Lysosomes are the main degradative compartment in cells and require an acidic luminal environment for correct function. Rahman et al. show that soluble adenylyl cyclase is required for localization of the V-ATPase proton pump to lysosomes and therefore lysosomal acidification and function.
Abstract
Lysosomes, the degradative organelles of the endocytic and autophagic pathways, function at an acidic pH. Lysosomes are acidified by the proton-pumping vacuolar ATPase (V-ATPase), but the molecular processes that set the organelle’s pH are not completely understood. In particular, pH-sensitive signaling enzymes that can regulate lysosomal acidification in steady-state physiological conditions have yet to be identified. Soluble adenylyl cyclase (sAC) is a widely expressed source of cAMP that serves as a physiological pH sensor in cells. For example, in proton-secreting epithelial cells, sAC is responsible for pH-dependent translocation of V-ATPase to the luminal surface. Here we show genetically and pharmacologically that sAC is also essential for lysosomal acidification. In the absence of sAC, V-ATPase does not properly localize to lysosomes, lysosomes fail to fully acidify, lysosomal degradative capacity is diminished, and autophagolysosomes accumulate.
INTRODUCTION
Lysosomes degrade extracellular material and intracellular components during endocytosis and autophagy, respectively. The hydrolases, which degrade the proteins, lipids, and polysaccharides, are optimally active in an acidic environment; thus, the pH of the lysosome lumen is maintained around 5 (Pillay et al., 2002). Acidification is mediated by the vacuolar ATPase (V-ATPase), which pumps protons into the lysosomal lumen in an ATP-dependent manner (Ohkuma et al., 1982; Breton and Brown, 2013). This transport generates a charge imbalance, which limits further proton translocation and lysosomal acidification. Several counter ion transport pathways have been identified that allow the lysosomes to dissipate this membrane potential (Pillay et al., 2002; Steinberg et al., 2010; Mindell, 2012; Xu and Ren, 2015). Although these functions provide a mechanism for acidification, they do not explain how lysosomes reach their appropriate pH; i.e., there remains a need for pH “sensors” to set lysosomal pH. A recently identified two-pore ion channel (TPC1) is a pH-sensitive sodium channel (Cang et al., 2014) essential for acidification during starvation-induced autophagy (Cang et al., 2013). However, TPC1’s role in acidification appears to be limited to starvation conditions; TPC1 knockout (KO) cells grown in the presence of sufficient nutrients have normal lysosomal pH (Cang et al., 2013). Therefore, there must be other pH-sensitive principles that regulate acidification during endosome–lysosome biogenesis.
Signaling cascades able to regulate acidification of lysosomes include the ubiquitous second messenger cAMP. In pathophysiological situations where lysosomes are insufficiently acidified, pharmacologically increasing intracellular cAMP acidifies them. For example, in retinal pigment epithelial cells (Liu et al., 2008) and fibroblasts (Coffey et al., 2014), mutations that cause lysosomal pH to be relatively alkaline can be rescued by exogenous addition of membrane-permeable cAMP. cAMP has also been proposed to mediate physiological acidification; the mean pH of lysosomes in resting microglia is ∼6, and signals that elevate intracellular cAMP acidify them (Majumdar et al., 2007). This cAMP-dependent acidification in microglia is mediated via PKA (Majumdar et al., 2007).
In mammalian cells, two distinct classes of adenylyl cyclase generate cAMP. In addition to the family of widely studied, G protein–regulated, hormonally responsive, transmembrane adenylyl cyclases (tmACs), there exists a molecularly and biochemically distinct soluble adenylyl cyclase (sAC; ADCY10). sAC is widely expressed, and unlike tmACs, it is directly regulated by bicarbonate (HCO3−) anions (Chen et al., 2000b; Kleinboelting et al., 2014). Because of ubiquitously expressed carbonic anhydrases (CAs), HCO3− is in rapid equilibrium with carbon dioxide (CO2) and protons (pH). Both proton movement through the cytoplasm (Stewart et al., 1999; Spitzer et al., 2002) and pH gradients in migrating cells (Martin et al., 2011; Tarbashevich et al., 2015) are dependent on CA activity; therefore, cytoplasmic pH changes, including transient ones, are reflected as local changes in HCO3− concentration. Thus, the CA-catalyzed CO2/HCO3/pH equilibrium allows HCO3−-sensing sAC to regulate biological functions in response to fluctuations in CO2 and/or pH (Tresguerres et al., 2011; Chang and Oude-Elferink, 2014; Levin and Buck, 2015).
We previously confirmed that sAC functions as a pH sensor (Tresguerres et al., 2010a; Levin and Buck, 2015). In pH-sensing epithelial cells, extracellular pH induces translocation of proton-pumping V-ATPase to the luminal surface. This extracellular pH sensing is essential for acid/base regulation and is dependent on intracellular sAC. In epididymis and the collecting duct of the kidney, luminal pH induces translocation of the V-ATPase to the acid-secreting surface (Breton and Brown, 2013). In these cells, sAC is found inside the cell, in a complex with V-ATPase (Păunescu et al., 2008a), and the pH-dependent V-ATPase translocation is mediated via sAC in a CA- and PKA-dependent manner (Pastor-Soler et al., 2003, 2008; Păunescu et al., 2008a, 2010). A similar mechanism involving sAC and V-ATPase is responsible for organismal pH regulation in marine invertebrates (Tresguerres et al., 2010b).
Because sAC is responsible for the pH-induced translocations of V-ATPase, we asked whether sAC also plays a role in the V-ATPase–dependent acidification of lysosomes. Here, we demonstrated that in the absence of sAC, V-ATPase does not properly colocalize with lysosomes, lysosomes do not fully acidify, their degradative activity is diminished, and autophagosomes (APs) accumulate. Thus, pH-sensitive sAC is an essential regulator of lysosomal acidification in mammalian cells.
MATERIALS AND METHODS
Cell lines and mouse
Mouse embryonic fibroblasts (MEFs) were immortalized from WT and sAC KO mice (Esposito et al., 2004; Hess et al., 2005) using the 3T3 method or SV40 large T antigen (Ramos-Espiritu et al., 2016). The cells were grown in DMEM supplemented with 1% penicillin/streptomycin, 10% fetal bovine serum, and 0.5% glutamine, and they were genetically and functionally authenticated in our laboratory. Primary neuronal cultures were prepared from cortices and hippocampi of embryonic day 15 (E15) sAC KO and WT littermate mouse embryos as described in Tampellini et al. (2009). sAC heterozygous male and female along with WT mice (The Jackson Laboratory) were bred to generate the embryos. Primary neurons were genetically authenticated by allele-specific PCR and used at 12 d in vitro (DIV) for all experiments. WT and sAC KO mice were studied at 15–18 mo together with age-matched controls. All cells were maintained at 37°C in 5% CO2 and were periodically checked for mycoplasma contamination. All animal work was performed with approval from the Institutional Animal Care and Use Committee of Weill Cornell Medical College (IACUC) and conforms to National Institutes of Health guidelines for the Care and Use of Laboratory Animals.
Antibodies and reagents
Mouse monoclonal antibodies to LC3 (catalog #0231-100/LC3-5F10; Nanotools) were used for immunoblotting (1:300) experiments. Anti-LAMP antibodies (LAMP-2: catalog #ABL-93, 1/200; LAMP-1: catalog #H4A3 1:200) used for immunofluorescence were purchased from the Developmental Studies Hybridoma Bank. Anti–V-ATPase subunit V1D (D-4; catalog #SC-166218; Santa Cruz Biotechnology, Inc.) was used at 1:200 for immunofluorescence and 1:500 for immunoblotting. Anti–V-ATPase subunit V1B2 (catalog #AB73404; Abcam) was used at 1:200 for immunofluorescence and 1:500 for immunoblotting. LysoTracker red DND-99 (500 nM), Dextran conjugated with Fluorescein and Tetramethylrhodamine, 70,000 mol wt (2.5 mg/ml), BODIPY-FL–Pepstatin A, and DQ-BSA (10 µg/ml) were purchased from Thermo Fisher Scientific. Rabbit polyclonal anti-GAPDH (catalog #14C10; 1:5,000) and anti-Actin (catalog #13E5; 1:5,000) were purchased from Cell Signaling Technology. Ammonium chloride (NH4Cl), bafilomycin A1, KT5720, leupeptin, E64D, Pepstatin A, and MG132 were purchased from Sigma-Aldrich. Sp-8-cpt-cAMP was purchased from Biolog Life Science Institute. We applied secondary antibodies conjugated to Alexa Fluor 488 or 546 (IF 1:1,000). The MTT assay kit was bought from Roche, and the experiment was performed according to the manufacturer’s protocol.
Cellular cAMP accumulation
Cellular cAMP accumulation was determined as performed previously (Bitterman et al., 2013). In brief, 2.5 × 105 WT or sAC KO MEFs were plated in each well of a 24-well plate in DMEM supplemented with 10% fetal bovine serum. The next day, cells were pretreated with 30 µM KH7 or DMSO (vehicle control) for 10 min, followed by the addition of 500 µM IBMX and 50 µM dipyridamole. After 5 min at 37°C, media was aspirated, and the cells were lysed with 200 µl of 0.1 M hydrochloric acid (HCl) per well. Intracellular cAMP content was determined using Correlate-EIA cAMP Direct Assay (Assay Designs, Inc.).
Lysosomal pH assay
The measurement of lysosomal pH by confocal microscopy is based on the use of the ratiometric assay of pH-sensitive fluorescein fluorescence to pH-insensitive rhodamine fluorescence as described previously (Majumdar et al., 2007). In brief, cells were incubated overnight (∼16 h) with 2.5 mg/ml dextran conjugated to both fluorescein and rhodamine (70,000 mol wt; D1951; Invitrogen) in complete growth media, including 25 mM HCO3−, and grown in 5% CO2. Cells were washed four times in complete media and chased for 4 h to allow the dextran to traffic to the lysosomes. The cells were then washed with medium 2 buffer (150 mM NaCl, 20 mM HEPES, pH 7.4, 1 mM CaCl2, 5 mM KCl, and 1 mM MgCl2) and were imaged on a heated stage at 37°C. 15–20 images were collected over a period of 5–10 min for each condition. The ratio of fluorescein to rhodamine fluorescence was determined for each individual lysosome. Images were analyzed using MetaMorph image processing software (Molecular Devices). For all experimental sets, cross-talk of the fluorophores was negligible. An independent standard curve was generated for each experimental day after fixing and equilibrating the fluorescein-rhodamine-dextran–loaded cells to a range of pH buffers. To accurately assess the acidic pH of normal lysosomes, the gain of the microscope was optimized with the calibration buffer at pH 5; this gain was maintained during data acquisition for all pH controls and for all experimental conditions. Preliminary experiments revealed that sAC inhibition required in excess of 6 h to see an effect. Where appropriate, cells were incubated with 30 µM KH7 for 10 h and/or 500 µΜ Sp-8-cpt-cAMP analogue for 1 h in the presence or absence of 50 µM KT5720, and cells were imaged as mentioned above. KH7 exhibits intrinsic fluorescence, but neither its excitation peak at 354 nm nor its emission peak at 395 nm interfered with the fluorescein and rhodamine fluorescence used to measure pH. In contrast, for the PKA experiments, we were limited to the use of the PKA inhibitor, KT5720, because H89 exhibited intrinsic fluorescence, which interfered with rhodamine quantitation.
Immunofluorescence
Immunofluorescence was performed as previously described (Tampellini et al., 2009). After treatment, cells were fixed in 4% PFA with 2% sucrose for 20 min, permeabilized, and blocked in PBS containing 2% normal goat serum, 0.1% saponin, and 1% BSA at room temperature for 1 h. After washing coverslips three times with PBST, coverslips were mounted with Fluoromount-G (SouthernBiotech). For V-ATPase subunits, cells were fixed in ice-cold methanol for 5 min, blocked as described above, and then incubated with the primary antibodies for 48 h. Cells were viewed using an LSM 510 laser-scanning confocal microscope (ZEISS) and a 63×, 1.4 NA Plan Apochromat objective equipped with HeNe 633 nm/HeNe 543 nm/Argon (458, 488 nm) imaging lasers. In dual-labeled experiments, channels were used sequentially to avoid bleed through. MetaMorph software 7.5 (Universal Imaging Co.) was used for quantitative analysis.
500 nM LysoTracker DND-99 was added for 30 min at 37°C to the cells. To quantify LysoTracker fluorescence, 50–70 cells were randomly imaged in total from three different coverslips for each independent experiment. Cells were viewed using an LSM 510 laser-scanning confocal microscope and a 63×, 1.4 NA Plan Apochromat objective equipped with HeNe 633 nm/HeNe 543 nm/Argon (458, 488 nm) imaging lasers. LysoTracker intensity, or the number of LysoTracker puncta, was measured and normalized to cytoplasmic area of the cell using MetaMorph software 7.5. Intensity threshold was set to remove background fluorescence.
Active Cathepsin D (CatD) was visualized by incubating cells in 5 µM BODIPY-FL–Pepstatin A for 1 h. Cells were then fixed with 4% PFA and visualized as described above. DQ-BSA degradation was evaluated in cells incubated overnight at 37°C with medium containing 10 µg/ml DQ-BSA followed by 4-h chase. For some experiments, after fixation, cells were counterstained with LAMP2 antibody overnight and visualized with Alexa-conjugated secondary antibody. For quantification of BODIPY-FL–Pepstatin A and DQ-BSA, three coverslips from each condition were analyzed, 20 cells per coverslip. Using the integrated morphometric analysis feature in MetaMorph, the number of puncta and fluorescence intensity of the puncta were measured.
Cellular protein degradation
Proteolysis in cultured cells was measured by pulse-chase experiments (Auteri et al., 1983; Kaushik and Cuervo, 2009). In brief, confluent cells were labeled with [3H]leucine (6 µCi/ml) for 48 h at 37°C to preferentially label long-lived proteins. After labeling, cells were extensively washed and maintained in complete growth medium (DMEM + 10% fetal bovine serum + 0.5% glutamine) with an excess of unlabeled leucine (2.8 mM; Sigma-Aldrich) to prevent reutilization of radiolabeled leucine. Aliquots of the medium were taken at different time points and precipitated with 10% trichloroacetic acid (TCA) and 0.5 mg/ml BSA. Proteolysis was measured as the percentage of acid-soluble radioactivity (amino acids and small peptides) divided by the initial acid insoluble radioactivity (protein).
Proteasome inhibition
WT and sAC KO MEFs were plated in 96-well plates (5 × 103 per well) and treated with the indicated concentrations of the proteasome inhibitor MG132. Cellular growth was determined after 24 h by MTT assay.
Protein extraction and Western blotting
Immunoblotting was performed according to standard protocol. In brief, to prepare samples for SDS-PAGE, cells were lysed in buffer containing 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.5% Tween-20 with protease and phosphatase inhibitors. Either 10 or 15% gels were run according to protein weight, and after electrophoresis, proteins were transferred onto 0.45 µm PVDF membranes (EMD Millipore). The membrane was incubated overnight in primary antibody and then incubated with HRP-conjugated secondary antibody. The blot was developed by ECL-kit (GE Healthcare). Visualization and band quantification were performed with a gel imager, ChemiDoc XRS system (Bio-Rad Laboratories). Protein concentration in cellular extracts was quantified using the DC Protein Assay (Bio-Rad Laboratories).
Electron microscopy
Brain sections were prepared for electron microscopy as previously described (Milner et al., 2011). In brief, the brains of 15–18-mo-old WT and sAC KO male mice were perfused with 3.75% acrolein and 2% paraformaldehyde in 0.1 M phosphate buffer. Coronal hippocampal sections (40 µm thick) were embedded in plastic, cut (70 nm thick) on an Ultratome, and collected on copper mesh grids. To prepare cells for electron microscopy, they were treated as required and fixed with 2% glutaraldehyde for 2 h at 4°C, followed by postfixation in 1% osmium tetroxide for 1 h at 4°C. The samples then were embedded in plastic, cut (70 nm thick), and collected on grids. Grids were counterstained with 5% uranyl acetate followed by Reynolds lead citrate (sections only) and viewed with a CM10 transmission electron microscope (Philips). Grids were photographed and analyzed by a person blind to the experimental condition. For quantitative analysis, images from 15 entire cells were photographed in each experimental group. Autophagic vacuoles (AVs) were identified using criteria described by Lee et al. (2010) and counted on electron micrographs (135,000 magnification). Cytosol area was determined using ImageJ (National Institutes of Health). Results were expressed as the number of vacuoles per square micrometer.
Statistical analysis
Statistical comparisons were made using two-tailed unpaired t tests with significance placed at P < 0.05. One set of MEFs grown separately in individual dishes were considered as one independent experiment (n = 1). A set of primary neuronal culture prepared from one mouse embryo was also considered as one independent experiment. Data are expressed as mean ± SEM. Statistical analysis was performed using Excel (Microsoft). Error bars represent ±SEM.
Online supplemental material
Fig. S1 shows the functional characterization of the sAC KO MEFs. Fig. S2 shows the controls for the ratiometric measurement of lysosomal pH by confocal microscopy using pH-sensitive fluorescein fluorescence and pH-insensitive rhodamine fluorescence. Fig. S3 shows that sAC regulates localization of V-ATPase subunit V1D to lysosomes. Fig. S4 shows that LysoTracker staining reflects organellar acidification in MEFs. Fig. S5 shows impaired lysosomal degradation in sAC KO MEFs. Fig. S6 shows that in the absence of functional sAC, APs accumulate in neurons and Huh7 cells.
RESULTS
Lysosomal acidification is dependent on sAC activity
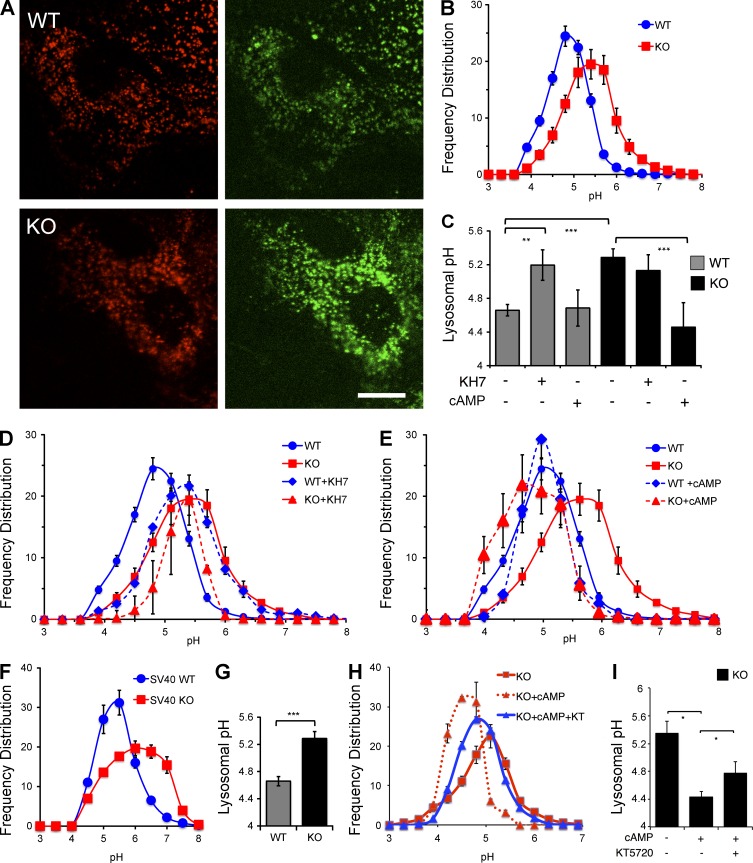
We derived MEF cell lines from sAC KO mice and from their WT littermates (Ramos-Espiritu et al., 2016). sAC KO 3T3 MEFs (hereafter referred to as sAC KO MEFs) generate less cAMP than WT MEFs, and unlike in WT MEFs, all the cAMP generated in sAC KO MEFs is insensitive to the sAC-specific inhibitor KH7 (Fig. S1). To quantify the pH of individual lysosomes in these MEF lines, we used ratiometric imaging of fluorescein (pH sensitive) and rhodamine (pH insensitive) dextran beads (Majumdar et al., 2007). The pH-sensitive nature of this dual-emission probe is demonstrated in Fig. S2. With decreasing pH, the pH-sensitive fluorescein (green) fluorescence is quenched, while the pH-insensitive rhodamine (red) fluorescence remains unchanged (Fig. S2, A and B); thus, the green/red ratio identifies each lysosome and determines its specific pH (Fig. S2 C). We confirmed the ability of this ratiometric method to measure changes in lysosomal pH by demonstrating alkalinization of lysosomes in WT cells perfused with 20 mM NH4Cl (Fig. S2, D and E). Using this quantitative method, lysosomal pH in WT MEFs was found to average 4.7 ± 0.1, consistent with previously published studies (Fig. 1, A–C; Ohkuma and Poole, 1978; Lee et al., 2010; Cang et al., 2013). In contrast, the mean pH of lysosomes in sAC KO MEFs was significantly higher, with their mean pH of 5.3 ± 0.1 (P < 0.001). In addition to having a higher mean pH, the distribution of pH values across all lysosomes was broader in sAC KO cells (Fig. 1 B).
Figure 1.
In the absence of sAC activity, lysosomal pH is elevated. (A) Representative images of WT or sAC KO 3T3 MEFs loaded with dextran beads conjugated to pH-insensitive rhodamine (red) and pH-sensitive fluorescein (green). Bar, 10 µm. (B) Frequency distribution of lysosomal pH measured as fluorescein/rhodamine ratios of individual lysosomes in WT and sAC KO 3T3 MEFs. Lysosomal pH values were determined from calibration curves generated from permeabilized cells in various pH buffers (Fig. S2). Number of lysosomes counted: WT = 3,141, KO = 3,559, n = 18 (from six independent experiment days). (C) Mean lysosomal pH in WT and sAC KO 3T3 MEFs in the absence or presence of KH7 or cAMP calculated from data shown in B, D, and E. (D) Frequency distribution of lysosomal pH in WT (WT+KH7) and sAC KO (KO+KH7) 3T3 MEFs treated with 30 µM KH7 for 10 h. Shown for comparison are the frequency distribution curves of WT and sAC KO MEFs from B. Number of lysosomes counted: WT+KH7 = 2,036, n = 12; KO+KH7 = 395, n = 3. (E) Frequency distribution of lysosomal pH in WT (WT+cAMP) and sAC KO (KO+cAMP) 3T3 MEFs treated with 500 µM Sp-8-cpt-cAMP for 60 min. Shown for comparison are the frequency distribution curves of WT and sAC KO MEFs from B. Number of lysosomes counted: KO+cAMP = 2,003, n = 12; WT+cAMP = 579, n = 3. (F) Frequency distribution of lysosomal pH in WT and sAC KO SV40 MEFs. Number of lysosomes counted: WT = 904, sAC KO =1,089, n = 6. (G) Mean lysosomal pH in WT and sAC KO SV40 MEFs calculated from data shown in F. (H) Frequency distribution of lysosomal pH in sAC KO 3T3 MEFs alone (KO), treated with 500 µM Sp-8-cpt-cAMP (KO+cAMP), or treated with 500 µM Sp-8-cpt-cAMP in the presence of 50 µM PKA inhibitor, KT 5720 (KO+cAMP+KT). Number of lysosomes counted: KO = 442, KO+cAMP = 250, KO+cAMP+KT = 1,024, n = 3 (from two independent experiment days). Data are distinct from the experiments shown in A–E. (I) Mean lysosomal pH in sAC KO 3T3 MEFs calculated from data shown in H. All values are given as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We confirmed the elevated lysosomal pH and altered distribution pattern was caused by loss of sAC in three ways; we pharmacologically inhibited sAC using the specific inhibitor, KH7 (Bitterman et al., 2013); we rescued the sAC KO phenotype by supplying cells with its product, cAMP; and we confirmed the elevated lysosomal pH in a sAC KO MEF line immortalized by an independent method (Ramos-Espiritu et al., 2016). Pharmacologically inhibiting sAC in WT cells altered lysosomal pH to a similar extent as the changes observed in sAC KO cells. Incubating WT cells with KH7 shifted the distribution and elevated the mean pH of lysosomes from 4.7 ± 0.1 to 5.2 ± 0.2 (P < 0.01). As expected, KH7 had no effect on sAC KO cells (Fig. 1, C and D). If the altered lysosomal pH in sAC KO MEFs is caused by the absence of sAC, addition of cAMP should rescue the phenotype. In microglia, elevation of intracellular cAMP acidified lysosomes within 45 min (Majumdar et al., 2007); therefore, we tested whether addition of cAMP for 1 h could affect lysosomal pH in sAC KO cells. Although supplying membrane-permeable cAMP did not affect lysosomal pH in WT cells, it reduced the mean pH in sAC KO lysosomes to 4.5 ± 0.3 (P < 0.001; Fig. 1, C and E). Finally, we confirmed that the elevated lysosomal pH in sAC KO 3T3 MEFs is not an artifact of their immortalization. Similar to the phenotype in sAC KO MEFs immortalized by the 3T3 method (Fig. 1, A–C), lysosomes in an independently derived line of sAC KO MEFs immortalized using SV40 large T antigen (SV40 KO MEFs; Ramos-Espiritu et al., 2016) had an elevated mean pH and a wider distribution of pH values relative to WT SV40-immortalized MEFs (Fig. 1, F and G). Thus, in the absence of sAC, lysosomal pH is elevated in a cAMP-dependent manner.
During activation of microglia, cAMP-dependent acidification of lysosomes is at least partially mediated via PKA (Majumdar et al., 2007). We took advantage of the ability to rescue lysosomal acidification in sAC KO MEFs to probe the mechanism by which sAC-generated cAMP acidifies lysosomes in normally growing fibroblasts. cAMP rescue was unaffected by the addition of cycloheximide (not depicted), demonstrating that cAMP’s effects on acidification are not dependent on new protein synthesis and consistent with cAMP acting via posttranslational signaling events. Inhibition of PKA using either a lower, more selective dose (10 µM; not depicted) or a saturating dose (50 µM) of the specific inhibitor KT5720 (IC50 for PKA = 3 µM; Davies et al., 2000) partially blocked the ability of cAMP to rescue lysosomal acidification in sAC KO cells (Fig. 1 H). In these experiments, the mean pH of lysosomes in sAC KO MEFs was 5.4 ± 0.2 (Fig. 1 I), and incubation in membrane-permeable cAMP for 1 h reduced the mean pH of sAC KO MEFs to 4.4 ± 0.1, which was nearly identical to the mean pH in WT MEFs (pH = 4.5 ± 0.2; not depicted). In contrast, the mean pH of lysosomes in sAC KO MEFs treated with cAMP in the presence of KT5720 for 1 h only reached 4.8 ± 0.1 (Fig. 1 I). Thus, at least part of cAMP’s ability to rescue the acidification defect in sAC KO MEFs is mediated via PKA, and there are possibly multiple cAMP-dependent pathways influencing lysosomal pH.
sAC regulates lysosomal acidification by ensuring proper V-ATPase localization to lysosomes
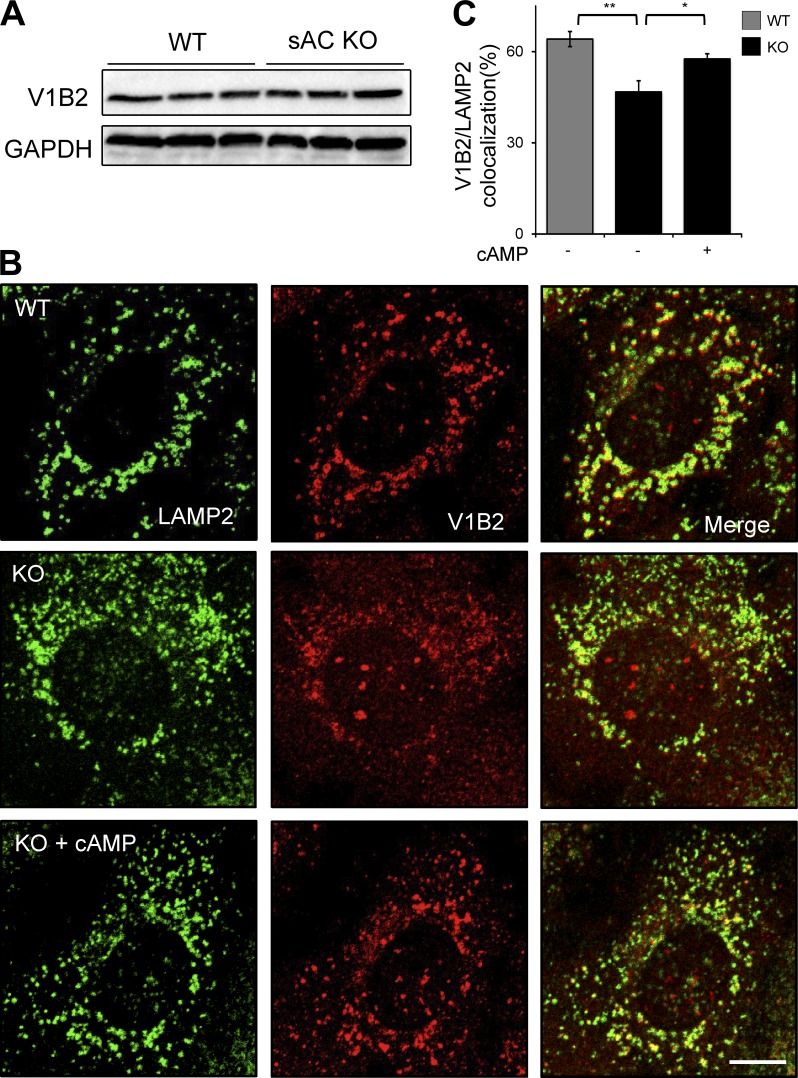
In pH-sensing epithelia, sAC-generated cAMP is responsible for translocation of the proton-pumping V-ATPase (Pastor-Soler et al., 2003; Păunescu et al., 2008b, 2010) via PKA (Pastor-Soler et al., 2008; Păunescu et al., 2010). Therefore, we asked whether sAC-dependent lysosomal acidification is dependent on proper localization of the V-ATPase. The V-ATPase is comprised of multiple subunits, and we assessed V-ATPase localization on lysosomes by double-immunofluorescence labeling of the lysosomal marker LAMP2 with either of two different cytoplasmic (V1) domain V-ATPase subunits, V1B2 or V1D. Total protein levels of both V1B2 (Fig. 2 A) and V1D (Fig. S3 A) were unchanged in WT versus sAC KO cells; thus, loss of sAC did not affect the amount of V-ATPase in cells. In WT cells, the majority of both V1B2 (Fig. 2, B and C) and V1D (Fig. S3 B) colocalized with LAMP2-positive vesicles. In contrast, in sAC KO cells, fewer LAMP2-positive vesicles were costained with the V-ATPase subunit antibodies (Fig. 2 C and Fig. S3 B), and both subunits were more diffusely distributed throughout the cytoplasm (Fig. 2 B and Fig. S3 B). To determine whether this altered staining pattern correlated with the acidification defect, we assessed V-ATPase subunit localization under conditions that rescued the lysosomal acidification in sAC KO cells. Addition of membrane-permeable cAMP for 1 h, which rescued the phenotypic defect in sAC KO cells (Fig. 1, C and E), restored LAMP2 colocalization with both V-ATPase subunits (Fig. 2, B and C; and Fig. S3 B). Thus, sAC-generated cAMP is responsible for both proper localization of V-ATPase subunits and acidification of lysosomes.
Figure 2.
sAC regulates V-ATPase localization to lysosomes. (A) Representative immunoblot of the V-ATPase subunit V1B2 along with GAPDH, which was used for loading control. Shown are whole cell extracts from three independently grown cultures of WT and sAC KO MEFs. n = 3. (B) Double-immunofluorescence labeling of V-ATPase V1B2 subunit (red) and LAMP2 (green) in WT, sAC KO, and sAC KO MEFs treated with 500 µM sp-8-cpt-cAMP for 1 h. Note the diffused cytosolic staining of V1B2 in KO cells. Bar, 10 µm. (C) Quantification of the colocalization between V1B2- and LAMP2-positive lysosomes. All values are given as mean percentage ± SEM. *, P < 0.05; **, P < 0.01.
Organelle acidification via sAC is a general phenomenon
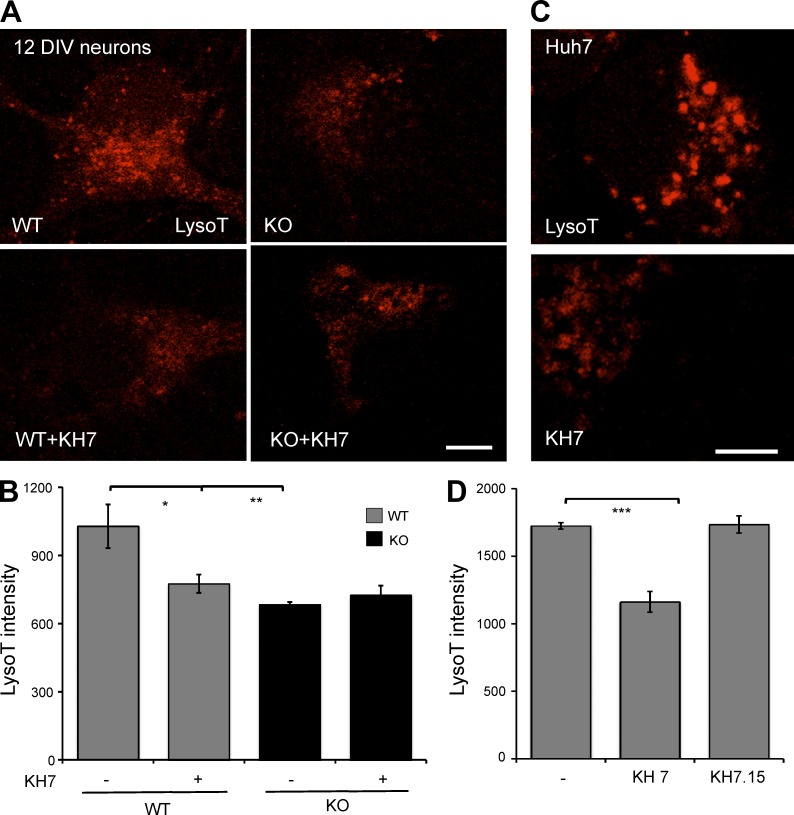
Lysosomal dysfunction and defective autophagy have been linked to age-related neurodegenerative diseases, which arise when neurons erroneously accumulate long-lived or misfolded proteins and damaged organelles (Menzies et al., 2015). Therefore, we asked whether sAC is essential for lysosomal acidification in neurons. To examine organelle acidification in primary neurons, which proved to be unsuitable for use with dextran conjugates, we took advantage of LysoTracker, a qualitative acidic pH indicator dye, which is enriched and strongly fluoresces in compartments that have a pH below 5 (Lee et al., 2010). We first confirmed that LysoTracker reproduced the lysosomal acidification defect observed in sAC KO MEFs. As a qualitative marker of organelle acidity, we limited our use of LysoTracker to compare overall fluorescence intensity or number of acidic puncta within a cell line before and after pharmacological treatment. In WT MEFs, the number of LysoTracker puncta (Fig. S4) and overall intensity (not depicted) was unaffected by cAMP addition. In contrast, cAMP increased the number of LysoTracker-positive puncta (Fig. S4) and overall intensity (not depicted) in sAC KO MEFs. Thus, LysoTracker staining reflects the ability of cAMP to rescue the lysosomal acidification defect in sAC KO cells demonstrated quantitatively above (Fig. 1, C and E). LysoTracker intensity of 12 DIV WT primary neurons was diminished when sAC was inhibited using KH7 (Fig. 3, A and B). These effects of KH7 were specifically mediated via inhibition of sAC activity because the same treatment was inert on 12 DIV sAC KO neurons. KH7, but not the inert structurally related KH7.15 (Wu et al., 2006), also diminished LysoTracker intensity in a human liver cancer cell line, Huh7 (Fig. 3, C and D). These data demonstrate that sAC regulates organellar acidification in different cell types and in mouse and human cells and is most likely a general phenomenon. These data also suggest that sAC’s role in organellar acidification may expand beyond acidifying endosomes into lysosomes. The quantitative dextran conjugate method is biased toward those lysosomes actively involved in the endocytotic pathway, whereas LysoTracker identifies all acidic vesicles.
Figure 3.
sAC is essential for organellar acidification in primary neurons and human Huh7 cells. (A) Representative images of staining with LysoTracker (red) in WT or sAC KO 12 DIV neurons grown in the absence or presence of 30 µM KH7 (3 h), with 100 nM LysoTracker included for the final 30 min. (B) Quantified LysoTracker intensity in neurons. (C) Representative images of staining with LysoTracker (red) in human Huh7 cells treated with 30 µM KH7 (or 30 µM KH7.15) for 10 h, with 100 nM LysoTracker included for the final 30 min. (D) Quantified LysoTracker intensity in Huh7 cells. For B and D, LysoTracker intensity was quantified using MetaMorph in multiple cells from three independent experiments. All values are given as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.002.
Lysosomal degradation is diminished in the absence of sAC
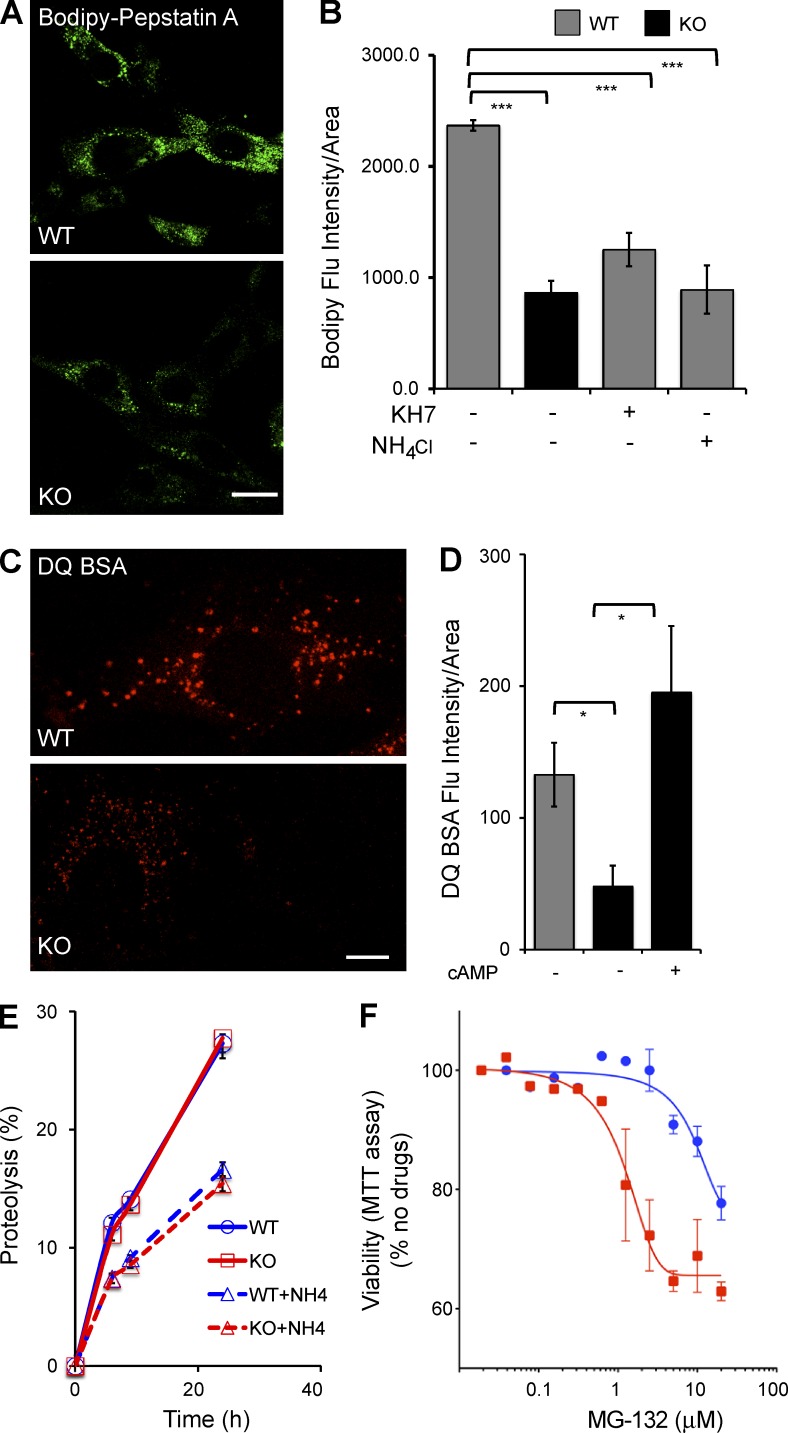
The lysosomal cathepsins and hydrolases that degrade proteins, lipids, and polysaccharides are optimally active in the acidic (i.e., pH < 5) lumen of the lysosome (Pillay et al., 2002). Because fewer lysosomes achieve the acidic pH necessary for optimal degradation in sAC KO cells (Fig. 1, A and B), we asked whether protein degradation was affected in sAC KO MEFs. We first examined the consequences of elevated lysosomal pH by assessing the activity of lysosomal cathepsins, specifically CatD. Fluorescently tagged BODIPY-FL–Pepstatin A binds to CatD only when the CatD active site is in an open state under acidic conditions (i.e., ∼pH 4.5; Chen et al., 2000a). As expected, in WT control cells, BODIPY-FL–Pepstatin A fluorescence intensity (Fig. 4, A and B) and number of puncta (Fig. S5 A) were decreased when lysosomal pH was chemically elevated by incubating cells in NH4Cl (Fig. 4 B and Fig. S5 A). Loss of sAC, both in sAC KO cells (Fig. 4, A and B) and in WT cells treated with KH7 (Fig. 4 B), decreased BODIPY-FL–Pepstatin A fluorescence intensity and puncta (Fig. S5 A) relative to WT cells. Thus, in the absence of sAC, there is diminished catalytically active CatD.
Figure 4.
Lysosomal proteolytic degradation defect in the absence of sAC. (A) Representative images of WT and sAC KO MEFs stained with BODIPY-FL–Pepstatin A. (B) BODIPY-FL–Pepstatin A intensity was quantified using MetaMorph in multiple cells from three independent experiments. All values are given as mean fluorescence intensity per cell area ± SEM. ***, P < 0.001. (C) Representative images of WT and sAC KO MEFs stained with 10 µg/ml DQ-BSA. (A and C) Bars, 10 µm. (D) DQ-BSA intensity was quantified using MetaMorph in multiple cells from three independent experiments. All values are given as mean ± SEM. *, P < 0.05. (E) Total Protein turnover is quantified as percent proteolysis (i.e., the percentage of acid-soluble radioactivity [amino acids and small peptides] divided by the initial acid-insoluble radioactivity [protein]) in WT and sAC KO MEFs after incorporation of [3H]leucine (n = 15, four independent experiment days). As control, proteolysis was assessed in the presence of NH4Cl in both WT (WT+NH4) and in sAC KO MEFs (KO+NH4). Error bars represent ±SEM. (F) WT (blue circles) and sAC KO (red squares) MEFs were plated in 96-well plates (5 × 103 per well) and treated with increasing concentrations of the proteasome inhibitor MG132 for 24 h. The percentage of growth was determined by MTT assay. Results are presented as percent viability relative to cells in the absence of any drug; data graphed are the mean of triplicate determinations (±SEM) of a representative experiment repeated three times.
We confirmed the diminished proteolytic activity in sAC KO lysosomes using DQ-BSA, a fluorogenic self-quenched probe. DQ-BSA is internalized via the endosomal pathway and is dequenched when proteolytically cleaved in the lysosome. sAC KO MEFs had lower DQ-BSA fluorescence intensity (Fig. 4, C and D) and fewer puncta (Fig. S5 B) relative to WT MEFs. Similar to the elevated lysosomal pH and defective V-ATPase trafficking, the impaired lysosomal degradation of DQ-BSA in sAC KO cells was rescued by exogenous cAMP. Treatment of KO cells with membrane-permeable cAMP increased DQ-BSA fluorescence intensity (Fig. 4 D) and puncta (Fig. S5 B). Interestingly, a subset of KO cells showed large DQ-BSA–positive puncta that colocalized with LAMP2-stained vesicles (Fig. S5 C). Large vesicles were not observed in WT cells. The nature of these vesicles remains under investigation.
Unlike the defect in lysosomal proteolysis in sAC KO cells, when we examined overall protein turnover in metabolic labeling experiments, the kinetics of protein turnover were indistinguishable between WT and sAC KO MEFs (Fig. 4 E), and both were similarly affected when turnover was pharmacologically inhibited using NH4Cl. In addition to lysosomal degradation, cells turn over proteins via the proteasome. When we attempted to use the proteasomal inhibitor MG132 to study lysosomal degradation in the absence of proteasomal degradation, we discovered that survival of sAC KO MEFs was more sensitive to MG132 than WT MEFs (Fig. 4 F). This increased dependence on proteasomal degradation for viability suggests proteasome-mediated proteolysis compensates for diminished lysosomal activity in sAC KO MEFs.
sAC KO cells accumulate AVs
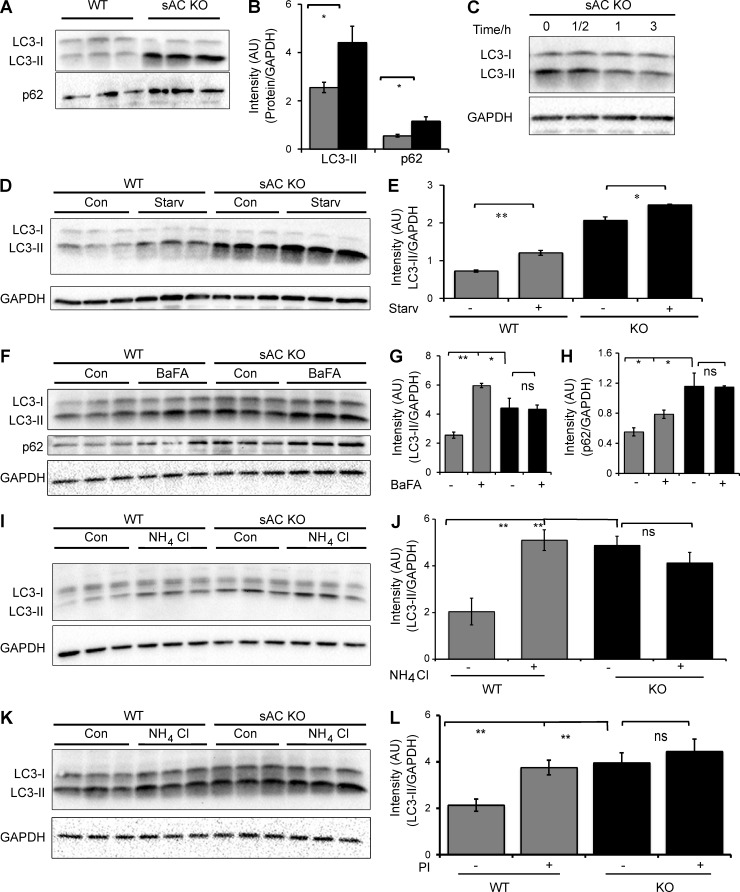
Macroautophagy (hereafter autophagy), the major lysosomal degradative pathway in cells, is responsible for degrading long-lived proteins, organelles, and protein aggregates (Klionsky, 2007; Mizushima, 2007). It involves sequestration of cytosolic regions into characteristic double-membrane APs that fuse with lysosomes (Yamamoto et al., 1998; Klionsky et al., 2012) to form single-membrane degradative autophagolysosomes (ALs). Because sAC KO cells have less acidic lysosomes (Fig. 1, A and B) and reduced lysosomal proteolytic capacity (Fig. 4), we hypothesized that the autophagic degradative system may also be impaired. The autophagic markers, LC3-II and p62 were more abundant in sAC KO MEFs (Fig. 5, A and B) and primary neurons from sAC KO mice (Fig. S6, A–C) compared with their WT counterparts. Similar to elevated lysosomal pH, mislocalization of V-ATPase, and decreased lysosomal proteolysis, this elevation of LC3-II in sAC KO MEFs was rescued by the addition of exogenous membrane-permeable cAMP (Fig. 5 C). And the time course of this rescue was consistent with the time course of cAMP-dependent acidification of microglia (Majumdar et al., 2007); it required >30 min and was complete after 1 h.
Figure 5.
APs accumulate in the absence of sAC activity. (A) Representative immunoblot of the autophagic markers LC3-II and p62 in three independent cultures of WT and sAC KO MEFs. This immunoblot is a reprobing of the blot shown in Fig. 2 A; therefore, the GAPDH control showed in Fig. 2 A is also relevant for this immunoblot. (B) Densitometric analysis of LC3-II and p62, normalized to GAPDH, in WT (gray bars) and sAC KO (black bars) MEFs. n = 6. (C) Representative immunoblot of LC3-II in sAC KO MEFs grown in the presence of 500 µM Sp-8-cpt-cAMP for the indicated time. GAPDH was used as loading control. (D) LC3-II immunoblot levels in WT and KO 3T3 MEFs after 6 h of serum starvation. GAPDH is used as loading control. (E) Densitometric analysis of LC3-II immunoblot from D normalized to GAPDH. Autophagic induction is unaltered in WT and sAC KO cells. Ratio of LC-II in starvation/control is 1.2 and 1.7 for WT and sAC KO, respectively. (F) LC3 and p62 immunoblot levels in WT and KO 3T3 MEFs with and without treatment with 100 nM Bafilomycin A1 (BafA) for 6 h. Protein levels of each protein were normalized to GAPDH as an internal control. (G) Densitometric analysis of LC3-II immunoblot from F normalized to GAPDH. n = 3. (H) Densitometric analysis of p62 immunoblot from F normalized to GAPDH. n = 3. (I) LC3 immunoblot of WT and KO 3T3 MEFs treated with 20 mM NH4Cl for 6 h. Protein levels were normalized to GAPDH as an internal control. (J) Densitometric analysis of LC3-II from I. n = 3–5 per experimental group. (K) LC3 immunoblot of WT and KO 3T3 MEFs treated with and without lysosomal protease inhibitors (PI) for 6 h (20 µM Leupeptin, 20 µM Pepstatin A, and 10 µM E64D). Protein levels were normalized to GAPDH as an internal control. (L) Densitometric analysis of LC3-II from K. n = 3–5 per experimental group. All values are given as mean ± SEM. *, P < 0.05; **, P < 0.01.
Inhibition of sAC activity using KH7 also increased LC3-II and p62 accumulation in WT neurons, but not in sAC KO neurons (Fig. S6, A–C), and in a time-dependent manner in Huh7 cells (Fig. S6 D). Interestingly, sAC KO neurons as well as WT neurons treated with KH7 (but not sAC KO neurons treated with KH7) also exhibit an increase in the soluble, cytoplasmic isoform of LC3, LC3-I (Fig. S6 A). This increase seems to reflect an up-regulation of total LC3, which may represent an additional compensation mechanism.
Autophagic markers will accumulate when there is increased AV formation or diminished lysosome-mediated clearance. As expected, when we induced autophagy by serum starvation, LC3-II levels increased in WT MEFs (Fig. 5, D and E), reflecting increased flux through the pathway. In response to serum starvation, LC3-II levels in sAC KO MEFs also increased, although in the KO cells the proportional increase over the already elevated level of LC3-II was less than in WT cells. Because sAC KO cells remain responsive to nutrient starvation, it suggests their accumulation of autophagic markers is not caused by constitutive induction. We next explored the effects of blocking AV clearance (a) with the V-ATPase inhibitor bafilomycin A1 (Fig. 5, F–H), (b) by chemically increasing lysosomal pH using NH4Cl (Fig. 5, I and J), or (c) by directly blocking lysosomal proteases (protease inhibitor cocktail [PI]; Fig. 5, K and L). Although all three treatments caused accumulation of the autophagic marker LC3-II (and p62) in WT cells, they had no effect in sAC KO cells. These data reveal sAC KO cells harbor a defect in AV clearance, consistent with their elevated pH and diminished proteolytic activity.
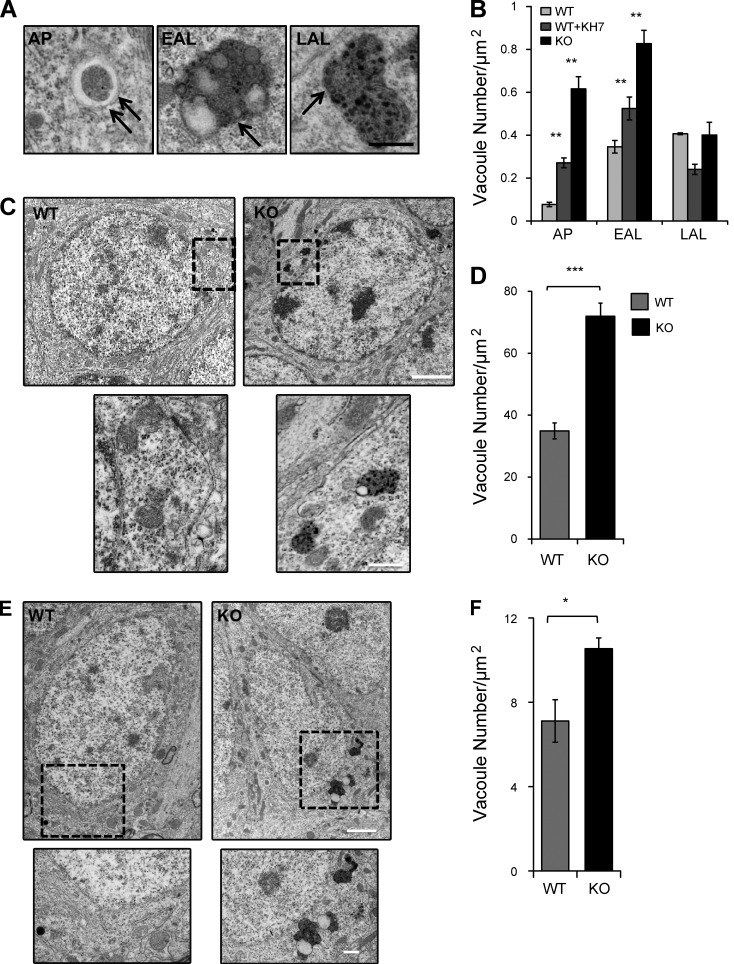
If the AV clearance defect is a direct consequence of diminished proteolytic activity in sAC KO cells, the absence of sAC should result in increased accumulation of APs and early-stage ALs (EALs; i.e., single membrane structures containing partially digested electron dense material) but not late-stage ALs (LALs; i.e., single membrane structures containing amorphous electron dense material; Fig. 6 A; Lee et al., 2010). Consistent with our hypothesis, ultrastructural analysis of sAC KO and WT MEFs treated with KH7 revealed an increase in both APs and EALs relative to untreated WT MEFs (Fig. 6 B). In WT MEFs treated with KH7, where sAC activity is acutely inhibited, this increase in APs and EALs was accompanied by a decrease in LALs, whereas in sAC KO MEFs LAL accumulation was unchanged relative to WT MEFs (Fig. 6 B). Therefore, our data suggest that the sAC KO lysosomal pH and proteolysis defects lead to impaired clearance of AVs.
Figure 6.
Electron-dense AVs accumulate in the absence of sAC activity. (A) AVs were subcategorized based on their morphology. AP: double membrane structures and/or double membrane structure containing undigested organelles; EAL: single membrane structures containing partially digested electron-dense material; LAL: single membrane structures containing amorphous electron-dense material. Double arrows represent double membrane, and single arrow represents single membrane. (B) Quantitative analysis of the number of AVs in WT, WT+KH7, and sAC KO MEFs. n = 10 cells/condition, two independent experiments. **, P < 0.02. (C) Representative electron micrographs of hippocampal dentate granule cells in WT and sAC KO aged mice. High-magnification views of the boxed areas below show examples of AVs in KO mice. (D) Quantitative analysis of the number of AVs in WT and sAC KO granular cells of the dentate gyrus. n = 3 mice/condition; n = 10 cells/mouse. ***, P < 0.002. (E) Representative electron micrographs of the pyramidal cells of the CA1 in WT and sAC KO aged mice. High-magnification views of the boxed areas below show examples of AVs in KO mice. Bars: (A, C [bottom], and E [bottom]) 500 nm; (C and E, top) 2 µm. (F) Quantitative analysis of the number of vacuoles in WT and sAC KO pyramidal cells of the CA1 of the hippocampus. n = 3 mice/condition; n = 10 cells/mouse. *, P < 0.05. All values are given as mean ± SEM.
sAC KO brains accumulate AVs
Accumulation of AVs is a hallmark of aging and neurodegenerative diseases (Bahr and Bendiske, 2002; Nixon et al., 2005). Alzheimer’s disease is a neurodegenerative disease that mostly affects the hippocampus. We used electron microscopy to quantitate AVs in the dentate gyrus (Fig. 6, C and D) and CA1 (Fig. 6, E and F) regions of the hippocampus from sAC KO and age-matched WT mice. In both regions, the granule cells of the dentate gyrus and the pyramidal cells of CA1, sAC KO mice accumulated more AVs than WT mice. Thus, accumulation of AVs is a general phenotype caused by loss of sAC.
DISCUSSION
Our results demonstrate that sAC activity is essential for proper lysosomal acidification. We find this to be a general principle, true in fibroblasts, neurons, and liver cells, as well as in rodents and humans. Although much has been learned about the channels and transporters contributing to acidification of lysosomes (Steinberg et al., 2010; Mindell, 2012; Xu and Ren, 2015), the pH sensors responsible for setting the pH of the lysosome’s lumen remain unknown. Because our data define pH-sensitive sAC as an essential regulator of endosomal–lysosomal acidification, it represents a likely candidate. In contrast to TPC1, which is a pH sensor essential for acidification during starvation-induced autophagy (Cang et al., 2013), sAC is the first pH-sensitive signaling enzyme that contributes to setting lysosomal pH under normal growth conditions.
The acidification defect in sAC KO cells is rescued by the addition of membrane-permeable cAMP within 1 h. Inhibiting PKA during this 1 h partially blocks rescue, implying that at least a subset of the cAMP-dependent steps contributing to acidification are PKA dependent. Acidification is achieved by the proton-pumping V-ATPase (Ohkuma et al., 1982; Breton and Brown, 2013), and several studies in diverse physiological systems demonstrate sAC mediates V-ATPase trafficking (Pastor-Soler et al., 2003; Păunescu et al., 2010; Tresguerres et al., 2010b) in a PKA-dependent manner (Pastor-Soler et al., 2008; Păunescu et al., 2010). We found V-ATPase localization to lysosomes is altered in sAC KO cells, and treatment with membrane-permeable cAMP, which rescues the lysosomal acidification defect in these cells, also restores normal V-ATPase localization. These data suggest that the sAC, PKA, V-ATPase trafficking pathway, which is essential in acid-sensing epithelia, also contributes to lysosomal acidification in normally growing cells.
In acid-secreting epithelia, intracellular sAC senses the extracellular pH in an adjacent lumen in a CA-dependent manner (Pastor-Soler et al., 2003, 2008; Păunescu et al., 2008a, 2010; Tresguerres et al., 2010b). It remains unclear whether cytoplasmic sAC might sense lysosomal luminal pH in an analogous way. In the yeast Saccharomyces cerevisiae, activity of the V-ATPase is regulated by a well-characterized, reversible dissociation/association of the cytoplasmic V1 and transmembrane V0 domains. Association of V1 and V0 domains on the yeast degradative acidic organelle is induced by elevated intracellular HCO3− (Dechant et al., 2010). Because the single adenylyl cyclase in yeast is HCO3− stimulated (Hess et al., 2014), similar to sAC, it is tempting to hypothesize that yeast adenylyl cyclase participates in the HCO3−-induced V1 assembly and that organellar acidification via HCO3−-regulated adenylyl cyclase activity might be evolutionarily conserved.
As a consequence of aging, postmitotic cells, such as neurons, have decreased lysosomal proteolytic activity; this change is thought to be caused by elevated lysosomal pH (Martinez-Vicente et al., 2005). Aberrant accumulation of AVs caused by lysosomal dysfunction, similar to the phenotype we observe in sAC KO mice, is a common phenotype shared by lysosomal storage disorders and age-related neurodegenerative diseases, such as Alzheimer’s disease, Parkinson’s disease, and frontotemporal dementia (Bahr and Bendiske, 2002). Interestingly, addition of cAMP to human Alzheimer’s disease fibroblasts reacidifies their lysosomes and rescues their autophagic defect (Coffey et al., 2014), suggesting sAC activation may have therapeutic potential for these pathophysiological conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Fred Maxfield and Lee Cohen-Gould for assistance with ratiometric imaging and electron microscopy; we thank Shafat Zaman for computer programming assistance; and we thank Drs. Fred Maxfield and Jonathan Zippin for comments on the manuscript.
This work was supported by the National Institutes of Health (grants R01GM107442, R01GM062328, and R01NS055255 to J. Buck and L.R. Levin and grant F31NS081930 to N. Rahman).
Drs. J. Buck and L.R. Levin own equity interest in CEP Biotech, which has licensed commercialization of a panel of monoclonal antibodies directed against sAC. The authors declare no further competing financial interests.
Author contributions: N. Rahman performed all experiments; L. Ramos-Espiritu generated the MEF lines; T.A. Milner supervised the brain electron microscopy; N. Rahman, J. Buck, and L.R. Levin designed experiments, interpreted data, and prepared the manuscript.
Angus C. Nairn served as editor.
Footnotes
Abbreviations used:
- AL
- autophagolysosome
- AP
- autophagosome
- AV
- autophagic vacuole
- CA
- carbonic anhydrase
- CatD
- Cathepsin D
- DIV
- day in vitro
- EAL
- early-stage AL
- KO
- knockout
- LAL
- late-stage AL
- MEF
- mouse embryonic fibroblast
- sAC
- soluble adenylyl cyclase
- V-ATPase
- vacuolar ATPase
References
- Auteri J.S., Okada A., Bochaki V., and Dice J.F.. 1983. Regulation of intracellular protein degradation in IMR-90 human diploid fibroblasts. J. Cell. Physiol. 115:167–174. 10.1002/jcp.1041150210 [DOI] [PubMed] [Google Scholar]
- Bahr B.A., and Bendiske J.. 2002. The neuropathogenic contributions of lysosomal dysfunction. J. Neurochem. 83:481–489. 10.1046/j.1471-4159.2002.01192.x [DOI] [PubMed] [Google Scholar]
- Bitterman J.L., Ramos-Espiritu L., Diaz A., Levin L.R., and Buck J.. 2013. Pharmacological distinction between soluble and transmembrane adenylyl cyclases. J. Pharmacol. Exp. Ther. 347:589–598. 10.1124/jpet.113.208496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S., and Brown D.. 2013. Regulation of luminal acidification by the V-ATPase. Physiology (Bethesda). 28:318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cang C., Zhou Y., Navarro B., Seo Y.J., Aranda K., Shi L., Battaglia-Hsu S., Nissim I., Clapham D.E., and Ren D.. 2013. mTOR regulates lysosomal ATP-sensitive two-pore Na+ channels to adapt to metabolic state. Cell. 152:778–790. 10.1016/j.cell.2013.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cang C., Bekele B., and Ren D.. 2014. The voltage-gated sodium channel TPC1 confers endolysosomal excitability. Nat. Chem. Biol. 10:463–469. 10.1038/nchembio.1522 [DOI] [PubMed] [Google Scholar]
- Chang J.C., and Oude-Elferink R.P.. 2014. Role of the bicarbonate-responsive soluble adenylyl cyclase in pH sensing and metabolic regulation. Front. Physiol. 5:42 10.3389/fphys.2014.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.S., Chen W.N., Zhou M., Arttamangkul S., and Haugland R.P.. 2000a Probing the cathepsin D using a BODIPY FL-pepstatin A: applications in fluorescence polarization and microscopy. J. Biochem. Biophys. Methods. 42:137–151. 10.1016/S0165-022X(00)00048-8 [DOI] [PubMed] [Google Scholar]
- Chen Y., Cann M.J., Litvin T.N., Iourgenko V., Sinclair M.L., Levin L.R., and Buck J.. 2000b Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science. 289:625–628. 10.1126/science.289.5479.625 [DOI] [PubMed] [Google Scholar]
- Coffey E.E., Beckel J.M., Laties A.M., and Mitchell C.H.. 2014. Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer’s disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience. 263:111–124. 10.1016/j.neuroscience.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies S.P., Reddy H., Caivano M., and Cohen P.. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351:95–105. 10.1042/bj3510095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant R., Binda M., Lee S.S., Pelet S., Winderickx J., and Peter M.. 2010. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 29:2515–2526. 10.1038/emboj.2010.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G., Jaiswal B.S., Xie F., Krajnc-Franken M.A., Robben T.J., Strik A.M., Kuil C., Philipsen R.L., van Duin M., Conti M., and Gossen J.A.. 2004. Mice deficient for soluble adenylyl cyclase are infertile because of a severe sperm-motility defect. Proc. Natl. Acad. Sci. USA. 101:2993–2998. (published erratum appears in Proc. Natl. Acad. Sci. USA 2004. 101:5180) 10.1073/pnas.0400050101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess K.C., Jones B.H., Marquez B., Chen Y., Ord T.S., Kamenetsky M., Miyamoto C., Zippin J.H., Kopf G.S., Suarez S.S., et al. 2005. The “soluble” adenylyl cyclase in sperm mediates multiple signaling events required for fertilization. Dev. Cell. 9:249–259. 10.1016/j.devcel.2005.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess K.C., Liu J., Manfredi G., Mühlschlegel F.A., Buck J., Levin L.R., and Barrientos A.. 2014. A mitochondrial CO2-adenylyl cyclase-cAMP signalosome controls yeast normoxic cytochrome c oxidase activity. FASEB J. 28:4369–4380. 10.1096/fj.14-252890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S., and Cuervo A.M.. 2009. Methods to monitor chaperone-mediated autophagy. Methods Enzymol. 452:297–324. 10.1016/S0076-6879(08)03619-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting S., Diaz A., Moniot S., van den Heuvel J., Weyand M., Levin L.R., Buck J., and Steegborn C.. 2014. Crystal structures of human soluble adenylyl cyclase reveal mechanisms of catalysis and of its activation through bicarbonate. Proc. Natl. Acad. Sci. USA. 111:3727–3732. 10.1073/pnas.1322778111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8:931–937. 10.1038/nrm2245 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A., et al. 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 8:445–544. 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Yu W.H., Kumar A., Lee S., Mohan P.S., Peterhoff C.M., Wolfe D.M., Martinez-Vicente M., Massey A.C., Sovak G., et al. 2010. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 141:1146–1158. 10.1016/j.cell.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin L.R., and Buck J.. 2015. Physiological roles of acid-base sensors. Annu. Rev. Physiol. 77:347–362. 10.1146/annurev-physiol-021014-071821 [DOI] [PubMed] [Google Scholar]
- Liu J., Lu W., Reigada D., Nguyen J., Laties A.M., and Mitchell C.H.. 2008. Restoration of lysosomal pH in RPE cells from cultured human and ABCA4−/− mice: pharmacologic approaches and functional recovery. Invest. Ophthalmol. Vis. Sci. 49:772–780. 10.1167/iovs.07-0675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A., Cruz D., Asamoah N., Buxbaum A., Sohar I., Lobel P., and Maxfield F.R.. 2007. Activation of microglia acidifies lysosomes and leads to degradation of Alzheimer amyloid fibrils. Mol. Biol. Cell. 18:1490–1496. 10.1091/mbc.E06-10-0975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Pedersen S.F., Schwab A., and Stock C.. 2011. Intracellular pH gradients in migrating cells. Am. J. Physiol. Cell Physiol. 300:C490–C495. 10.1152/ajpcell.00280.2010 [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M., Sovak G., and Cuervo A.M.. 2005. Protein degradation and aging. Exp. Gerontol. 40:622–633. 10.1016/j.exger.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Menzies F.M., Fleming A., and Rubinsztein D.C.. 2015. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 16:345–357. 10.1038/nrn3961 [DOI] [PubMed] [Google Scholar]
- Milner T.A., Waters E.M., Robinson D.C., and Pierce J.P.. 2011. Degenerating processes identified by electron microscopic immunocytochemical methods. Methods Mol. Biol. 793:23–59. 10.1007/978-1-61779-328-8_3 [DOI] [PubMed] [Google Scholar]
- Mindell J.A. 2012. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 74:69–86. 10.1146/annurev-physiol-012110-142317 [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873. 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- Nixon R.A., Wegiel J., Kumar A., Yu W.H., Peterhoff C., Cataldo A., and Cuervo A.M.. 2005. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 64:113–122. 10.1093/jnen/64.2.113 [DOI] [PubMed] [Google Scholar]
- Ohkuma S., and Poole B.. 1978. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. USA. 75:3327–3331. 10.1073/pnas.75.7.3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuma S., Moriyama Y., and Takano T.. 1982. Identification and characterization of a proton pump on lysosomes by fluorescein-isothiocyanate-dextran fluorescence. Proc. Natl. Acad. Sci. USA. 79:2758–2762. 10.1073/pnas.79.9.2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Soler N., Beaulieu V., Litvin T.N., Da Silva N., Chen Y., Brown D., Buck J., Levin L.R., and Breton S.. 2003. Bicarbonate-regulated adenylyl cyclase (sAC) is a sensor that regulates pH-dependent V-ATPase recycling. J. Biol. Chem. 278:49523–49529. 10.1074/jbc.M309543200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Soler N.M., Hallows K.R., Smolak C., Gong F., Brown D., and Breton S.. 2008. Alkaline pH- and cAMP-induced V-ATPase membrane accumulation is mediated by protein kinase A in epididymal clear cells. Am. J. Physiol. Cell Physiol. 294:C488–C494. 10.1152/ajpcell.00537.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Păunescu T.G., Da Silva N., Russo L.M., McKee M., Lu H.A., Breton S., and Brown D.. 2008a Association of soluble adenylyl cyclase with the V-ATPase in renal epithelial cells. Am. J. Physiol. Renal Physiol. 294:F130–F138. 10.1152/ajprenal.00406.2007 [DOI] [PubMed] [Google Scholar]
- Păunescu T.G., Jones A.C., Tyszkowski R., and Brown D.. 2008b V-ATPase expression in the mouse olfactory epithelium. Am. J. Physiol. Cell Physiol. 295:C923–C930. 10.1152/ajpcell.00237.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Păunescu T.G., Ljubojevic M., Russo L.M., Winter C., McLaughlin M.M., Wagner C.A., Breton S., and Brown D.. 2010. cAMP stimulates apical V-ATPase accumulation, microvillar elongation, and proton extrusion in kidney collecting duct A-intercalated cells. Am. J. Physiol. Renal Physiol. 298:F643–F654. 10.1152/ajprenal.00584.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay C.S., Elliott E., and Dennison C.. 2002. Endolysosomal proteolysis and its regulation. Biochem. J. 363:417–429. 10.1042/bj3630417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Espiritu L., Diaz A., Nardin C., Saviola A.J., Shaw F., Plitt T., Yang X., Wolchok J., Pirog E.C., Desman G., et al. 2016. The metabolic/pH sensor soluble adenylyl cyclase is a tumor suppressor protein. Oncotarget. 10.18632/oncotarget.10056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer K.W., Skolnick R.L., Peercy B.E., Keener J.P., and Vaughan-Jones R.D.. 2002. Facilitation of intracellular H(+) ion mobility by CO2/HCO3− in rabbit ventricular myocytes is regulated by carbonic anhydrase. J. Physiol. 541:159–167. 10.1113/jphysiol.2001.013268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg B.E., Huynh K.K., Brodovitch A., Jabs S., Stauber T., Jentsch T.J., and Grinstein S.. 2010. A cation counterflux supports lysosomal acidification. J. Cell Biol. 189:1171–1186. 10.1083/jcb.200911083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A.K., Boyd C.A., and Vaughan-Jones R.D.. 1999. A novel role for carbonic anhydrase: cytoplasmic pH gradient dissipation in mouse small intestinal enterocytes. J. Physiol. 516:209–217. 10.1111/j.1469-7793.1999.209aa.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tampellini D., Rahman N., Gallo E.F., Huang Z., Dumont M., Capetillo-Zarate E., Ma T., Zheng R., Lu B., Nanus D.M., et al. 2009. Synaptic activity reduces intraneuronal Aβ, promotes APP transport to synapses, and protects against Aβ-related synaptic alterations. J. Neurosci. 29:9704–9713. 10.1523/JNEUROSCI.2292-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarbashevich K., Reichman-Fried M., Grimaldi C., and Raz E.. 2015. Chemokine-dependent pH elevation at the cell front sustains polarity in directionally migrating zebrafish germ cells. Curr. Biol. 25:1096–1103. 10.1016/j.cub.2015.02.071 [DOI] [PubMed] [Google Scholar]
- Tresguerres M., Buck J., and Levin L.R.. 2010a Physiological carbon dioxide, bicarbonate, and pH sensing. Pflugers Arch. 460:953–964. 10.1007/s00424-010-0865-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresguerres M., Parks S.K., Salazar E., Levin L.R., Goss G.G., and Buck J.. 2010b Bicarbonate-sensing soluble adenylyl cyclase is an essential sensor for acid/base homeostasis. Proc. Natl. Acad. Sci. USA. 107:442–447. 10.1073/pnas.0911790107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresguerres M., Levin L.R., and Buck J.. 2011. Intracellular cAMP signaling by soluble adenylyl cyclase. Kidney Int. 79:1277–1288. 10.1038/ki.2011.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.Y., Zippin J.H., Huron D.R., Kamenetsky M., Hengst U., Buck J., Levin L.R., and Jaffrey S.R.. 2006. Soluble adenylyl cyclase is required for netrin-1 signaling in nerve growth cones. Nat. Neurosci. 9:1257–1264. 10.1038/nn1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., and Ren D.. 2015. Lysosomal physiology. Annu. Rev. Physiol. 77:57–80. 10.1146/annurev-physiol-021014-071649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., and Tashiro Y.. 1998. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23:33–42. 10.1247/csf.23.33 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.