Abstract
Cytolytic pore-forming toxins are important for the virulence of many disease-causing bacteria. How target cells molecularly respond to these toxins and whether or not they can mount a defense are poorly understood. By using microarrays, we demonstrate that the nematode Caenorhabditis elegans responds robustly to Cry5B, a member of the pore-forming Crystal toxin family made by Bacillus thuringiensis. This genomic response is distinct from that seen with a different stressor, the heavy metal cadmium. A p38 mitogen-activated protein kinase (MAPK) kinase and a c-Jun N-terminal-like MAPK are both transcriptionally up-regulated by Cry5B. Moreover, both MAPK pathways are functionally important because elimination of either leads to animals that are (i) hypersensitive to a low, chronic dose of toxin and (ii) hypersensitive to a high, brief dose of toxin such that the animal might naturally encounter in the wild. These results extend to mammalian cells because inhibition of p38 results in the hypersensitivity of baby hamster kidney cells to aerolysin, a pore-forming toxin that targets humans. Furthermore, we identify two downstream transcriptional targets of the p38 MAPK pathway, ttm-1 and ttm-2, that are required for defense against Cry5B. Our data demonstrate that cells defend against pore-forming toxins by means of conserved MAPK pathways.
Cytolytic pore-forming toxins (PFTs) comprise ≈25% of all known bacterial protein toxins and act by forming pores at the plasma membrane of target cells (reviewed in refs. 1–4). Prominent examples of PFTs that attack human cells include cholesterol-dependent cytolysins (e.g., Streptococcus pyogenes streptolysin O), repeats in toxin cytolysins (e.g., Escherichia coli α-hemolysin), Aeromonas hydrophila aerolysin, Staphylococcus aureus α-toxin, and Vibrio cholerae hemolysin. The pores formed by these toxins can vary in effective diameter from 1–2 nm [e.g., α-toxin, hemolysin, aerolysin, and Bacillus thuringiensis (Bt) Crystal (Cry) toxins (see below)] to 25–30 nm (e.g., streptolysin O).
Three-domain Cry toxins made by Bt are also PFTs, but these target insects and nematodes (5). Mammalian cells are not affected by these toxins because they lack cellular receptors that allow binding (6). Over 100 phylogenetically related three-domain Cry toxins are known (5, 7). X-ray crystallographic and structure-function data indicate that, for Cry toxins, pore-formation is associated with the N-terminal domain (domain I) that is comprised of seven conserved α-helices, two of which, α4 and α5, are likely to form the pore (7).
Although PFTs are important to the virulence of many pathogenic bacteria, there is little understanding, apart from physiological data, as to how cells respond to these toxins and whether they mount a defense. The role of PFTs is unlikely to be rapid cell lysis, because most mammalian cells do not rapidly swell and lyse when treated with low (physiological) concentrations of PFTs but rather stay viable for many hours (the toxins are, however, cytolytic at higher concentrations) (1). Interestingly, different cell types treated with different PFTs can display similar physiological responses, e.g., increases in cytosolic Ca2+ and vacuolization, implying some conserved and possibly concerted responses (1–4). However, the functional significance of these responses is uncertain; they could promote pathogenesis or cellular defenses or promote both or neither. Responses to PFTs at the molecular level, e.g., activation of targets through signal transduction cascades, and the roles of these responses in coping with PFTs also are poorly understood.
Here, we use microarrays to characterize the genomic response of Caenorhabditis elegans to Cry5B toxin, a member of the three-domain α-helical pore-forming Cry toxin family. We show that two mitogen-activated protein kinase (MAPK) pathways [p38 and c-Jun N-terminal kinase (JNK)-like] are transcriptionally up-regulated by the toxin, that both of these MAPK pathways provide a significant cellular defense against the toxin, and that this defense is conserved in mammalian cells attacked by a PFT. We use this system to further identify two downstream transcriptional targets of the p38 MAPK pathway that help mediate the defense against PFTs.
Materials and Methods
C. elegans Maintenance. C. elegans N2 Bristol was maintained by using standard techniques (8). The following strains were used in this study: LGI, glp-4(bn2); LGII, rrf-3(pk1426); LGIV, kgb-1(um3) and jnk-1(gk7); and LGX, sek-1(km4). All C. elegans assays were carried out at 20°C. Unless otherwise noted, standard NG plates were used. Images of C. elegans were captured on an Olympus BX-60 microscope by using a ×10/0.25 numerical aperture objective and a DVC camera with a ×0.5 camera mount and 81.4-msec exposure times.
Cry5B and Cadmium (Cd) Plate Assays. E. coli strain JM103 carrying empty vector pQE9 or inducible Cry5B and Cry21A in pQE9 are described in ref. 9. Toxin-expressing plates were prepared as follows. A saturated overnight culture was diluted 1:10 in LB containing 50 μg/ml carbenicillin, grown for 1 h at 37°C, and induced with 50 μM isopropyl β-d-thiogalactoside for an additional 3 h at 30°C. For 100% Cry5B plates, 30 μl of Cry5B bacteria was spread on 60-mm C. elegans high-growth plates with 100 μM isopropyl β-d-thiogalactoside and 50 μg/ml carbenicillin (when 100-mm plates were used, 100 μl of bacteria was spread). For 10% Cry5B plates, one part Cry5B-expressing bacteria was diluted with nine parts empty vector containing bacteria before plating (at the same OD600). Cry21A plates were similarly prepared. The lawns were grown overnight at 25°C, and the plates were used within 1–2 days. For Cd experiments, CdCl2 from a 0.5 M stock was added to ENG media before pouring. The plates were spread with 30 μl of E. coli OP50, incubated at 25°C overnight, and used the next day. For each experiment, >10 synchronized L4-staged animals were added to each lawn. Each experiment was independently replicated at least three times.
Quantitative Growth Assays with Cry5B and Cd. Assays were carried out as described in ref. 10, except that the toxin source was purified Cry5B (11) or CdCl2, OP50 was added at an optical density of 0.2–0.25 OD600, and 30–40 L1 larvae were used per well.
Preparation of RNA for Microarrays. Synchronous populations of glp-4(bn2) animals were grown to L4 stage on high-growth plates seeded with OP50, removed in sterile water, washed once, and then split into two populations: Half were seeded onto plates spread with JM103 E. coli expressing Cry5B, and half were seeded onto plates spread with JM103 containing empty vector alone. For Cd assays, half the population was plated on 1 mM Cd plates (with OP50), and the other half was plated on identical plates lacking the CdCl2. After 3 h, the animals were washed off the assay plates with water, pelleted, washed with 5 ml of water, and pelleted again. Total RNA was extracted and isolated and further purified by using Qiagen (Valencia, CA) RNeasy columns. Microarray experiments and real-time PCR methodologies are discussed in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
RNA Interference (RNAi). RNAi feeding was done by using rrf-3(pk1426) (12). This strain shows normal sensitivity to Cry5B (Fig. 7, which is published as supporting information on the PNAS web site). The feeding protocol was adapted from ref. 13 with the following modifications. L4 hermaphrodites were pipetted onto the RNAi feeding plates (0.1 mM isopropyl β-d-thiogalactoside/25 μg/ml carbenicillin), allowed to feed for 30–32 h, moved to new plates, allowed to lay eggs for 2–4 h, and removed. Phenotypes were assessed in these progeny by using plate assays described above. ttm-1 and ttm-2 RNAi feeding clones were obtained from the Ahringer library (13). The feeding clone for pmk-1 was made by amplifying cDNA corresponding to nucleotides 2,435–4,302 of cosmid B0218 and subcloning them into the L4440 feeding vector, followed by transformation into E. coli HT115.
Aerolysin Experiments. Baby hamster kidney cells were cultured as described in ref. 14, grown to confluence, and incubated or not with 10 μM SB203580 for 30 min in culture medium in the CO2 incubator. The cells were then treated for either 45 sec or 3 min with proaerolysin at different concentrations, rinsed, further incubated for 6 or 48 h in the incubator, and finally incubated with 2 μg/ml propidium iodide. Cells were either visualized by fluorescence microscopy or trypsinized, pelleted, and resuspended in PBS/1% FCS for fluorescence-activated cell sorter analysis.
Results
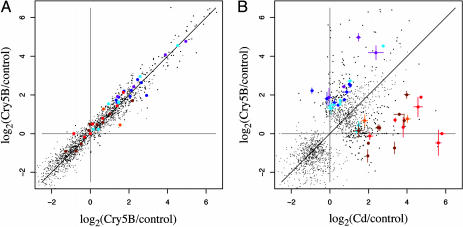
Genomic Response of C. elegans to Bt Toxin, Cry5B. To study the global effects of a PFT in the context of an intact animal, we used commercially available C. elegans (Affymetrix, Santa Clara, CA) complete transcriptome microarrays. The presence and abundance of C. elegans transcripts were determined by using RNA isolated from animals fed for 3 h on E. coli lawns expressing Cry5B and RNA isolated from animals fed the same E. coli strain transformed with empty vector alone (no toxin control). Three hours was chosen as an early time point by which intoxication of the nematodes is evident based on their behavior but by which visible intestinal damage at the compound microscope level is not. Data from three independent trials from each of Cry5B and vector only conditions were combined and analyzed (see supporting information). The experiment was conducted by using glp-4(bn2) animals that lack a germ line but that otherwise show a normal response to Cry5B (see supporting information) because the intestine, the target tissue of Cry5B, will comprise a large portion of these animals. Just over 1,000 genes are reproducibly up- and down-regulated in response to Cry5B (Fig. 1A and Table 2, which is published as supporting information on the PNAS web site).
Fig. 1.
Behavior of 1,670 probe sets significantly induced by one or both of Cry5B and Cd treatment. A few strongly down-regulated genes were omitted to improve plot resolution for up-regulated genes. (A) The fold induction of the probe sets for two of the independent Cry5B trials plotted against each other. The plot demonstrates excellent correlation between the two trials. (B) The fold induction of the probe sets for Cd trials (average of three trials) and for Cry5B (averaged over three trials) plotted against each other. Although the fold induction of some probe sets is similar, other probe sets (located off the diagonal) show significant differences, indicative of different responses to Cry5B and Cd. Some genes that show >2-fold difference between the two conditions were grouped based on their ontology and are shown in color. A few genes preferentially induced by Cd are glutathione S-transferase genes (brown; ref. 9), heat-shock genes (red; ref. 7), and known stress-induced genes (orange; ref. 2). A few genes preferentially induced by Cry5B include pathogen or pathogen-associated induced genes (purple; ref. 4) and cytoskeletal and cell adhesion genes (dark blue; ref. 9). Shown in light blue are transcription factor genes; two are preferentially induced by Cd, and six others are preferentially induced by Cry5B.
To determine how specific this response was, a similar experiment was performed in which glp-4(bn2) animals were treated with the heavy metal Cd by using a dose that kills C. elegans at a rate comparable to Cry5B (data not shown). The RNA isolated from three independent experiments for each of experimental (Cd) and control (no Cd) conditions was used to generate cRNA targets for Affymetrix microarrays and analyzed as for Cry5B. Although an equally large number of genes were regulated by the heavy metal (Table 3, which is published as supporting information on the PNAS web site), the responses to Cry5B and Cd have significant differences (Fig. 1B). One caveat of these comparisons is that the nonpathogenic E. coli strains used for the two experiments are different and the media for the Cry5B experiment contained antibiotics and isopropyl β-d-thiogalactoside (see Materials and Methods). However, in each case, the nontoxin control is appropriately matched. Although it is possible that these alterations influence some genes, we believe that they are not substantially influencing the overall differences in the responses (e.g., that general stress-related genes are markedly more induced by Cd and that pathogenesis/cytoskeletal/transcription factor genes are markedly more induced by Cry5B) (Fig. 1B).
Two MAPK Pathways Protect Animals Against Cry5B Toxin. The list of Cry5B-induced genes included several that mediate signal transduction, including the gene R03G5.2 or sek-1. SEK-1 is a MAPK kinase (MAPKK) that is immediately upstream of the C. elegans p38 MAPK, PMK-1, and immediately downstream of the MAPKK kinase, NSY-1 (15, 16). Because the p38 pathway is important for innate immunity in eukaryotes, including C. elegans (see Discussion), the sek-1 gene was of immediate interest. Real-time PCR analysis confirmed that sek-1 RNA is induced by Cry5B (Table 1).
Table 1. Regulation of toxin-induced genes in wild type and sek-1 (km4).
| Fold toxin induced
|
|||
|---|---|---|---|
| Transcript | sek-1 genotype* | Affy† | RT-PCR |
| sek-1 | + | 3.7 | 4.8 |
| - | 1.1 | 1.0 | |
| kgb-1 | + | 2.6 | 3.6 |
| - | 2.4 | 2.9 | |
| ttm-1 | + | 4.7 | 5.3 |
| - | 1.1 | 1.3 | |
| ttm-2 | + | 3.5 | 3.5 |
| - | 1.4 | 1.8 | |
+, glp-4(bn2); -, glp-4(bn2);sek-1(km4).
Average induction seen on microarray experiments.
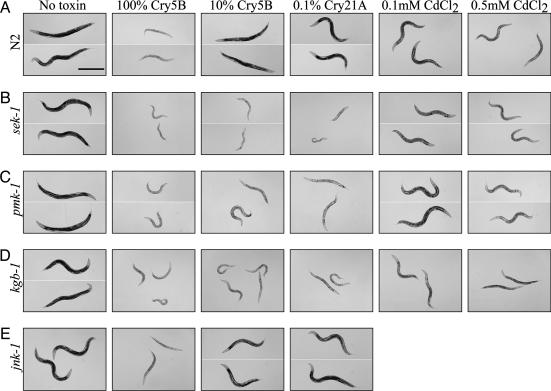
To test whether this induced MAPKK gene, sek-1, is functionally relevant, we fed animals lacking the sek-1 gene [deletion allele sek-1(km4)] high and low doses of Cry5B toxin expressed in E. coli. In the absence of toxin, sek-1(km4) animals develop normally and generally are as healthy as wild-type animals, as previously reported (16) (Fig. 2 A and B, no toxin). When fed high doses of Cry5B, both wild-type and sek-1(km4) animals become intoxicated; they rapidly become lethargic, turn pale, and degenerate over the course of 1–2 days (Fig. 2 A and B), although sek-1(km4) animals appear to become more intoxicated. When fed lower doses of Cry5B, wild-type animals are much healthier; they are large, have good coloration, move well, and do not degenerate (Fig. 2 A). In contrast, sek-1(km4) animals are still severely intoxicated even by this low dose of toxin (Fig. 2B). This result extends to the other genes in the same MAPK pathway. Animals lacking the p38 MAPK in this pathway (pmk-1) also are hypersensitive to Cry5B (Fig. 2C) as are animals lacking the upstream MAPKK kinase nsy-1 (not shown). The protection conferred by this p38 pathway against Cry5B is specific in at least two ways. First, animals lacking another p38 MAPK, pmk-3, are not hypersensitive to Cry5B (data not shown). Second, animals mutant for genes in the sek-1–pmk-1 pathway are not overly hypersensitive to the heavy metal Cd in this assay (Fig. 2 B and C). Because animals lacking sek-1 are more readily intoxicated by Cry5B, we conclude that one wild-type function of sek-1 is to protect C. elegans against this toxin.
Fig. 2.
sek-1–pmk-1 (p38) and kgb-1 (JNK-like) MAPK pathways protect C. elegans from Cry5B. As indicated above each column, L4 animals were plated on the lawns of E. coli containing empty vector (no toxin), high levels of Cry5B toxin (100% Cry5B), low levels of Cry5B toxin (10% Cry5B), low levels of Cry21A toxin (0.1% Cry21A), moderate levels of CdCl2 (0.5 mM CdCl2), and low levels of Cd (0.1 mM CdCl2). Representative animals are shown after 40 h (for no toxin and Bt toxins) or 48 h (for Cd) of feeding at each condition. Each row corresponds to a different genotype. (A) N2 (wild type). (B) sek-1(km4) deletion allele. (C) pmk-1 RNAi (by means of feeding on double-stranded pmk-1 RNA). (D) kgb-1(um3) deletion allele. (E) jnk-1(gk7) deletion allele. All nematodes are shown at the same magnification. (Scale bars here and in other figures, 0.5 mm.)
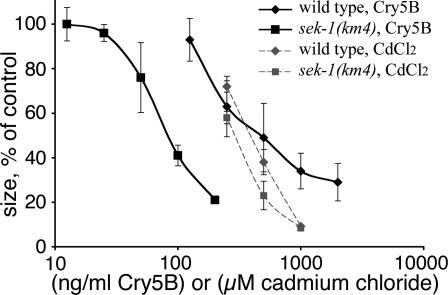
Quantitatively, sek-1(km4) animals are an order of magnitude more sensitive to Cry5B than wild-type animals (Fig. 3). In contrast, sek-1(km4) animals display only slightly increased sensitivity to Cd in this assay (<2-fold; Fig. 3). Hypersensitivity of p38 pathway mutants is not restricted to one Cry toxin. Animals lacking the sek-1 MAPKK or the p38 MAPK pmk-1 also are hypersensitive to nematicidal Cry21A toxin (Fig. 2 A–C). Furthermore, animals lacking sek-1 are hypersensitive to Cry5B when the Gram-positive bacterium Bacillus subtilis is used as the food source instead of E. coli (data not shown), demonstrating that hypersensitivity is associated with the toxin and not the bacterium used.
Fig. 3.
Quantitative growth-based toxicity assays by using wild-type and sek-1(km4) mutant animals at various doses of Cry5B and Cd (plotted on a logarithmic scale). L1-staged animals were exposed to indicated doses of stressors in microtiter wells, and after 60 h, their cross-sectional areas were measured. The ordinate axis shows the size of nematodes relative to no-toxin controls run in parallel. Each point represents ≈60 animals from three independent experiments. SE bars are indicated.
Two JNK-family MAPK genes, kgb-1 and kgb-2, are also induced by Cry5B on the microarrays. Animals mutant for kgb-1 [deletion allele kgb-1(um3)] are healthy in the absence of toxin at the temperature assayed (20°C; Fig. 2D). However, like sek-1 mutant animals, we find that kgb-1(um3) animals are hypersensitive to low doses of Cry5B and Cry21A (Fig. 2D). As with sek-1, kgb-1(um3) animals are still hypersensitive to the toxin when B. subtilis is used as the food source (data not shown). kgb-1 mutant animals also appear hypersensitive to low doses of Cd (Fig. 2D), as has been reported (17), suggesting that this pathway has broader functions than the p38 pathway in controlling stress responses. No mutant allele for kgb-2 was available, and RNAi of kgb-2 did not give rise to animals hypersensitive to Cry5B. We have confirmed that not all JNK-like MAPKs are required for protection because animals that lack a different JNK-like MAPK, jnk-1, are not hypersensitive to Cry5B (Fig. 2E).
MAPK Pathways Protect Against a Short but High Toxin Pulse. Both Bt and many free-living nematodes coexist in the soil. Because these nematodes ingest bacteria as a food source and because Bt has evolved Cry toxins that kill diverse nematodes (9), it seems likely that nematodes and Bt encounter each other in the ecosystem. We speculate that, in the soil, nematodes might transiently come across Bt and its associated crystals, ingest some, and then move away. We therefore hypothesized that a physiologically relevant form of defense for soil nematodes against Cry toxins might involve survival to a short, high dose of ingested toxin.
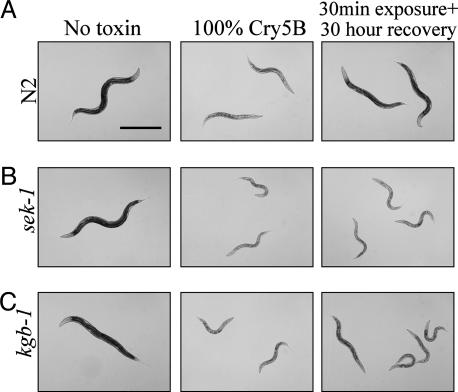
To test this hypothesis, animals were fed 100% Cry5B-expressing E. coli for 30 min and then transferred to plates containing nontoxic E. coli. Wild-type animals tolerate this pulse of toxin well and 1 day later are healthy (Fig. 4A). In contrast, sek-1 or kgb-1 mutant animals cannot tolerate this short exposure to toxin (Fig. 4 B and C). These animals degenerate even in the absence of toxin and are intoxicated 30 h later. These results, clear for both mutants, are exceptionally dramatic for sek-1 because sek-1(km4) mutants fed a pulse of toxin are as intoxicated as animals that fed on toxin continually. Similar results were obtained for pmk-1 RNAi and nsy-1 mutants (data not shown).
Fig. 4.
MAPK mutant animals are hypersensitive to a short pulse of toxin. Column 1 shows no toxin controls. Column 2 shows L4 animals that were placed on 100% Cry5B-expressing E. coli lawns and imaged 30 h later. Column 3 shows L4 animals that were placed on 100% Cry5B-expressing E. coli lawns for 30 min, removed to nontoxin-expressing E. coli lawns, and imaged 30 h later. Each row shows representative animals for each genotype at the same magnification. (A) Wild type (N2). (B) sek-1(km4) deletion allele. (C) kgb-1(um3) deletion allele.
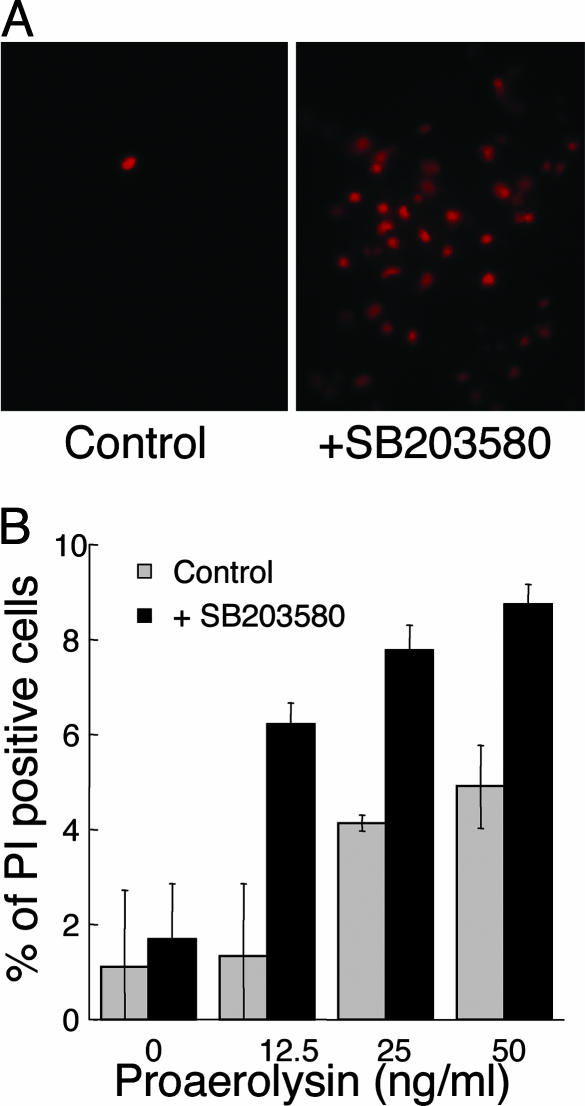
p38 MAPK Pathways Protect Mammalian Cells Against a Bacterial PFT. Because MAPK pathways are well conserved between C. elegans and mammals, we hypothesized that MAPK pathways might protect mammalian cells against PFTs as well. To test this hypothesis, baby hamster kidney cells were pretreated with the p38-specific inhibitor SB203580 for 30 min and then treated with a short pulse (either 45 sec or 3 min) of the PFT proaerolysin made by the human pathogen A. hydrophila. In agreement with our results in C. elegans, inhibition of p38 in mammalian cells results in hypersensitivity to the PFT proaerolysin (Fig. 5A). At all concentrations of proaerolysin, a 2- to 5-fold increase in the number of cells undergoing cell death is evident when p38 is inhibited (Fig. 5B).
Fig. 5.
A p38 MAPK pathway protects mammalian cells against the PFT aerolysin. (A) Baby hamster kidney cells were incubated in either media (Left) or media plus p38 inhibitor SB203580 (Right) for 30 min and then incubated for 45 sec with 20 ng/ml proaerolysin. After being washed, cells were cultured for 28 h and stained with propidium iodine (PI, a marker for cell death). An increased number of dying cells is evident in the presence of the p38 inhibitor (both panels contain similar numbers of cells in a confluent lawn not shown). (B) Quantitation of cell death. Baby hamster kidney cells were prepared as above without or with SB203580, treated with proaerolysin for 3 min, cultured for 6 h, and stained with propidium iodine. The percentage of propidium iodine-positive cells is shown. Shown is the average number of propidium iodine-positive cells for three independent trials with SE bars.
Identification of Downstream Targets of the p38 Protection Pathway. An important effect of MAPK pathways is transcriptional activation of downstream target genes. The robust genomic response seen in nematodes to Cry5B suggested that we might be able to identify functional downstream targets of the MAPK pathway by using microarrays. We therefore compared the genomic responses of C. elegans to Cry5B with and without the p38 MAPK pathway intact to identify genes whose up-regulation depends on the pathway. Animals from two strains, glp-4(bn2);sek-1(+) and glp-4(bn2);sek-1(km4), were fed Cry5B-expressing or non-Cry5B-expressing E. coli and their RNA was harvested after 3 h, processed, and hybridized to Affymetrix arrays. By comparing these responses, we identified >100 potential target genes whose up-regulation by Cry5B on microarrays shows statistical dependence on the presence of a functional p38 pathway (data not shown). We screened through many of these genes by RNAi to identify any that mutate to a toxin-hypersensitive phenotype and identified at least two of these that do: Y39E4A.2 and F26G1.4. As confirmed by real-time PCR, Y39E4A.2 and F26G1.4 (called ttm-1 and ttm-2, respectively; see below) are both induced by toxin and require p38 MAPKK for full induction (Table 1). Reducing the products of either Y39E4A.2 or F26G1.4 by RNAi gives rise to animals that are healthy in the absence of toxin but hypersensitive to low, chronic doses of Cry5B (Fig. 6). Hypersensitivity to Cry5B seen by reduction of Y39E4A.2 and F26G1.4 is robust but not as severe as with loss of sek-1. Interestingly, RNAi of Y39E4A.2 leads to hypersensitivity to Cd as well (Fig. 6B; see Discussion). We call these functionally protective genes, respectively, ttm-1 and ttm-2 for toxin-regulated targets of MAPK, in recognition that their response to toxin depends on an intact MAPK pathway.
Fig. 6.
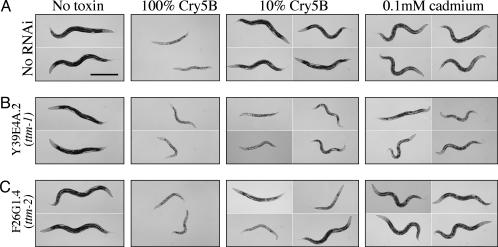
ttm-1 and ttm-2 genes are required for defense against Cry5B. rrf-3(pk1426) L4 animals were fed double-stranded RNA-producing bacteria containing empty vector (no RNAi control) (A), a Y39E4A.2 insert (ttm-1)(B), or a F26G1.4 insert (ttm-2)(C). Nematodes were then transferred to E. coli plates expressing no toxin, 100% Cry5B, 10% Cry5B, or 0.1 mM CdCl2. Representative nematodes are shown after 40 h (control, Cry5B) or 48 h (Cd). RNAi of ttm-1 leads to high penetrance of animals hypersensitive to toxin (B, 10% Cry5B column). RNAi of ttm-2 leads to animals reproducibly but variably hypersensitive to toxin (C, 10% Cry5B column).
Discussion
Our results demonstrate that PFTs elicit a robust transcriptional response from target cells and that at least part of this response is functionally important for defense. After 3 h of exposure to Cry5B (a member of the three-domain Bt Cry toxin family) 1,003 C. elegans genes are significantly induced or repressed relative to controls. Two of the induced genes encode different MAPK pathway components: sek-1, a p38 MAPKK, and kgb-1, a JNK-like MAPK. The transcriptional response of C. elegans to Cry5B can be qualitatively discriminated from the response seen to the heavy metal Cd, which elicits a generalized stress response as indicated by the preferential induction of heat-shock proteins and glutathione S-transferase genes. Conversely, four genes induced by pathogens or pathogen-associated factors in other systems are preferentially up-regulated by Cry5B, consistent with the idea that C. elegans identifies Cry5B as part of a pathogenic attack. We note that although pore-forming activity for Cry5B has not yet been demonstrated, all members of the three-domain Cry toxin family are generally accepted to be PFTs (5), because, for example, the domain I pore-forming α-helices are well conserved in these toxins, including in Cry5B.
Loss of either sek-1 MAPKK or the downstream p38 MAPK gene results in C. elegans that are hypersensitive to low, chronic doses of Cry5B or to high, short doses of toxin. Dramatically, we are able extend these results to mammals by demonstrating that inhibition of the p38 pathway in hamster cells results in hypersensitivity to a pulse of aerolysin, a PFT associated with the human pathogen A. hydrophila.
These results thus demonstrate for the first time, to our knowledge, that (i) animal cells have a defense against PFTs, and (ii) the defense depends on the conserved p38 pathway. The p38 pathway plays an important role in mammalian innate immunity (18). In C. elegans, this pathway provides an innate immune defense against Pseudomonas aeruginosa, Salmonella enterica, and S. aureus because C. elegans mutants lacking an intact p38 pathway are killed more readily by these pathogens (16, 19, 20). The current study differs from these by focusing on one bacterial toxin rather than an entire bacterium. By showing that inhibition of p38 leads to hypersensitivity of C. elegans to Bt Cry toxins and hypersensitivity of mammalian cells to aerolysin, we demonstrate that p38-mediated defensive mechanisms exist in these organisms to help counter the effects of a single virulence factor, a PFT. Hence, we discovered a role for the p38 pathway in the protection against PFTs.
We have found that a second MAPK pathway also is required for protection against Cry toxins because loss of the JNK-like MAPK kgb-1 leads to hypersensitivity to Cry5B and Cry21A toxins. Like p38 MAPKs, JNK MAPKs are important for innate immune responses in mammals and, more recently, have been shown to be important for innate immune defense of C. elegans against P. aeruginosa infection (18, 21). Why two MAPK pathways, p38 and JNK, are both required for protection against Cry toxins is not clear. Perhaps the function of one pathway is to activate the other, although we find no evidence that sek-1 and kgb-1 regulate each other transcriptionally (Table 1 and data not shown). Posttranslational regulation of one pathway by the other also is possible. Alternatively, it is possible that the two pathways act independently to promote protection.
Our data suggest that protection provided by the two MAPK pathways has real consequences for survival of the nematode in the wild. Wild-type C. elegans exposed briefly to Bt toxin in the soil are likely to crawl away, recuperate, and give rise to progeny, whereas animals lacking sek-1 or kgb-1 exposed briefly to Bt toxin might never recover. Thus, these pathways may have evolved to allow the species to propagate under the adverse conditions in the natural environment.
A key and often challenging step in further understanding MAPK pathways is the identification of downstream targets that are activated by the MAPK and that perform the actual protective function. Analysis of microarrays by using a sek-1 mutant followed by RNAi have led to the identification of two targets of the MAPK pathway, ttm-1 and ttm-2. Both targets are transcriptionally induced by Cry5B toxin, both require the sek-1 MAPK pathway for their full induction, and both are required for protection of the animal against the toxin. Although protein data are forthcoming, i.e., we cannot be sure that protein levels are also induced, these data suggest that activation of MAPK pathway leads directly or indirectly to increased production of these proteins that in turn leads to protection and defense against the toxin. That RNAi of either ttm-1 or ttm-2 leads to less hypersensitivity than that caused by loss of sek-1 could mean (i) that elimination of ttm-1 and ttm-2 by RNAi is incomplete and/or (ii) that the MAPK pathway also activates other genes in response to Cry5B, each of which provides incremental protection against the toxin.
The identity of one of these genes is particularly revealing; the protein encoded by ttm-1 shows significant amino acid identity to cation efflux channels and is conserved in mammals [e.g., 34% amino acid identity to the human zinc transporter, ZnT-3 (22)]. We speculate that one consequence of pore formation might be an increase in cytosolic levels of cytotoxic cations that could be alleviated by up-regulation of an efflux transporter. If such a model were correct, ttm-1 might also play a role in protection against Cd (e.g., by pumping it out of the cell). Indeed, examination of our microarray data indicates that ttm-1 is induced 2.6-fold upon exposure of nematodes to Cd. Furthermore, RNAi of ttm-1 leads to Cd hypersensitivity. These data are consistent with ttm-1 playing a role in removing cytotoxic cations from the cytosol, either when coming from high levels in the diet or when coming into the cell by means of the action of PFTs.
Our microarray and mutational data together indicate that animal cells have a functionally sophisticated response to bacterial PFTs. That the results with Cry5B and C. elegans involve conserved innate immune pathways and extend to aerolysin interactions with mammalian cells suggest that what is learned from studying Cry toxins and C. elegans will continue to yield important insights into how PFTs interact with their targets and how target cells protect themselves against the toxins.
Supplementary Material
Acknowledgments
We thank Drs. Karen Bennett and Fred Ausubel for providing C. elegans mutants; Wayne Hsu, Christine Plotkin, Steffney Rought, and Pinyi Du for technical assistance; and Dr. Larry Bischof for critical reading of the manuscript. Strains also were provided by the C. elegans Genetics Center (funded by the National Institutes of Health National Center for Research Resources). The microarray work was done at the University of California at San Diego Genomics Core Laboratory. This work was supported by National Science Foundation Grant MCB-9983013 and grants from the Burroughs–Wellcome Foundation and the Beckman Foundation (to R.V.A.). J.C. is a holder of a Canada Research Chair.
Abbreviations: MAPK, mitogen-activated protein kinase; MAPKK, MAPK kinase; PFT, pore-forming toxin; Cry, Crystal; Bt, Bacillus thuringiensis; JNK, c-Jun N-terminal kinase; RNAi, RNA interference.
References
- 1.van der Goot, F. G. (2003) in Bacterial Protein Toxins, eds. Burns, D. L., Barbieri, J. T., Iglewski, B. H. & Rappuoli, R. (Am. Soc. Microbiol., Washington, DC), pp. 189–202.
- 2.Alouf, J. E. (2003) Folia Microbiol. (Praha) 48, 5–16. [DOI] [PubMed] [Google Scholar]
- 3.Alouf, J. E. (2001) Curr. Top. Microbiol. Immunol. 257, 1–14. [PubMed] [Google Scholar]
- 4.Huffman, D. L., Bischof, L. J., Griffitts, J. S. & Aroian, R. V. (2004) Int. J. Med. Microbiol. 293, 599–607. [DOI] [PubMed] [Google Scholar]
- 5.de Maagd, R. A., Bravo, A., Berry, C., Crickmore, N. & Schnepf, H. E. (2003) Annu. Rev. Genet. 37, 409–433. [DOI] [PubMed] [Google Scholar]
- 6.Betz, F. S., Hammond, B. G. & Fuchs, R. L. (2000) Regul. Toxicol. Pharmacol. 32, 156–173. [DOI] [PubMed] [Google Scholar]
- 7.de Maagd, R. A., Bravo, A. & Crickmore, N. (2001) Trends Genet. 17, 193–199. [DOI] [PubMed] [Google Scholar]
- 8.Brenner, S. (1974) Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei, J. Z., Hale, K., Carta, L., Platzer, E., Wong, C., Fang, S. C. & Aroian, R. V. (2003) Proc. Natl. Acad. Sci. USA 100, 2760–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffitts, J. S., Huffman, D. L., Whitacre, J. L., Barrows, B. B., Marroquin, L. D., Muller, R., Brown, J. R., Hennet, T., Esko, J. D. & Aroian, R. V. (2003) J. Biol. Chem. 278, 45594–45602. [DOI] [PubMed] [Google Scholar]
- 11.Griffitts, J. S., Whitacre, J. L., Stevens, D. E. & Aroian, R. V. (2001) Science 293, 860–864. [DOI] [PubMed] [Google Scholar]
- 12.Simmer, F., Tijsterman, M., Parrish, S., Koushika, S. P., Nonet, M. L., Fire, A., Ahringer, J. & Plasterk, R. H. (2002) Curr. Biol. 12, 1317–1319. [DOI] [PubMed] [Google Scholar]
- 13.Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003) Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 14.Abrami, L., Fivaz, M., Glauser, P. E., Parton, R. G. & van der Goot, F. G. (1998) J. Cell Biol. 140, 525–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka-Hino, M., Sagasti, A., Hisamoto, N., Kawasaki, M., Nakano, S., Ninomiya-Tsuji, J., Bargmann, C. I. & Matsumoto, K. (2002) EMBO Rep. 3, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, D. H., Feinbaum, R., Alloing, G., Emerson, F. E., Garsin, D. A., Inoue, H., Tanaka-Hino, M., Hisamoto, N., Matsumoto, K., Tan, M. W. & Ausubel, F. M. (2002) Science 297, 623–626. [DOI] [PubMed] [Google Scholar]
- 17.Mizuno, T., Hisamoto, N., Terada, T., Kondo, T., Adachi, M., Nishida, E., Kim, D. H., Ausubel, F. M. & Matsumoto, K. (2004) EMBO J. 23, 2226–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong, C., Davis, R. J. & Flavell, R. A. (2002) Annu. Rev. Immunol. 20, 55–72. [DOI] [PubMed] [Google Scholar]
- 19.Aballay, A., Drenkard, E., Hilbun, L. R. & Ausubel, F. M. (2003) Curr. Biol. 13, 47–52. [DOI] [PubMed] [Google Scholar]
- 20.Sifri, C. D., Begun, J., Ausubel, F. M. & Calderwood, S. B. (2003) Infect. Immun. 71, 2208–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, D. H., Liberati, N. T., Mizuno, T., Inoue, H., Hisamoto, N., Matsumoto, K. & Ausubel, F. M. (2004) Proc. Natl. Acad. Sci. USA 101, 10990–10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmiter, R. D., Cole, T. B., Quaife, C. J. & Findley, S. D. (1996) Proc. Natl. Acad. Sci. USA 93, 14934–14939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.