Abstract
Although the majority of animals and plants, including humans, are dominated by the diploid phase of their life cycle, extensive diversity in ploidy level exists among eukaryotes, with some groups being primarily haploid whereas others alternate between haploid and diploid phases. Previous theory has illuminated conditions that favor the evolution of increased or decreased ploidy but has shed little light on which species should be primarily haploid and which primarily diploid. Here, we report a discovery that emerged from host–parasite models in which ploidy levels were allowed to evolve: selection is more likely to favor diploidy in host species and haploidy in parasite species. Essentially, when parasites must evade a host's immune system or defense response, selection favors parasitic individuals that express a narrow array of antigens and elicitors, thus favoring haploid parasites over diploid parasites. Conversely, when hosts must recognize a parasite before mounting a defensive response, selection favors hosts with a broader arsenal of recognition molecules, thus favoring diploid hosts over haploid hosts. These results are consistent with the predominance of haploidy among parasitic protists.
The grand challenge of evolutionary biology is to account for the enormity of biological diversity. This diversity extends to the genomic level, where the number of genomic copies (the ploidy level) varies among species and even over time within species. Although humans and most animals are diploid (with two copies of each gene except during a brief gamete stage), a large number of protists, fungi, algae, and nonvascular plants are primarily haploid (with one copy of each gene) or alternate between haploid and diploid phases (1). Previous theories for the evolution of ploidy level have focused on the influence of mutations that are either deleterious or beneficial (2). These analyses predict that higher ploidy levels are favored when deleterious alleles are partially recessive, because individuals with more than one allele per locus mask these deleterious effects (3, 4). Conversely, lower ploidy levels are favored when beneficial alleles are partially recessive, because the beneficial effects of the mutations are fully revealed in haploids (5). A weakness of these theories is that they have not generated predictions that are easily tested for which species should be haploid and which diploid.
Generating testable predictions has been hampered by the fact that there is no general consensus for how the pattern of dominance should vary among species. Coevolutionary interactions between hosts and parasites (6–8) do, however, generate strong a priori predictions for the patterns of dominance exhibited by traits mediating infection and resistance. We show that, as a consequence, the evolutionary forces acting on the ploidy level of hosts and parasites differ substantially.
To investigate the relationship between species interactions and ploidy evolution, we integrated host–parasite coevolution into existing models of life-cycle evolution. We modeled host–parasite coevolution by using the three forms of genetic interaction that are thought to be the most prevalent (Table 1). In the gene-for-gene (GFG) model (9), avirulence alleles in the parasite produce signal molecules that elicit a defense response in hosts carrying an appropriate resistance allele, whereas parasites carrying virulence alleles are able to infect hosts with either resistance or susceptible alleles (10). GFG interactions are prevalent in plant–pathogen interactions and are generally characterized by host resistance alleles that are dominant and parasite virulence alleles that are recessive (7, 11). Because costs of resistance and virulence have been demonstrated in some GFG systems (12, 13), we also incorporated such fitness costs. The matching alleles model (MAM) (1, 14, 15) is predicated on a system of self/nonself recognition. Hosts can successfully defend against attack by any parasite whose genotype does not match their own. Such recognition systems have been observed in invertebrates (16) and vertebrates (17), where the maturation of the immune system involves the elimination of MHC molecules that bind self-peptides, thus creating the potential for parasites to infect hosts via molecular mimicry. Finally, in the inverse MAM (18) hosts can recognize and defend against any parasite carrying matching alleles. The inverse MAM is the primary mode of action of the vertebrate MHC system, where each host allele has a unique amino acid motif that allows it to bind to a particular suite of parasitic antigens (19, 20). In all three models, we assume that successful infection reduces host fitness but increases parasite fitness (Table 1). Thus we consider only organisms that decrease host fitness through infection to be parasites.
Table 1. An interaction between a host and a parasite results in either infection (I) or resistance (R) depending on the genotypes of the interacting species (A or a for haploids, AA, Aa or aa for diploids).
| Host
|
|||
|---|---|---|---|
| Parasite | A or AA | Aa | a or aa |
| A or AA | {I,I,R} | {I,I,R} | {I,R,I} |
| Aa | {R,R,R} | {R,I,R} | {I,R,R} |
| a or aa | {R,R,I} | {R,I,R} | {I,I,R} |
Each vector represents the outcome of a species interaction under the following three models. GFG interaction: Infection reduces host fitness by γh; resistance reduces parasite fitness by γp. Matching alleles interaction: Infection reduces host fitness by ξh; resistance reduces parasite fitness by ξp. Inverse matching alleles interaction: Infection reduces host fitness by αh; resistance reduces parasite fitness by αp.
To study the evolution of ploidy we assumed that a single ploidy locus controlled the probability that an organism is diploid during infection (4, 5) (Supporting Text and Fig. 2, which are published as supporting information on the PNAS web site). All organisms were assumed to reproduce sexually by random mating, but the probability that meiosis happens soon after the union of gametes (haploids) or before the production of gametes (diploids) was influenced by the ploidy locus. For simplicity we considered the evolution of ploidy in only one of the species at a time and refer to this as the focal species. The ploidy of the nonfocal species was assumed to be diploid (except for a transient haploid gamete stage), although qualitatively identical results were obtained when the nonfocal species was haploid. Interactions between species were assumed to occur at random such that the probability of the focal species interacting as a haploid vs. diploid was directly proportional to the probability of being at that ploidy level. When interactions between species did occur, they were assumed to be mediated by a single locus following the rules imposed by either the GFG model, MAM, or inverse MAM (Table 1).
We took two approaches to analyzing the model. In the first, we assumed that recombination between the ploidy and selected locus was sufficiently frequent relative to the strength of selection to prevent substantial disequilibria from evolving. This allowed us to derive very general conditions for the evolution of ploidy in the focal species by using quasi-linkage equilibrium approximations (21, 22). Because previous theory has shown that tight linkage can favor the evolution of haploidy (4), however, we also used numerical simulations of the exact recursions to evaluate the robustness of our analytical predictions.
Our analytical results demonstrate that a strong difference exists in the ploidy level that ultimately evolves in parasites vs. hosts (see Supporting Text). Selection often favored the evolution of diploidy among hosts, but nearly universally favored the evolution of haploidy among parasites (Table 2).
Table 2. Selectively favored ploidy level for different coevolutionary models.
| Model | Parasite (%) | Host (%) |
|---|---|---|
| GFG | Haploid (100) | Diploid (100) |
| GFG with costs | Haploid* (96.3) | Haploid† (75.9) |
| Diploid* (3.7) | Diploid† (24.1) | |
| Matching alleles | Haploid (100) | Haploid (100) |
| Inverse matching alleles | Haploid (100) | Diploid (100) |
Predictions based on the QLE approximation are summarized (in text), followed by the percentage of simulations in which the predicted ploidy level increased in frequency (in parentheses).
Among parasites in the GFG model with costs, the analysis predicts that haploidy should rise in frequency during periods when resistant alleles are common (during peaks within the cycle) and should decline when resistant alleles are rare (during troughs within the cycle). Simulations demonstrate that the overall tendency is for haploidy to spread among parasites.
Among hosts in the GFG model with costs, the analysis predicts that haploidy should rise in frequency during periods when virulent alleles are common (during peaks within the cycle) and should decline when virulent alleles are rare (during troughs within the cycle). Simulations indicate that there is no overall tendency and that both haploid and diploid life cycles can evolve, depending on the parameters.
These results may be understood intuitively. When parasites must evade a host's immune system or defense response, selection will favor parasitic individuals that express a narrow array of antigens and elicitors, thus favoring haploid parasites over diploid parasites. Conversely, when hosts must recognize a parasite, selection will favor hosts with a broader arsenal of recognition molecules, thus favoring diploid hosts over haploid hosts. The exceptions to these general rules can also be understood intuitively. In the MAM, haploidy is favored among hosts, because hosts bearing fewer alleles are less likely to be mimicked by a parasite. Furthermore, when costs are added to the GFG model, there are periods of time when the dominant resistance alleles are selected against (when virulence is common among parasites) and when the recessive virulence alleles are selected against (when resistance is rare among hosts), during which the evolutionary forces acting on ploidy levels reverse.
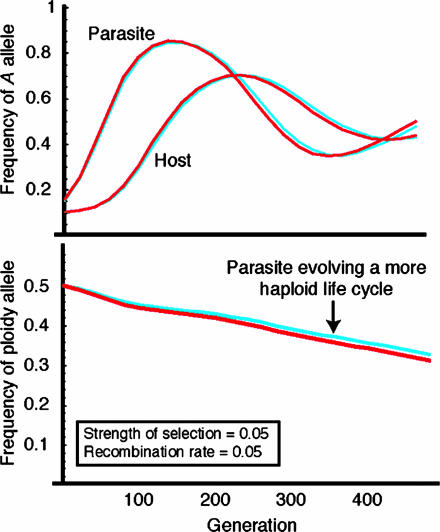
To evaluate whether our analytical results are robust when the key assumption of frequent recombination relative to selection is violated, we numerically iterated the exact recursions (see Supporting Text). For each genetic model of coevolution we considered all combinations of the following selection intensities (0.005, 0.05, and 0.50) and recombination rates (0.005, 0.05, and 0.50), running three simulations for each parameter combination from randomly chosen initial allele frequencies. Furthermore, because selection on ploidy levels is expected to cycle in the GFG model with costs, we iterated the GFG model under two levels for costs of resistance and virulence (5% and 25% of the selection intensity) to evaluate the total selective force. In no case did the numerical and analytical predictions for ploidy evolution differ qualitatively, even as the recombination rate was reduced to 0.005 and the strength of selection increased to 0.50 (Table 2). Furthermore, the match between the analytical solution and the simulations was quite accurate even when recombination rates and survival differences were similar in magnitude (Fig. 1).
Fig. 1.
Comparison of quasi-linkage equilibrium predictions (blue) and exact numerical results (red) for the inverse MAM. (Upper) Predicted values of allele frequencies at the loci governing coevolutionary interactions in the two species. (Lower) The predicted frequency of an allele that increases the probability that a parasite is diploid.
In the GFG model with costs, haploidy was often favored in the hosts (75.9% of simulation runs), in contrast to the model without costs. This result arises because parasite virulence alleles often reach and maintain high frequencies in the simulations, a scenario shown by our analytical results to favor the evolution of host haploidy. In contrast, adding costs had virtually no impact on parasite ploidy evolution. The frequency of haploid parasites increased in 96.3% of the simulations with costs compared to 100% of the simulations in the absence of costs. Thus, the simulations confirm our central result: coevolutionary interactions virtually always favor the spread of haploidy among parasites, in contrast to hosts where diploidy is often favored.
These striking results make good sense in light of previous theory, which predicts that diploidy should evolve if new favorable mutations are dominant (5). For the common genetic scenarios of resistance and infection that we have considered, alleles that increase host resistance tend to be dominant whereas alleles that increase the parasite's ability to infect the host tend to be recessive. Our results, while based on standing genetic variation rather than new mutations, fit neatly into these existing theoretical predictions.
Although we have focused on the evolution of ploidy levels, the reasoning underlying our results suggests that parasitic lifestyles should more easily evolve in haploids than diploids, as parasitic individuals in haploid populations will be able to infect a larger fraction of host individuals. Similarly, selection should favor gene duplication and coexpression of resistance genes in hosts [as has been observed in both the R gene family in plants (23, 24) and the MHC and Ig gene families in animals (25)]. In contrast, gene duplication of antigen or elicitor genes should be selected against in parasites unless mechanisms exist to limit expression to few members of the gene family. Indeed, such mechanisms have been repeatedly observed. For example, among protistan parasites, Trypanosomes typically express only one of thousands of variant surface glycoprotein genes (26); Giardia express only one of 30–150 variant-specific surface protein genes (27); ciliates also express only one of many genes encoding surface antigens (28).
To assess the relationship between host–parasite interactions and ploidy levels, we surveyed data on heterotrophic protists to determine whether a correlation exists between protists engaging in parasitic lifestyles and ploidy level (Table 3 and Supporting Data Set, which is published as supporting information on the PNAS web site). Although both haploid and diploid protists engage in parasitism, parasitic protists are approximately three to four times as likely to be haploid as are nonparasitic protists. Specifically, haploids account for 2,573 of the 4,041 total species of protists with parasitic lifestyles. In contrast, haploids account for only 1,465 of the 8,749 total species of protists with nonparasitic lifestyles (Table 3).
Table 3. Survey of ploidy levels among heterotrophic protists.
| Diploid parasite (≈ 1,468-1,963 spp.) | Haploid parasite (≈ 2,573-3,068 spp.) | Diploid nonparasite (≈ 7,284 spp.) | Haploid nonparasite (≈ 1,465 spp.) |
|---|---|---|---|
| Some ciliates | All apicomplexa | Most ciliates | All cellular slime molds |
| Some diplomonads | Some dinoflagellates | Most diplomonads | Most dinoflagellates† |
| Some lobose amoebae | All phytomyxids | All heliozoa | Most hypermastigotes |
| Some oomycetes | Some trichomonads | Some hypermastigotes | Some oxymonads |
| Some trypanosomes | Some trypanosomes | Most lobose amoebae | Most trichomonads |
| Some oomycetes | |||
| All opalinids* | |||
| Some oxymonads |
Phototrophic protists were excluded. Also not included are a number of protists with mitotic divisions in both haploid and diploid phases (including acellular slime molds, foraminifera, radiolaria, and some heterotrophic haptophytes and cryptomonads), which are typically not parasitic. References and further details are available in Supporting Text and Supporting Data Set.
Although often classified as parasites, opalinids are more likely commensals, because there is no evidence that they harm their hosts (30).
Approximately half of the dinoflagellates are photoauxotrophic, but the remainder are heterotrophic.
Even when a parasite is genetically diploid, it might not be functionally diploid with respect to interactions with its host. A tantalizing example is Trypanosoma brucei. Whereas the majority of its genome is diploid, most of the genomic regions in which the variant surface glycoproteins reside are haploid and lack homologous chromosomal segments (29). This finding is perhaps the best evidence that the selective forces described by our model are acting on parasites, favoring haploidy either in full or in part.
We have demonstrated that host–parasite dynamics favor haploidy more often in parasites than in hosts. This finding is consistent with the greater species diversity of parasitic protists than nonparasitic protists among haploids but not diploids (Table 3). The parasitic haploid groups include the agriculturally important pathogens Plasmodiophora brassicae (club root disease), Spongospora subterranean (powdery scab of potato) and the human pathogens Plasmodium spp. (malaria), Toxoplasma gondii (Toxoplasmosis), and Trichomonas vaginalis (a prevalent sexually transmitted disease). Despite this generally favorable accord between theory and data, several caveats must be mentioned. Most importantly, the correlation between haploidy and parasitic lifestyles (Table 3) is based on raw species numbers and might not be robust to phylogenetic correction (see Supporting Text). Furthermore, ploidy levels are not definitively known for most protists. Our categorization of species as parasitic or nonparasitic must also be regarded as tentative, because fitness effects on the host are often unmeasured. Furthermore, it is misleading to view protists solely as potential parasites, as they often act as hosts themselves to a variety of viruses and bacteria, frustrating the simple application of our model. Finally, our model is based on the simplest forms of genetic interactions among species. As molecular data accumulate shedding light on host–pathogen interactions and the form of dominance of antigens, elicitors, and receptors, our model may be further refined to improve our understanding of the evolutionary forces acting on genomic copy number.
Supplementary Material
Acknowledgments
We thank Aneil Agrawal, Mark Dybdahl, Larry Hufford, Curt Lively, Andy Peters, Barrie Robison, and Michael Whitlock for helpful comments. Funding was provided by National Science Foundation Grant DEB-0343023 (to S.L.N.) and the Natural Sciences and Engineering Research Council (Canada) (S.P.O.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GFG, gene-for-gene; MAM, matching alleles model.
References
- 1.Bell, G. (1982) The Masterpiece of Nature: The Evolution and Genetics of Sexuality (Univ. of California, Berkeley).
- 2.Mable, B. K. & Otto, S. P. (1998) BioEssays 20, 453–462. [Google Scholar]
- 3.Perrot, V., Richerd, S. & Valero, M. (1991) Nature 351, 315–317. [DOI] [PubMed] [Google Scholar]
- 4.Otto, S. P. & Goldstein, D. B. (1992) Genetics 131, 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orr, H. A. & Otto, S. P. (1994) Genetics 136, 1475–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burdon, J. J. (1987) Oecologia 73, 257–267. [DOI] [PubMed] [Google Scholar]
- 7.Burdon, J. J. (1997) in The Gene-for-Gene Relationship in Plant–Parasite Interactions, eds. Crute, I. R., Holub, E. B. & Burdon, J. J. (CAB International, New York), pp. 245–262.
- 8.Cartea, M. E., Malvar, R. A., Vales, M. I., Butron, A. & Ordas, A. (2001) J. Econ. Entomol. 94, 277–283. [DOI] [PubMed] [Google Scholar]
- 9.Flor, H. H. (1956) Adv. Genet. 8, 29–54. [Google Scholar]
- 10.Thompson, J. N. & Burdon, J. J. (1992) Nature 360, 121–125. [Google Scholar]
- 11.De Wit, P. J. G. M. (1987) in Fungal Infection of Plants, eds. Pegg, G. F. & Ayres, P. G. (Cambridge Univ. Press, Cambridge), pp. 1–24.
- 12.Thrall, P. H. & Burdon, J. J. (2003) Science 299, 1735–1737. [DOI] [PubMed] [Google Scholar]
- 13.Tian, D., Traw, M. B., Chen, J. Q., Kreitman, M. & Bergelson, J. (2003) Nature 423, 74–77. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton, W. D. (1980) Oikos 35, 282–290. [Google Scholar]
- 15.Peters, A. D. & Lively, C. M. (1999) Am. Nat. 154, 393–405. [DOI] [PubMed] [Google Scholar]
- 16.Grosberg, R. K. & Hart, M. W. (2000) Science 289, 2111–2114. [DOI] [PubMed] [Google Scholar]
- 17.Klein, J. & O'Huigin, C. (1994) Philos. Trans. R. Soc. London B 346, 351–357; discussion 357–358. [DOI] [PubMed] [Google Scholar]
- 18.Frank, S. A. (1997) in Metapopulation Biology, eds. Hanski, I. & Gilpen, M. E. (Academic, San Diego), pp. 325–352.
- 19.Hughes, A. L., Hughes, M. K., Howell, C. Y. & Nei, M. (1994) Philos. Trans. R. Soc. London B 346, 359–366; discussion 366–367. [DOI] [PubMed] [Google Scholar]
- 20.Frank, S. A. (2002) Immunology and the Evolution of Infectious Disease (Princeton Univ. Press, Princeton). [PubMed]
- 21.Nagylaki, T. J. (1993) Genetics 134, 627–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirkpatrick, M., Johnson, T. & Barton, N. (2002) Genetics 161, 1727–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beynon, J. L. (1997) in The Gene-For-Gene Relationship in Plant–Parasite Interactions, eds. Crute, I. R., Holub, E. B. & Burdon, J. J. (CAB International, New York), pp. 359–378.
- 24.Young, N. D. (2000) Curr. Opin. Plant Biol. 3, 285–290. [DOI] [PubMed] [Google Scholar]
- 25.Nei, M., Gu, X. & Sitnikova, T. (1997) Proc. Natl. Acad. Sci. USA 94, 7799–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donelson, J. E. (1995) J. Biol. Chem. 270, 7783–7786. [DOI] [PubMed] [Google Scholar]
- 27.Svard, S. G., Meng, T. C., Hetsko, M. L., McCaffery, J. M. & Gillin, F. D. (1998) Mol. Microbiol. 30, 979–989. [DOI] [PubMed] [Google Scholar]
- 28.Kusch, J. & Schmidt, H. J. (2001) J. Membr. Biol. 180, 101–109. [DOI] [PubMed] [Google Scholar]
- 29.Melville, S. E., Leech, V., Navarro, M. & Cross, G. A. (2000) Mol. Biochem. Parasitol. 111, 261–273. [DOI] [PubMed] [Google Scholar]
- 30.Margulis, L., Corlis, J. O., Melkonian, M. & Chapman, D. J., eds. (1990) Handbook of Protoctista: The Structure, Cultivation, Habitats, and Life Histories of Eukaryotic Microorganisms and Descendants Exclusive of Animals, Plant, and Fungi (Jones & Bartlett, Boston).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.