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rauh I, Mettenleiter TC. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J Virol. 1991;65:5348–5356. doi: 10.1128/jvi.65.10.5348-5356.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang CH, Yuan J, Qin HY, Luo Y, Cong X, Li Y, Chen J, Li S, Sun Y, Qiu HJ. A novel gE-deleted pseudorabies virus (PRV) provides rapid and complete protection from lethal challenge with the PRV variant emerging in Bartha-K61-vaccinated swine population in China. Vaccine. 2014;32:3379–3385. doi: 10.1016/j.vaccine.2014.04.035. [DOI] [PubMed] [Google Scholar]
- 18.Wu R, Bai C, Sun J, Chang S, Zhang X. Emergence of virulent pseudorabies virus infection in northern China. J Vet Sci. 2013;14:363–365. doi: 10.4142/jvs.2013.14.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu X, Zhou Z, Hu D, Zhang Q, Han T, Li X, Gu X, Yuan L, Zhang S, Wang B, Qu P, Liu J, Zhai X, Tian K. Pathogenic pseudorabies virus, China, 2012. Emerg Infect Dis. 2014;20:102–104. doi: 10.3201/eid2001.130531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan Q, Li Z, Nan X, Wu Y, Li Y. Isolation and identification of pseudorabies virus. Chin J Prev Vet Med. 1987;3:10–11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRV strains isolated from Guangdong province of southern China during 2013–2014
Reference strains used in this work