Abstract
The p38 and JNK classes of mitogen-activated protein kinases (MAPKs) have evolutionarily conserved roles in the control of cellular responses to microbial and abiotic stresses. The mechanisms by which crosstalk between distinct p38 and c-Jun N-terminal kinase (JNK) MAPK pathways occurs with resultant integration of signaling information have been difficult to establish, particularly in the context of whole organism physiology. In Caenorhabditis elegans a PMK-1 p38 MAPK pathway is required for resistance to bacterial infection, and a KGB-1 JNK-like MAPK pathway has recently been shown to mediate resistance to heavy metal stress. Here, we show that two components of the KGB-1 pathway, MEK-1 MAPK kinase (MAPKK), a homolog of mammalian MKK7, and VHP-1 MAPK phosphatase (MKP), a homolog of mammalian MKP7, also regulate pathogen resistance through the modulation of PMK-1 activity. The regulation of p38 and JNK-like MAPK pathways mediating immunity and heavy metal stress by common MAPKK and MKP signaling components suggests pivotal roles for MEK-1 and VHP-1 in the integration of diverse stress signals contributing to pathogen resistance in C. elegans. In addition, these data point to mechanisms in multicellular organisms by which signals transduced by distinct MAPK pathways may be subject to physiological integration at the level of regulation of MAPK activity by MAPKKs and MKPs.
Mitogen-activated protein kinase (MAPK) signaling pathways serve as transducers of extracellular stimuli that allow cellular adaptation to changes in environment. MAPK pathways are highly evolutionarily conserved and play key roles in many diverse physiological processes, including development, growth and proliferation, stress responses, and immunity. MAPK activation generally involves the phosphorylation of Thr and Tyr residues in a signature T–X–Y activation domain motif by a dual-specificity MAPK kinase (MAPKK) (1). Whereas the genetic characterization of MAPK activation in yeast has shown a one-to-one correspondence between a particular MAPKK and a cognate MAPK (2), studies of MAPK activation in multicellular organisms suggest that at least two MAPKKs can function as activators of a MAPK (1), although the functional significance of the ability of multiple MAPKKs to activate a single MAPK remains unclear. Negative regulatory elements of MAPK signaling have been less well characterized. Many classes of phosphatases, including Ser/Thr phosphatases, Tyr phosphatases, and dual-specificity MAPK phosphatases (MKPs), have been implicated in inhibition of MAPK signaling pathways (3).
The MAPK signaling cassette represents perhaps the most ancient of evolutionarily conserved pathways of immunity, being conserved from plants to mammals (4, 5). Two classes of MAPKs, the c-Jun N-terminal kinase (JNK) and p38 MAPK, function as key mediators of stress and immune signaling in mammals (1, 5). The MKK4 and MKK7 MAPKKs have been shown to activate JNK (6–13), and the MKK3 and MKK6 MAPKKs serve as the major activators of p38 MAPK (14–21). Genetic analysis in mice suggests nonredundant distinct roles for MKK4 and MKK7 in JNK activation in response to different cellular stimuli (13). Similar studies of p38 MAPK activation by MKK3 and MKK6 suggest distinct tissue-specific roles, but MKK3, MKK6, and even MKK4 may function redundantly in p38 MAPK activation as well (21).
The mechanisms of negative regulation of p38 and JNK MAPK in vivo have been characterized in less detail. In Drosophila, the puckered gene encodes a MKP that has been shown to play a key role in regulating JNK activity in dorsal closure (22). In contrast to the extensive genetic studies of MAPKK activators of p38 and JNK MAPK in mice, no such studies have been conducted on corresponding MKPs, and the relative magnitudes of the contributions of MAPKKs and MKPs to the balanced regulation of MAPKs have been difficult to assess. In addition, although p38 and JNK MAPKs are responsive to similar stimuli in mammalian cells, information flow through each pathway has been thought to be distinct, and points of crosstalk and signal integration between stress-activated MAPK pathways await identification.
Phylogenetic analysis of MAPK signaling pathways indicates a high degree of conservation of p38 and JNK MAPK signaling components between Caenorhabditis elegans and mammals (23, 24). We recently established a functionally conserved role for the C. elegans PMK-1 p38 MAPK pathway in innate immunity (25). From a forward genetic screen for mutants with enhanced susceptibility to pathogen-induced killing (Esp phenotype), we characterized a requirement for a NSY-1 MAPKKK/SEK-1 MAPKK/PMK-1 MAPK pathway (homologous to the mammalian ASK-1-MKK3/6-p38 MAPK signaling cassette) in mediating resistance to bacterial pathogens (25). The nsy-1 and sek-1 mutants have enhanced susceptibility to pathogens but are viable and fertile, with pleiotropies limited to an egg-laying defect and aberrant neuronal cell fate determination in development (26, 27). The minimal pleiotropies of C. elegans PMK-1 p38 MAPK pathway mutants contrast with the embryonic lethality of homozygous deletions of p38 MAPK pathway components in Drosophila melanogaster and mice (28, 29) and permit the genetic analysis of putative regulatory components in the immune response of an intact C. elegans organism.
Here, we describe the characterization of regulatory MAPKK and MKP components of the PMK-1 p38 MAPK pathway in the C. elegans immune response. We show that the MKK3/6 homolog SEK-1 is essential for PMK-1 activation, but that, unexpectedly, a C. elegans MKK7 homolog, MEK-1, previously identified as being required for heavy metal stress (30), is also required for the full physiological activation of PMK-1. In addition, we show that the MKP VHP-1, a homolog of mammalian MKP7, negatively regulates PMK-1 function in pathogen defense. We discuss implications with respect to potential roles for MEK-1 and VHP-1 in mediating crosstalk and signal integration through p38 and JNK MAPK pathways.
Materials and Methods
Mutant strains of C. elegans were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis) or generated in the Matsumoto laboratory. Mutant alleles used in this study, which are described in detail in refs. 27, 30, and 31, are all deletions in which large segments of each gene encoding catalytic regions have been deleted and thus are expected to represent null alleles. Pathogen killing assays with Pseudomonas aeruginosa strain PA14 were carried out as described in refs. 25 and 32. Immunoblotting of lysates derived from synchronized populations of young adult worms with anti-phospho-p38 Ab (Promega), anti-PMK-1 (K.M. laboratory), and anti-β-tubulin (E7, Developmental Studies Hybridoma Bank, Iowa City, IA), was performed as described in ref. 25. RNA interference (RNAi) by feeding of Escherichia coli HT115 carrying a plasmid with sequences corresponding to pmk-1 (using pDK177) (25), vhp-1 (using clone 44D3 from Julie Ahringer, Wellcome Trust, University of Cambridge, Cambridge, U.K.) (33) and aforementioned candidate genes encoding dual-specificity phosphatase domains (clones from the Ahringer library) (33) was performed by dropping synchronized populations of L1 larvae onto lawns seeded with corresponding bacterial strains as described in ref. 25. For pathogen killing assays involving feeding RNAi, worms were transferred from HT115 to PA14 and scored accordingly.
Results and Discussion
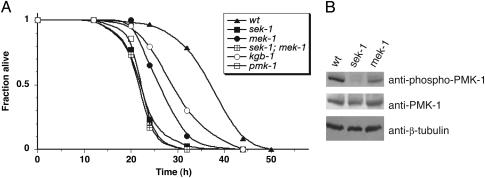
Requirement for MEK-1 and SEK-1 MAPKKs in PMK-1 p38 MAPK Activation in C. elegans Immunity. The sek-1 gene encodes a C. elegans homolog of mammalian MKK3 and MKK6 classes of MAPKKs (27). Previously, we reported that loss-of-function mutations in sek-1, as well as nsy-1, encoding an upstream MAPKKK, conferred enhanced susceptibility to killing by pathogens (Esp phenotype), and markedly diminished levels of PMK-1 activation were observed in sek-1 and nsy-1 mutant whole worm lysates relative to WT lysates. RNAi of pmk-1, an ortholog of p38 MAPK, also resulted in a strong Esp phenotype (25). As expected from these prior studies, we found that the pmk-1(km25) mutant (31) also exhibited a pronounced Esp phenotype equivalent to that of the sek-1(km4) mutant (Fig. 1A).
Fig. 1.
Requirement for MEK-1 and SEK-1 in the activation of PMK-1 in C. elegans immunity. (A) Pathogen susceptibility of C. elegans mek-1(ks54), sek-1(km4), mek-1(ks54);sek-1(km4), pmk-1(km25), and kgb-1(km21) mutants. At least 40 L4-stage worms were allowed to feed on lawns of P. aeruginosa strain PA14 and were scored as dead when they no longer responded to touch as described in ref. 32. Representative plots of multiple experiments are shown. All mutant alleles are deletions that are predicted to be null alleles. (B) Immunoblot analysis of PMK-1 activation in young adult whole worm lysates of mek-1(ks54), sek-1(km4), and WT worms by using Abs specific for the doubly phosphorylated activated form of PMK-1 (anti-phospho-p38). Anti-β-tubulin was used as a loading control.
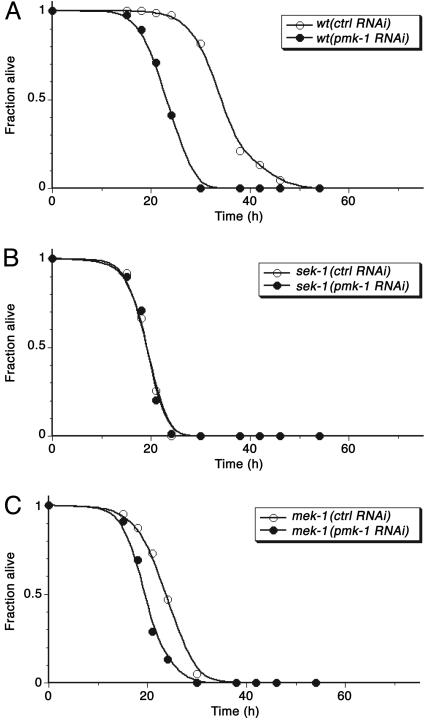
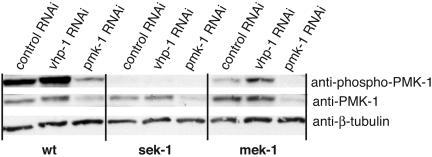
The mek-1 gene encodes a C. elegans homolog of the mammalian MKK7 class of MAPKKs, and a deletion allele, mek-1(ks54), was previously noted to confer enhanced sensitivity to heavy metal stress (30). We found that the mek-1(ks54) mutant had an Esp phenotype, although not as strong as that of the sek-1 mutant (Fig. 1 A). The mek-1(ks54);sek-1(km4) double mutant was observed to have an Esp phenotype equivalent to that of sek-1(km4) alone or pmk-1(km25) alone (Fig. 1 A), suggesting that mek-1 might act in the same pathway as sek-1, upstream of pmk-1. Indeed, immunoblot analysis of PMK-1 activation in mek-1(ks54) lysates revealed that levels of PMK-1 activation are diminished substantially compared with WT levels, although not undetectable as in sek-1(km4) lysates (Fig. 1B). These data suggest that SEK-1 is essential for PMK-1 activation but that full activation of PMK-1 requires the additional activity of MEK-1. Consistent with the correlation of Esp phenotype with levels of PMK-1 activation, there is no enhancement of the Esp phenotype of the sek-1(km4) mutant when subjected to pmk-1 RNAi, whereas there is some enhancement of the Esp phenotype of the mek-1(ks54) mutant subjected to pmk-1 RNAi (Fig. 2). The effect of RNAi of pmk-1 on relative levels of PMK-1 activation in WT, sek-1, and mek-1 backgrounds also was analyzed directly by immunoblotting and corroborated the effects of RNAi of pmk-1 on the Esp phenotype (Fig. 3).
Fig. 2.
Effect of RNAi of pmk-1 on the pathogen susceptibility of WT (A), sek-1(km4) (B), and mek-1(ks54) (C) worms. Synchronized populations of L1 larvae for each strain were propagated on E. coli HT115 carrying either vector control (L4440) or pDK177 (pmk-1) as described previously and transferred to lawns of P. aeruginosa strain PA14 at the L4 larval stage. At least 40 worms were scored for each assay and scored as dead when they no longer responded to touch. Representative plots of multiple experiments are shown.
Fig. 3.
Modulation of PMK-1 activation by MEK-1 and SEK-1 MAPKKs and VHP-1 MKP: correlation of pathogen susceptibility with levels of PMK-1 activation. Immunoblot analysis of lysates derived from WT and sek-1(km4) and mek-1(ks54) mutants subjected to RNAi by feeding with bacterial strains expressing double-stranded RNA corresponding to the sequence of control (L4440 vector only), pmk-1 (pDK177), or vhp-1 (Ahringer 44D3) genes. Activated levels of PMK-1 were detected by using an Ab (anti-phospho-p38) specific for the doubly phosphorylated form of PMK-1.
These data suggest that both MEK-1 and SEK-1 are required for full activation of PMK-1 under physiological conditions. These data do not suggest that the requirement for two MAPKKs in PMK-1 activation results from the action of each MAPKK separately in distinct tissues because no PMK-1 activation is observed in the absence of SEK-1. A potential direct role for MEK-1 may be to function in the coordinated or synergistic activation of PMK-1 in conjunction with SEK-1. Prior studies from mammalian systems suggested the specificity of MKK7 for JNK MAPK (9–11), but our data suggest that the MKK7 homolog MEK-1 is required for full activation of PMK-1 p38 MAPK under physiological conditions.
SEK-1 seems to be sufficient to activate PMK-1 in the absence of MEK-1, but not to maximal levels that confer WT levels of pathogen resistance. Studies of JNK activation by MKK4 and MKK7 suggest that MKK4 and MKK7 sometimes may function synergistically in the coordinated activation of JNK (6, 34–36). Although studies of p38 MAPK activation have not provided evidence of coordinated or synergistic activation involving MKK3, MKK4, or MKK6, our results suggest that MKK7 might serve an adjunct role in augmenting the activity of the principal p38 MAPK kinases, MKK3 and MKK6.
Because MEK-1 is a homolog of MKK7, an activator of JNK MAPK, we anticipated that in addition to PMK-1, MEK-1 might act on additional JNK MAPK targets. JNK-1 had been reported in ref. 37 as the downstream target of MEK-1, but we were unable to reproduce the reported heavy metal sensitivity phenotype of the jnk-1(gk7) mutant (31). As we reported previously, the jnk-1(gk7) mutant also did not exhibit an Esp phenotype (25). Mizuno et al. (31) recently found that kgb-1, encoding a JNK-like MAPK (38), is the downstream target of mek-1 in mediating heavy metal stress. We observed that the kgb-1(km21) mutant also had an Esp phenotype, although not as strong as that of mek-1(ks54), sek-1(km4), or pmk-1(km25) (Fig. 1 A).
Consistent with their involvement in general stress responses, we observe that both mek-1(ks54) and kgb-1(km21) mutants exhibit compromised lifespan when propagated on E. coli strain OP50 (data not shown). The observed diminished longevity may be secondary to reduced resistance to oxidative stress that may contribute to the aging process, as well as the activity of E. coli as a weak pathogen. Such diminished stress sensitivity likely contributes significantly to the observed Esp phenotype of the kgb-1(km21) mutant on P. aeruginosa PA14 and also may contribute to the Esp phenotype of the mek-1(ks54) mutant.
MEK-1 seems to act upstream of both PMK-1 and KGB-1 MAPKs, although the considerably stronger Esp phenotype of pmk-1(km25) relative to kgb-1(km21), the stronger Esp phenotype of mek-1(ks54) compared with kgb-1(km21), and the comparable Esp phenotypes of the sek-1(km4), sek-1(km4);mek-1(ks54), and pmk-1(km25) mutants all suggest that the major target of MEK-1 with respect to immunity is PMK-1. In addition, we did not observe diminished levels of PMK-1 activation in kgb-1(km21) lysates compared with WT lysates (data not shown) in contrast to the markedly diminished levels observed in mek-1(ks54) lysates (Fig. 1B), ruling out the possibility that PMK-1 levels are secondarily affected downstream of MEK-1–KGB-1 signaling.
Although we cannot exclude the possibility that PMK-1 levels are affected indirectly by MEK-1, for example, through the activation of a different MAPK by MEK-1 or by more complex interactions involving common scaffolding components, these data demonstrate that distinct MAPK pathways converging on PMK-1 and KGB-1 have a common component, MEK-1, which is required for intact signaling through each pathway and serves as a potential mediator of crosstalk between immune and stress responses. Further biochemical dissection of PMK-1 activation by SEK-1 and MEK-1 may illuminate the detailed mechanisms through which these MAPKKs modulate PMK-1 activity.
VHP-1 Dual-Specificity MAPK Phosphatase Negatively Regulates PMK-1 p38 MAPK in C. elegans Immunity. We undertook a reverse genetic approach to identify negative regulators of PMK-1 p38 MAPK activation in C. elegans. Specifically, we analyzed the effect of RNAi of each of the following nine genes predicted to encode proteins with a conserved dual-specificity phosphatase domain on the Esp phenotype of the mek-1(ks54) mutant: C05B10.1, ZK757.2, F26A3.4, F13D11.3, F08B1.1, C04F12.8, C16A3.2, F28C6.8, and C24F3.2. We anticipated that inactivation of phosphatases negatively regulating PMK-1 might result in increased levels of PMK-1 activation that might confer increased resistance to infection. Specifically, in the mek-1(ks54) mutant background partially defective in PMK-1 activation, phosphatase inactivation might be expected to reverse diminished levels of PMK-1 activation and thus suppress the Esp phenotype of mek-1(ks54).
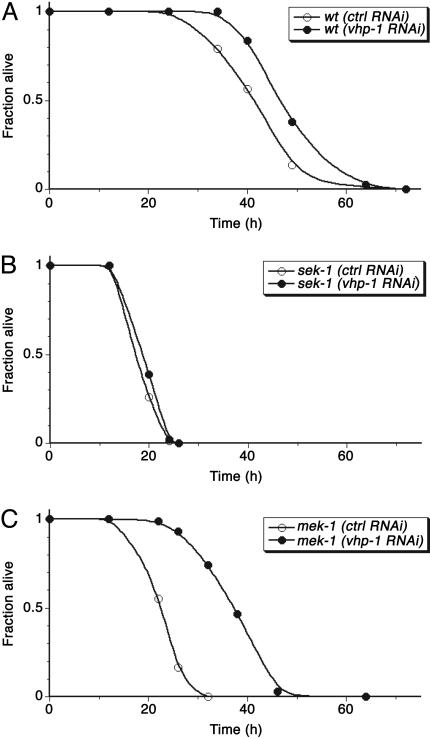
One of the nine genes tested by RNAi had a marked effect on the Esp phenotype of the mek-1(ks54) mutant. The C. elegans gene F08B1.1, also termed vhp-1, is predicted to encode a MKP with homology to mammalian MKP7, which has been shown to dephosphorylate both p38 and JNK MAPKs (39). RNAi of vhp-1 was observed to suppress the pathogen susceptibility of the mek-1(ks54) mutant (Fig. 4). RNAi of vhp-1 also resulted in slightly increased pathogen resistance in the WT background but had no effect on the Esp phenotype of the sek-1 mutant (Fig. 4). These data are consistent with negative regulation of PMK-1 by VHP-1. In particular, VHP-1 inhibition would be expected to have no effect on the sek-1(km4) mutant, in which there is no PMK-1 activation.
Fig. 4.
Effect of RNAi of vhp-1 on the Esp phenotype of WT (A) and sek-1(km4) (B) and mek-1(ks54) (C) mutants. Synchronized populations of L1 larvae for each strain were propagated on E. coli HT115 carrying either vector control (L4440) or Ahringer clone 44D3 (vhp-1) as described previously and transferred to lawns of P. aeruginosa strain PA14 at the L4 larval stage. At least 40 worms were scored for each assay and scored as dead when they no longer responded to touch. Representative plots of multiple experiments are shown.
Direct detection of levels of PMK-1 activation from whole worm lysates derived from WT and sek-1(km4) and mek-1(ks54) mutants subjected to RNAi of vhp-1 demonstrated correlation of pathogen susceptibility with levels of PMK-1 activation (Fig. 3). In particular, RNAi of vhp-1 was observed to partially revert the diminished PMK-1 activation present in the mek-1(ks54) mutant. These data strongly suggest that VHP-1 acts to negatively regulate PMK-1 activation in C. elegans immunity and underscore the correlation between levels of PMK-1 activation and organism resistance to bacterial pathogens. Of note, Mizuno et al. (31) have shown that the null phenotype of the vhp-1 mutant is L3 larval arrest, a phenotype that is suppressed by the mek-1 mutation. The slightly increased survival of WT worms subjected to vhp-1 RNAi (Fig. 4A) likely represents a partial loss-of-function phenotype, in which the worms do not become arrested at the L3 larval stage but in which resultant increased PMK-1 activation contributes to enhanced survival on pathogens.
Integration of Immune and Stress Response Pathways Through MEK-1 and VHP-1. Our genetic and biochemical analyses of mek-1 and vhp-1 function, carried out in the whole organism, suggest that these two genes play critical roles in the modulation of C. elegans pathogen resistance. The expression patterns of mek-1 (30) and vhp-1 (31), deduced from the expression of GFP gene fusions, overlap with previously reported tissue expression for sek-1 and pmk-1 that included the intestine (27, 40), the primary site of bacterial infection in our experimental system. The correlation of PMK-1 p38 MAPK activity with whole organism pathogen sensitivity suggests a molecular mechanism of positive and negative regulation of C. elegans immunity through the fine-tuning of PMK-1 activity.
MEK-1 had previously been implicated in mediating heavy metal stress in C. elegans (30), and, more recently, Mizuno et al. (31) established that MEK-1 and VHP-1 are also components of a KGB-1 JNK-like MAPK pathway involved in heavy metal stress. The requirement for KGB-1 and MEK-1 in mediating resistance to heavy metal stress suggests that hypersensitivity to general stress may contribute to the Esp phenotypes of the kgb-1(km21) and mek-1(ks54) mutants, although we provide strong evidence that MEK-1 modulates PMK-1 activity as well. The involvement of VHP-1 in the dephosphorylation of both KGB-1 (31) and PMK-1 leaves open the possibility of more complex interactions governing the effect of MEK-1 on each pathway. For example, diminished activation of KGB-1 could result in increased recruitment of VHP-1 to PMK-1 instead, thus indirectly leading to the apparent diminished activation of PMK-1 observed in the mek-1(ks54) mutant.
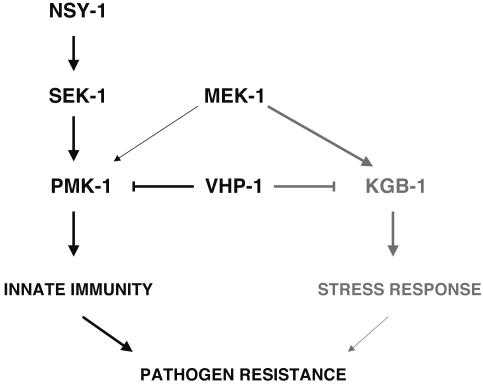
Although the detailed mechanisms of regulation of distinct pathways contributing to crosstalk remain to be elucidated, these genetic and biochemical data obtained from intact organisms suggest mechanisms of signal integration in the whole organism by means of the crossregulation of distinct signaling pathways, in this case between stress response and immune MAPK pathways in C. elegans. There is precedent from studies in yeast for integration of signals transduced by MAPK pathways through negative crossregulation of parallel pathways, as is the case for the ability of the Hog1 pathway to repress the pheromone response pathway (41). Our data point to a pivotal role for MEK-1 and VHP-1, and MAPKKs and MKPs in general, in the crossregulation and integration of signals from extracellular stresses, including pathogen attack, that generate specific responses through downstream MAPKs in multicellular organisms (Fig. 5).
Fig. 5.
Integration of distinct PMK-1 p38 MAPK innate immunity and KGB-1 JNK-like MAPK stress response pathways by common MEK-1 MAPKK and VHP-1 MAPK phosphatase signaling components.
Acknowledgments
We thank Gary Ruvkun and members of his laboratory for expert advice and Julie Ahringer for the RNAi library. Some of the strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Center for Research Resources. D.H.K. is supported by a Howard Hughes Medical Institute Postdoctoral Fellowship for Physicians, a National Institutes of Health K08 Career Development Award, and a Burroughs Wellcome Fund Career Award in the Biomedical Sciences. N.T.L. is supported by a National Science Foundation postdoctoral research fellowship in microbial biology. This work was supported by National Institutes of Health Grant GM48707 (to F.M.A.) and a grant from the Advanced Research on Cancer from the Ministry of Education, Culture, and Science of Japan (to K.M.).
Abbreviations: JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; MAPKK, MAPK kinase; MKP, MAPK phosphatase; Esp, enhanced susceptibility to pathogens; RNAi, RNA interference.
References
- 1.Kyriakis, J. M. & Avruch, J. (2001) Physiol. Rev. 81, 807–869. [DOI] [PubMed] [Google Scholar]
- 2.Schaeffer, H. J. & Weber, M. J. (1999) Mol. Cell. Biol. 19, 2435–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamura, S., Hanada, M., Ohnishi, M., Katsura, K., Sasaki, M. & Kobayashi, T. (2002) Eur. J. Biochem. 269, 1060–1066. [DOI] [PubMed] [Google Scholar]
- 4.Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., Boller, T., Ausubel, F. M. & Sheen, J. (2002) Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- 5.Dong, C., Davis, R. J. & Flavell, R. A. (2002) Annu. Rev. Immunol. 20, 55–72. [DOI] [PubMed] [Google Scholar]
- 6.Wada, T., Nakagawa, K., Watanabe, T., Nishitai, G., Seo, J., Kishimoto, H., Kitagawa, D., Sasaki, T., Penninger, J. M., Nishina, H., et al. (2001) J. Biol. Chem. 276, 30892–30897. [DOI] [PubMed] [Google Scholar]
- 7.Ganiatsas, S., Kwee, L., Fujiwara, Y., Perkins, A., Ikeda, T., Labow, M. A. & Zon, L. I. (1998) Proc. Natl. Acad. Sci. USA 95, 6881–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang, D., Tournier, C., Wysk, M., Lu, H. T., Xu, J., Davis, R. J. & Flavell, R. A. (1997) Proc. Natl. Acad. Sci. USA 94, 3004–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holland, P. M., Suzanne, M., Campbell, J. S., Noselli, S. & Cooper, J. A. (1997) J. Biol. Chem. 272, 24994–24998. [DOI] [PubMed] [Google Scholar]
- 10.Moriguchi, T., Toyoshima, F., Masuyama, N., Hanafusa, H., Gotoh, Y. & Nishida, E. (1997) EMBO J. 16, 7045–7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foltz, I. N., Gerl, R. E., Wieler, J. S., Luckach, M., Salmon, R. A. & Schrader, J. W. (1998) J. Biol. Chem. 273, 9344–9351. [DOI] [PubMed] [Google Scholar]
- 12.Tournier, C., Whitmarsh, A. J., Cavanagh, J., Barrett, T. & Davis, R. J. (1999) Mol. Cell. Biol. 19, 1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tournier, C., Dong, C., Turner, T. K., Jones, S. N., Flavell, R. A. & Davis, R. J. (2001) Genes Dev. 15, 1419–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han, J., Lee, J. D., Jiang, Y., Li, Z., Feng, L. & Ulevitch, R. J. (1996) J. Biol. Chem. 271, 2886–2891. [DOI] [PubMed] [Google Scholar]
- 15.Moriguchi, T., Kuroyanagi, N., Yamaguchi, K., Gotoh, Y., Irie, K., Kano, T., Shirakabe, K., Muro, Y., Shibuya, H., Matsumoto, K., et al. (1996) J. Biol. Chem. 271, 13675–13679. [DOI] [PubMed] [Google Scholar]
- 16.Raingeaud, J., Whitmarsh, A. J., Barrett, T., Derijard, B. & Davis, R. J. (1996) Mol. Cell. Biol. 16, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enslen, H., Raingeaud, J. & Davis, R. J. (1998) J. Biol. Chem. 273, 1741–1748. [DOI] [PubMed] [Google Scholar]
- 18.Wysk, M., Yang, D. D., Lu, H. T., Flavell, R. A. & Davis, R. J. (1999) Proc. Natl. Acad. Sci. USA 96, 3763–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, H. T., Yang, D. D., Wysk, M., Gatti, E., Mellman, I., Davis, R. J. & Flavell, R. A. (1999) EMBO J. 18, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka, N., Kamanaka, M., Enslen, H., Dong, C., Wysk, M., Davis, R. J. & Flavell, R. A. (2002) EMBO Rep. 3, 785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brancho, D., Tanaka, N., Jaeschke, A., Ventura, J. J., Kelkar, N., Tanaka, Y., Kyuuma, M., Takeshita, T., Flavell, R. A. & Davis, R. J. (2003) Genes Dev. 17, 1969–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Blanco, E., Gampel, A., Ring, J., Virdee, K., Kirov, N., Tolkovsky, A. M. & Martinez-Arias, A. (1998) Genes Dev. 12, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caffrey, D. R., O'Neill, L. A. & Shields, D. C. (1999) J. Mol. Evol. 49, 567–582. [DOI] [PubMed] [Google Scholar]
- 24.Plowman, G. D., Sudarsanam, S., Bingham, J., Whyte, D. & Hunter, T. (1999) Proc. Natl. Acad. Sci. USA 96, 13603–13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, D. H., Feinbaum, R., Alloing, G., Emerson, F. E., Garsin, D. A., Inoue, H., Tanaka-Hino, M., Hisamoto, N., Matsumoto, K., Tan, M. W., et al. (2002) Science 297, 623–626. [DOI] [PubMed] [Google Scholar]
- 26.Sagasti, A., Hisamoto, N., Hyodo, J., Tanaka-Hino, M., Matsumoto, K. & Bargmann, C. I. (2001) Cell 105, 221–232. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka-Hino, M., Sagasti, A., Hisamoto, N., Kawasaki, M., Nakano, S., Ninomiya-Tsuji, J., Bargmann, C. I. & Matsumoto, K. (2002) EMBO Rep. 3, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzanne, M., Irie, K., Glise, B., Agnès, F., Mori, E., Matsumoto, K. & Noselli, S. (1999) Genes Dev. 13, 1464–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ihle, J. N. (2000) Cell 102, 131–134. [DOI] [PubMed] [Google Scholar]
- 30.Koga, M., Zwaal, R., Guan, K. L., Avery, L. & Ohshima, Y. (2000) EMBO J. 19, 5148–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizuno, T., Hisamoto, N., Terada, T., Kondo, T., Adachi, M., Nishida, E., Kim, D. H., Ausubel, F. M. & Matsumoto, K. (2004) EMBO J. 23, 2226–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan, M. W., Mahajan-Miklos, S. & Ausubel, F. M. (1999) Proc. Natl. Acad. Sci. USA 96, 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003) Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 34.Lawler, S., Fleming, Y., Goedert, M. & Cohen, P. (1998) Curr. Biol. 8, 1387–1390. [DOI] [PubMed] [Google Scholar]
- 35.Fleming, Y., Armstrong, C. G., Morrice, N., Paterson, A., Goedert, M. & Cohen, P. (2000) Biochem. J. 352, 145–154. [PMC free article] [PubMed] [Google Scholar]
- 36.Kishimoto, H., Nakagawa, K., Watanabe, T., Kitagawa, D., Momose, H., Seo, J., Nishitai, G., Shimizu, N., Ohata, S., Tanemura, S., et al. (2003) J. Biol. Chem. 278, 16595–16601. [DOI] [PubMed] [Google Scholar]
- 37.Villanueva, A., Lozano, J., Morales, A., Lin, X., Deng, X., Hengartner, M. O. & Kolesnick, R. N. (2001) EMBO J. 20, 5114–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, P., Leung-Chiu, W. M., Montgomery, R., Orsborn, A., Kuznicki, K., Gressman-Coberly, E., Mutapcic, L. & Bennett, K. (2002) Dev. Biol. 251, 333–347. [DOI] [PubMed] [Google Scholar]
- 39.Tanoue, T., Yamamoto, T., Maeda, R. & Nishida, E. (2001) J. Biol. Chem. 276, 26629–26639. [DOI] [PubMed] [Google Scholar]
- 40.Berman, K., McKay, J., Avery, L. & Cobb, M. (2001) Mol. Cell. Biol. Res. Commun. 4, 337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Rourke, S. M. & Herskowitz, I. (1998) Genes Dev. 12, 2874–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]