p190RhoGAP (p190A) is a negative regulator of RhoA and localizes to membrane protrusions, where its GAP activity is required for directional migration. Here, Binamé et al. identify the protrusion-localization sequence in p190A and show that cancer-associated mutations in this region affect p190A localization and function as well as tumor cell migration.
Abstract
Spatiotemporal regulation of RhoGTPases such as RhoA is required at the cell leading edge to achieve cell migration. p190RhoGAP (p190A) is the main negative regulator of RhoA and localizes to membrane protrusions, where its GTPase-activating protein (GAP) activity is required for directional migration. In this study, we investigated the molecular processes responsible for p190A targeting to actin protrusions. By analyzing the subcellular localization of truncated versions of p190A in hepatocellular carcinoma cells, we identified a novel functional p190A domain: the protrusion localization sequence (PLS) necessary and sufficient for p190A targeting to leading edges. Interestingly, the PLS is also required for the negative regulation of p190A RhoGAP activity. Further, we show that the F-actin binding protein cortactin binds the PLS and is required for p190A targeting to protrusions. Lastly, we demonstrate that cancer-associated mutations in PLS affect p190A localization and function, as well as tumor cell migration. Altogether, our data unveil a new mechanism of regulation of p190A in migrating tumor cells.
Introduction
Cell migration plays key roles in embryonic development, immunity, angiogenesis, and tumor metastasis. Efficient cell locomotion requires polarized processes: membrane protrusions at the front side and retraction at the trailing side. This occurs through the coordinated regulation of actin dynamics and integrin-mediated adhesion to the substratum. At the leading edge, the actin-based protrusions lamellipodia and filopodia, respectively, flattened protrusions and microspikes, contribute to cell movement (Ridley, 2011). Downstream of membrane-bound receptors, the RhoGTPases have emerged as major regulators of the formation of F-actin–rich protrusions. RhoGTPases associate with plasma membrane under their GTP-bound form and function by facilitating the formation of effector complexes at the right time and place. Spatiotemporal analysis of the process revealed that RhoA plays a role in the onset of the protrusion, whereas Rac1 and Cdc42 are involved in the reinforcement and stabilization of the newly expanded protrusion (Machacek et al., 2009). In addition, the reciprocal balance between these GTPases activity determines cell movement. Indeed, Rac1 promotes cellular protrusion, which counteracts RhoA signaling. RhoGTPase activation is tightly regulated by the coordinated action of guanine nucleotide exchange factors (GEFs), which facilitate GTP loading and GTPase-activating proteins (GAPs), which promote GTPase inactivation by enhancing GTP hydrolysis.
p190RhoGAP (also known as ARHGAP35 or GRLF1 and hereafter called p190A) is an important regulator of RhoA activity involved in the antagonism between RhoA and Rac1 at cell protrusions (Herbrand and Ahmadian, 2006; Bustos et al., 2008). p190A was first described as a tyrosine phosphorylated protein in v-Src–transformed cells (Ellis et al., 1990; Settleman et al., 1992). This phosphorylation promotes the association of p190A with p120RasGAP and its recruitment to the plasma membrane (McGlade et al., 1993; Bryant et al., 1995; Hu and Settleman, 1997; Roof et al., 1998). p190A is responsible for RhoA inactivation upon integrin signaling in fibroblasts (Nakahara et al., 1998; Arthur et al., 2000; Bradley et al., 2006; Bass et al., 2008). It also plays a central role in axon outgrowth and neural development (Brouns et al., 2000, 2001) and is a mechanosensitive master switch to determine lineage-type specification in the cardiac tissue (Kshitiz et al., 2014).
To date, only few studies are available on the function of p190A in cancer. Early studies demonstrated that p190A inhibition results in transformation of NIH/3T3 fibroblasts, whereas the overexpression of its GAP domain inhibits Ras-dependent transformation (Wang et al., 1997). This tumor-suppressor function was confirmed in oligodendroglioma and pancreatic cancer (Wolf et al., 2003; Kusama et al., 2006). However, high expression of p190A mRNA is associated with advanced state of lung carcinoma, and its expression in lung adenocarcinoma and breast carcinoma correlates with cell proliferation, migration, and invasion, arguing for an oncogenic role (Shen et al., 2008; Notsuda et al., 2013). Recently, ARHGAP35, the gene encoding p190A, was reported highly significantly mutated in 15% of endometrial tumors and 2% of global cancers (Lawrence et al., 2014). In this large-scale study, the nonsense mutations support a tumor suppressor function of p190A. Surprisingly, among the large number of missense mutations, none of them target the phosphorylated amino acids implicated in the regulation of p190A (Roof et al., 1998; Tatsis et al., 1998; Haskell et al., 2001; Jiang et al., 2008; Lévay et al., 2009). To get insight into the impact of cancer-associated mutations on p190A function, we performed a structure/function analysis of the protein. This approach led to the identification of a sequence sufficient to ensure p190A targeting to cell protrusions, such as lamellipodia. Moreover, a construct of p190A deleted of the identified protrusion localization sequence (p190AΔPLS) is mislocalized and cannot target to these actin-based structures. Functional characterization of p190AΔPLS revealed that deletion of the PLS increased the RhoGAP activity of p190A in cells. We further identified cortactin as a binding partner of the PLS and demonstrated its requirement for p190A targeting to lamellipodia’s edge. Finally, we identified and characterized cancer-associated mutations in the PLS that affect p190A in a similar way to PLS deletion, unveiling a new regulatory mechanism of this RhoGAP in tumor pathophysiology.
Results
Identification of the PLS, the domain responsible for the localization of p190A to actin-based protrusions
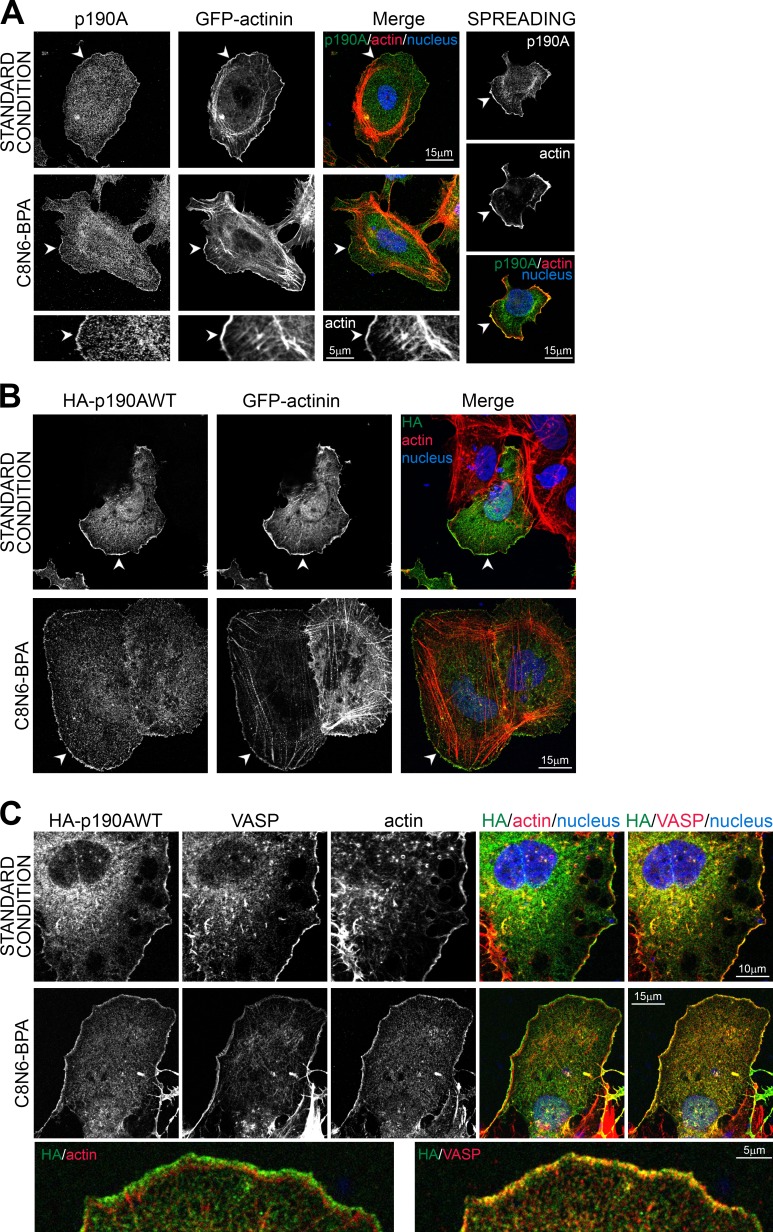
In human hepatocellular carcinoma Huh7 cells, endogenous p190A localizes to cell edges such as membrane ruffles and lamellipodia (Fig. 1 A), as previously described in other cell types (Tsubouchi et al., 2002; Bass et al., 2008; Guegan et al., 2008). In standard conditions in which cells were plated overnight on glass coverslips, p190A colocalizes with cortical F-actin at the cell periphery. To unambiguously show the localization of p190A to actin-rich protrusions, we induced lamellipodium outgrowth by short-term spreading on fibronectin or by chemical treatment with the lamellipodial growth promoter C8N6-branched polyamine (BPA; Nedeva et al., 2013). Endogenous p190A colocalizes along lamellipodium’s edge with actin and α-actinin, an F-actin cross-linking protein found at the leading edge of migrating cells (Langanger et al., 1984; Knight et al., 2000). When expressed in cells, wild-type (WT) HA-tagged p190A (HA-p190AWT) localized similarly to the endogenous protein in standard and stimulated conditions (Fig. 1 B). Moreover, costaining with anti–vasodilator-stimulated phosphoprotein (VASP) antibodies revealed that HA-tagged p190A localized at the very edge of lamellipodia (Fig. 1 C).
Figure 1.
p190A localizes to membrane protrusions in Huh7 cells. (A) As a standard condition, Huh7 cells were plated on glass coverslips and then fixed after overnight culture. Lamellipodium growth was either stimulated by spreading on fibronectin for 30 min (top) or by treatment with 100 µM C8N6-BPA (middle). Huh7 cells were transfected with α-actinin–GFP used as a lamellipodium marker, fixed, and stained for p190A (green), F-actin (red), and nuclei (blue). Bottom panel shows enlargement of C8N6-BPA condition. (B) Huh7 cells were transfected with HA-p190AWT and α-actinin–GFP, fixed, and stained for HA tag (green), F-actin (red), and nuclei (blue). (C) Huh7 cells were transfected with HA-p190AWT, fixed, and stained for HA tag (green), VASP (red) or F-actin (red), and nuclei (blue). Bottom panel shows enlargement of C8N6-BPA condition. (A and B) Arrowheads show membrane ruffles with p190A/HA-p190AWT localization.
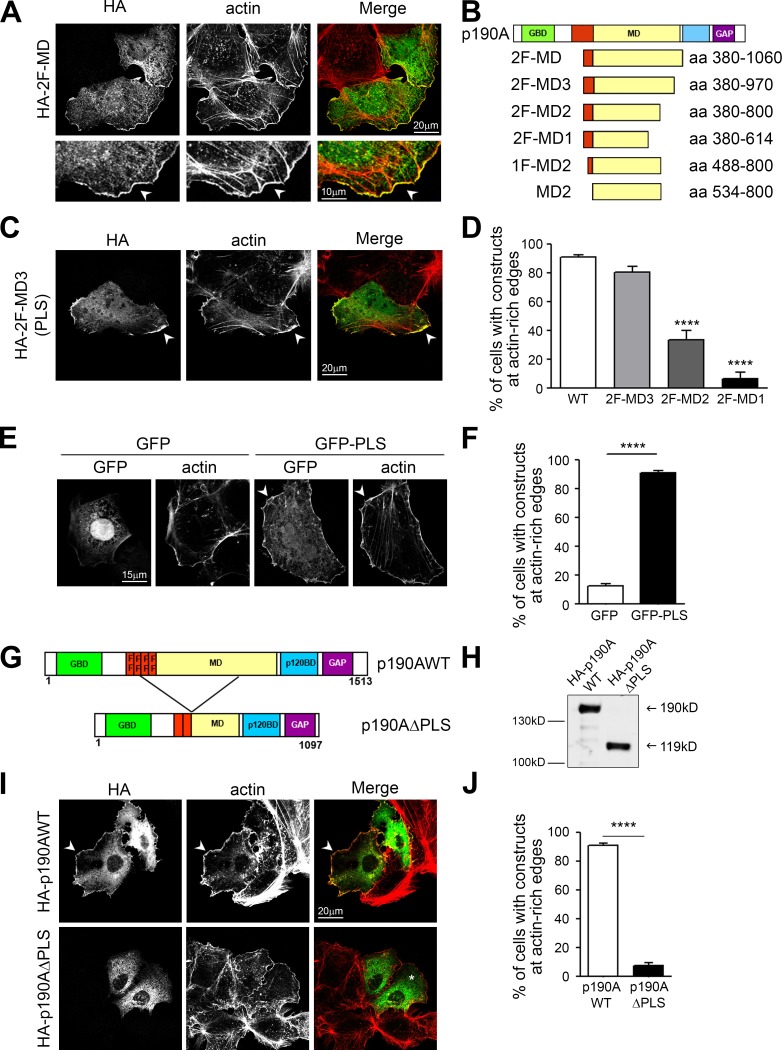
p190A is a large multidomain protein including an N-terminal GTP-binding segment (GBD) followed by four FF domains (protein–protein-interacting modules harboring two strictly conserved phenylalanine residues), a middle domain (MD), a p120RasGAP binding domain (p120BD), and the C-terminal RhoGAP domain (GAP). To functionally characterize p190A domains, we generated 12 HA-tagged truncated versions of p190A overlapping the different domains as described in Table S3. Each construct was then transfected into Huh7 cells, and actin remodeling and recombinant protein localization were analyzed by immunofluorescence microscopy (Fig. S1). Due to its RhoGAP activity, overexpression of HA-p190AWT in cells induces stress fiber loss (Ridley et al., 1993). The same pattern was observed for each construct containing the GAP domain (Fig. S1; p120BD-GAP, GAP, ΔGBD, and ΔGBD-2F). On the contrary, all of the constructs without this catalytic domain left the stress fibers unaffected (Fig. S1; Δp120BD-GAP, GBD-4F, 4F, 4F-MD, 2F-MD-p120BD, p120BD, 2F-MD, and 2F), confirming that the GAP domain alone is sufficient to promote actin cytoskeleton reorganization. This analysis also led to the identification of a construct composed of the two last FF domains and the MD of p190A (2F-MD) as the minimal p190A protein that colocalizes with F-actin at membrane protrusions (Fig. 2 A; Fig. S1; and Table S3). This enrichment at actin-rich edges was not observed for many other soluble HA-tagged truncated constructs, ruling out artifactual staining attributable to an increased membrane thickness in this region (Fig. S1). To determine the minimal PLS, N- and C-terminal truncations of the 2F-MD construct were performed (Fig. 2 B). A deletion of 90 aa from the C-terminal end of the 2F-MD construct did not alter its membrane targeting (2F-MD3; Fig. 2, C and D). However, further C-terminal truncation dramatically decreased its localization to actin-rich edges (Fig. 2, B and D; and Fig. S2). Furthermore, truncation from the N-terminal end (1F-MD2 and MD2) altered the specificity of the localization (Fig. S2). Indeed, the suppression of the FF domains led to the nonspecific localization of p190A mutants to all F-actin structures, including stress fibers and focal adhesions (Fig. S2). We can therefore hypothesize that the MD mediates the localization of p190A to polymerized actin, whereas the FF domains coupled to the MD specify actin-rich protrusion targeting. Hence, our results demonstrate that the 2F-MD3 construct, corresponding to aa 380–971, is the minimal domain sufficient to target p190A to membrane protrusions and is therefore p190A PLS.
Figure 2.
Identification of a domain targeting p190A to cell protrusions. (A) A region of p190A containing the two last FF motifs and the MD (2F-MD, 380–1,060 aa) was generated (see Table S3). Huh7 cells were transfected with HA-2F-MD, fixed, and stained for HA (green) and F-actin (red). Bottom panels show membrane outgrowth at higher magnification. Arrowheads show cell edge where p190A constructs and F-actin colocalize. (B) Schematic representation of N and C terminus truncations of 2F-MD. (C) Immunostaining of Huh7 cells transfected with HA-2F-MD3, fixed, and stained as in A. (D) Quantification of the number of cells presenting a localization of p190A constructs at actin-rich edges. Values are expressed as the mean ± SEM (n > 200 for each construct; three to four independent experiments). ****, P < 0.0001 when compared with p190AWT condition, by ANOVA followed by Tukey’s multiple-comparison test. (E) Huh7 cells were transfected with GFP or GFP-PLS, fixed, and stained for actin. Arrowheads point out colocalization of p190A constructs and F-actin. (F) Quantification of cells showing localization of GFP or GFP-PLS at actin-rich edges. Values are expressed as the mean ± SEM (n = 360; three independent experiments). P-value from the unpaired t test is indicated. ****, P < 0.0001. (G) Schematic representation of the p190AΔPLS protein compared with the full-length p190A protein (p190AWT). (H) Western blot analysis of Huh7 cells transiently expressing the recombinant proteins HA-p190AWT and HA-p190AΔPLS. (I) Huh7 cells were transfected with HA-p190AWT or HA-p190AΔPLS, fixed, and immunostained for HA (green) and F-actin (red). Arrowheads show actin-rich edges with construct localization; * indicates the cytoplasmic localization of p190AΔPLS. (J) Quantification of (I), indicating percentage of cells showing HA-p190AWT or HA-p190AΔPLS at actin-rich edges. Values are expressed as the mean ± SEM (n = 715; three independent experiments). P-value from the unpaired t test is indicated. ****, P < 0.0001.
PLS is necessary and sufficient to target p190A to actin-based protrusions
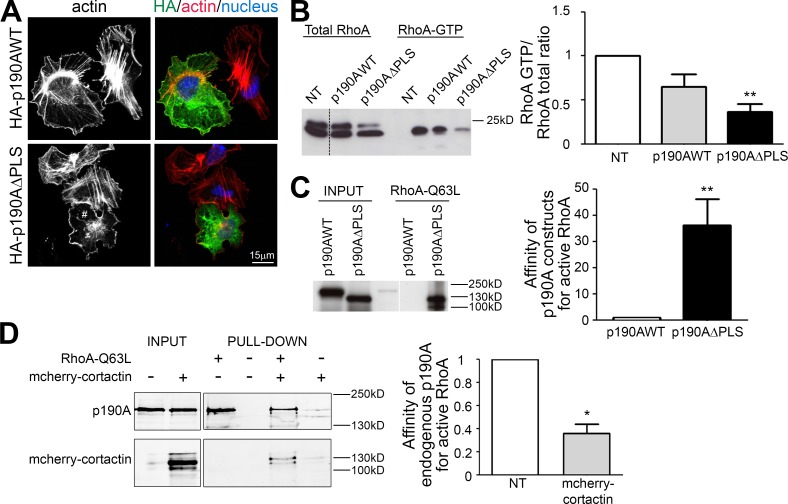
Considering this domain as a functional PLS would imply its ability to target irrelevant proteins to cell leading edges. To test this, the PLS of p190A was fused to GFP, and the localization of the fusion protein was determined by confocal microscopy. Compared with GFP alone, which showed cytoplasmic and nuclear localizations, GFP-PLS chimera was strongly targeted to actin-rich edges (Fig. 2, E and F). Hence, the PLS of p190A is sufficient to target proteins to the cell periphery where ruffles and lamellipodia are formed. To demonstrate that the PLS domain is required for p190A localization to membrane protrusions, a p190AΔPLS mutant, corresponding to the protein lacking the PLS domain, was constructed and expressed in Huh7 cells (Fig. 2, G and H). Although HA-p190AWT strongly localized to cell edges (mean 91.3 ± 1.1%), the HA-p190AΔPLS mutant was not targeted to these structures (mean 7.7 ± 1.8%; Fig. 2, I and J).
We next investigated whether p190A constructs localize to Rac1-induced lamellipodia. To this end, we used porcine aortic endothelial (PAE) cells expressing a constitutively active form of Rac1 (Rac1G12V) in an IPTG-inducible manner (PAE-V12Rac1; Welch et al., 1998). As expected, IPTG-driven expression of V12Rac1 induced spreading of the majority of PAE cells (Fig. S3, A and B). These cells form continuous lamellipodia covering the entire cell periphery, where p190AWT colocalizes with F-actin (Fig. S3 C). The expression of PLS and p190AΔPLS in these cells confirmed that the PLS localized to the lamellipodia, whereas p190A devoid of this domain did not (Fig. S3 C). Overall, our results demonstrate that the PLS is the minimal domain necessary and sufficient for p190A localization to ruffles and lamellipodia.
p190A interacts with cortactin via the PLS
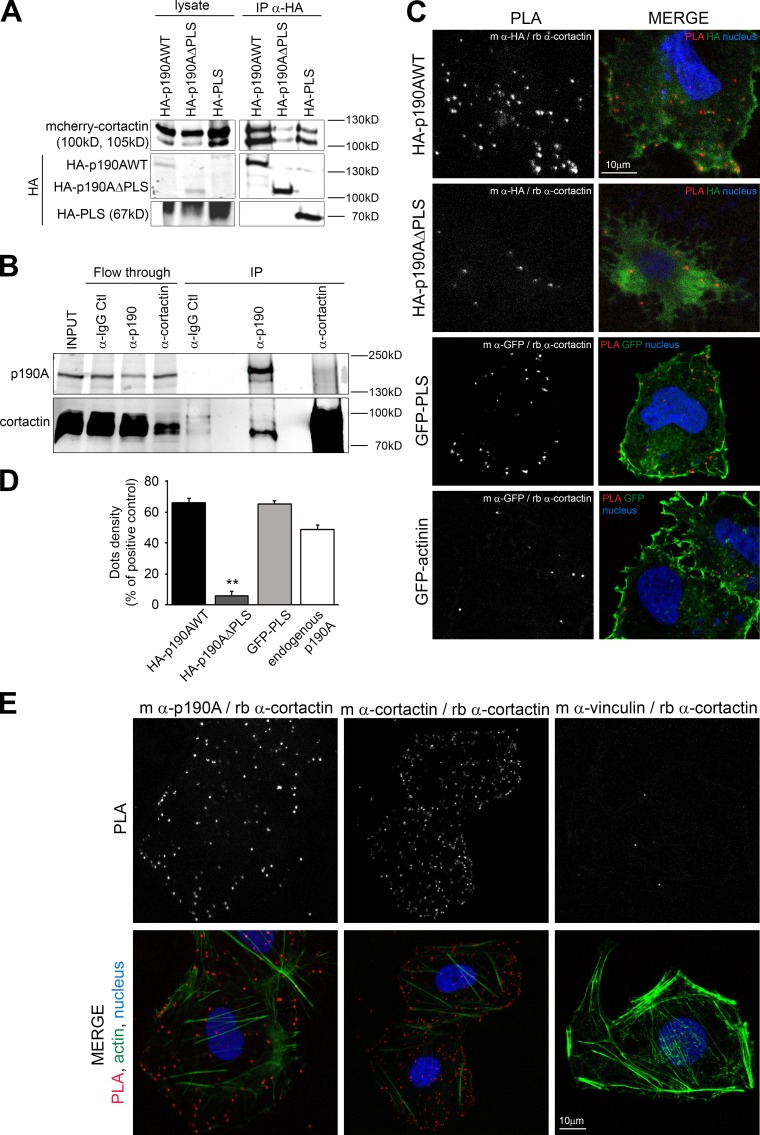
To further characterize the molecular mechanisms responsible for PLS function, we searched for PLS binding partners. As PLS targets proteins to membrane protrusions, we first tested a potential interaction of the PLS with phospholipids. Phospholipid binding assays were performed using purified GST-PLS on PIP strip membranes. Unlike PLC-γ1–PH, which corresponds to the pleckstrin homology domain of the PLC-γ1 and Rgd1, a yeast RhoGAP protein (Prouzet-Mauléon et al., 2008), PLS was not able to interact with phospholipids (Fig. S4 A). We then performed GST pulldown assays using GST-PLS as bait in Huh7 cell lysates. Candidate partner proteins were screened by immunoblotting based on shared characteristics with p190A, such as association with F-actin and localization to membrane protrusions. This approach successfully identified cortactin as a specific partner of PLS, contrary to the actin-related protein 2 (Arp2), which did not bind to GST-PLS (Fig. S4 B). Cortactin is a crucial regulator of actin cytoskeleton dynamics involved in lamellipodial persistence through the binding of the Arp2/3 complex and F-actin (Bryce et al., 2005). We confirmed this interaction in Huh7 cells by coimmunoprecipitating transiently expressed mcherry-cortactin with HA-tagged PLS and p190AWT. In the same conditions, p190AΔPLS displayed a dramatically reduced binding to cortactin (Fig. 3 A). The interaction between p190A and cortactin was also confirmed on endogenous proteins by coprecipitating cortactin using anti-p190 antibody in Huh7 cells (Fig. 3 B). To visualize the interaction between cortactin and p190A in the cell, we used the proximity ligation assay (PLA). As shown in Fig. 3 (C and D), PLS-containing constructs HA-p190AWT and GFP-PLS displayed dots of interaction with cortactin in the whole cell body with an increased density at the plasma membrane. α-Actinin–GFP used as negative control and HA-p190AΔPLS displayed only background dots (Fig. 3, C and D). To avoid any potential artifact from expression difference between constructs, PLA was then tested using antibodies directed against endogenous p190A and cortactin. As positive and negative controls, antibodies directed, respectively, against two epitopes of cortactin or two noninteracting proteins present at the cell periphery, cortactin and vinculin, were used. As expected, the positive control produced many dots in the whole cell with a greater density at membrane ruffles where cortactin localizes, whereas the negative control produced only few background dots (Fig. 3 E). PLA performed with endogenous p190A and cortactin revealed dots in the whole cell but preferentially at the cell edges (Fig. 3 E, left). Quantification of dots density (Fig. 3 D) confirmed the close association of p190A and cortactin and the requirement of PLS in this association.
Figure 3.
p190A interacts with cortactin via the PLS domain. (A) Huh7 cells transfected with both mcherry-cortactin and indicated HA-tagged p190A plasmids were used to perform immunoprecipitation (IP) with anti-HA beads. Presence of cortactin is analyzed by immunoblot using an anti-cortactin antibody. (B) Coimmunoprecipitation was performed on Huh7 lysates with IgG control (Ctl), anti-p190A, and anti-cortactin antibodies. PLA was performed on Huh7 cells transfected (C) or not (E) with the indicated constructs. Cells were stained for nuclei (Hoechst, blue). The green staining represents, as shown on each panel, the HA staining, GFP tag, or F-actin staining. The dots in the PLA panel represent locations where interaction between indicated proteins (stained using primary antibodies from mouse [m] or rabbit [rb] species) occurs; dots density is quantified in D. Values are expressed as the mean ± SEM of three independent experiments (n = 9 cells per condition). P-value from the ANOVA test is indicated. **, P < 0.01.
Cortactin is required for p190A targeting to actin-based protrusions
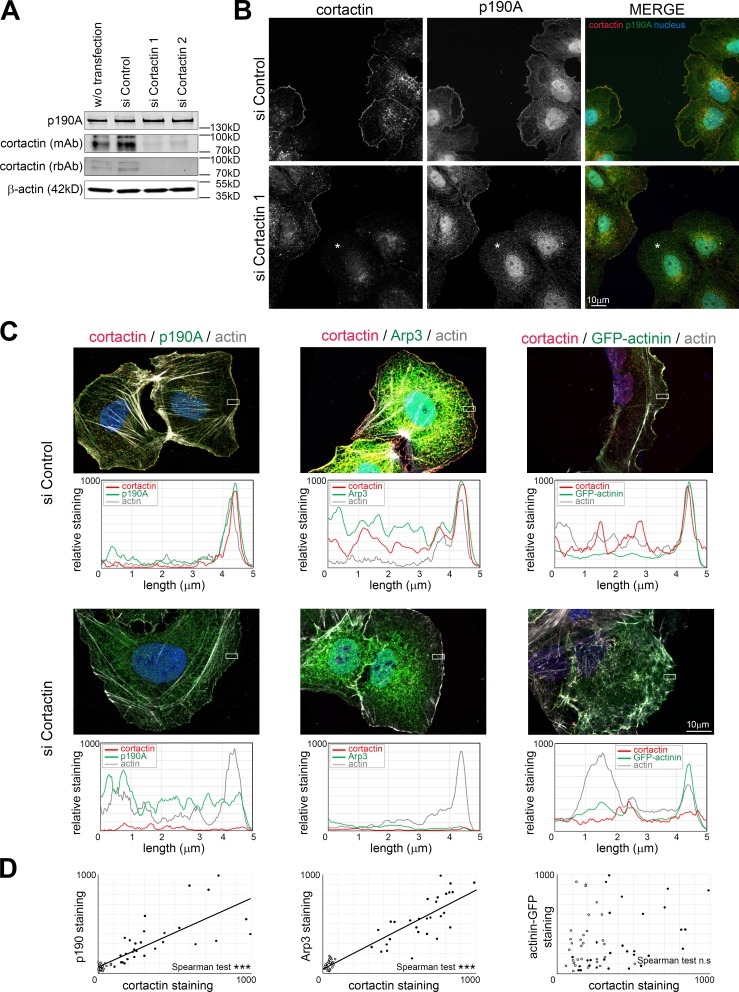
To test whether cortactin is required for p190A targeting to membrane protrusions, we studied the impact of cortactin silencing on p190A subcellular localization. To study this mechanism in a well-defined structure, we induced lamellipodia with C8N6-BPA in Huh7 cells. Silencing cortactin with two different siRNAs produced a similar and dramatic decrease of cortactin expression without affecting p190A level (Fig. 4 A). Lamellipodia induced in Huh7 cells transfected with control siRNA presented both cortactin and p190A at their edge (Fig. 4 B). When siRNA targeting cortactin is transfected, cells exhibiting the most efficient silencing of cortactin displayed a defect in p190A staining at lamellipodia’s edge. We quantified the effect of cortactin knockdown on the localization of p190A at the cell periphery (Fig. 4, C and D; and Fig. S4 C). Arp3, a known interacting partner of cortactin, and α-actinin were used as positive and negative controls, respectively. Measurement of relative staining intensities in cells treated with control siRNA showed peaks of actin, cortactin, Arp3, and α-actinin–GFP at the lamellipodium’s edge, as expected. Downregulation of cortactin erased the peak of cortactin without affecting actin or α-actinin–GFP expression at the cell edge. However, it reduced the peak of Arp3 and p190A. These results, together with the highly significant correlation found between cortactin staining and both Arp3 and p190A at lamellipodium’s edge (Fig. 4 D and Fig. S4 C), confirm the role of cortactin in Arp3 recruitment (Weaver et al., 2002) and reveal the requirement of cortactin for p190A targeting (Fig. 4 C).
Figure 4.
Cortactin recruits p190A to the lamellipodium’s edge. Huh7 cells were transfected with control siRNA (siControl) or siRNA targeting cortactin (siCortactin1 and 2) and treated with 100 µM C8N6-BPA to induce lamellipodia growth. (A) The expression of endogenous p190A and cortactin was assayed by immunoblotting with an anti-p190A antibody and two antibodies from different species (mAb from mouse and rbAb from rabbit) against cortactin. β-Actin is used as a loading control. (B) Immunofluorescence staining of cortactin (red) and endogenous p190A (green) was performed on Huh7 cells transfected with siControl or siCortactin1. Asterisk indicates cells with absence of cortactin staining. (C) Huh7 cells transfected with siControl or siCortactin1 were immunostained as indicated on each panel. Graphs under each panel represent the quantification of the relative staining of cortactin (red), actin (gray), and p190A, Arp3, or α-actinin–GFP (green) determined inside the white rectangle of 2 × 5 µm. Results are expressed as mean value of the height. (D) Correlation of cortactin staining with p190A, Arp3, and α-actinin–GFP at the lamellipodium edge. Mean values of membrane areas of 2 × 0.28 µm are plotted on the graph. Closed and open circles represent measurement in cells transfected with control siRNA and siCortactin1, respectively (n = 30/condition). Linear regression is presented when Spearman correlation is significant. ***, P < 0.001; n.s, not significant.
PLS is required for the autoinhibition of p190A GAP activity
To functionally characterize the PLS domain, we analyzed the impact of its deletion on the RhoGAP activity of p190A. Because RhoA activity controls stress fibers and focal adhesion maturation and disassembly (Binamé et al., 2010), we analyzed the effect of p190A constructs expression on actin cytoskeleton (Fig. 5 A). 3 h after plating on a highly adhesive substrate of fibronectin, untransfected cells displayed many stress fibers, cells expressing HA-p190AWT less fibers, and cells expressing HA-p190AΔPLS no fibers at all (Figs. 5 A and 6 D for quantification), suggesting that the expression of this latter construct resulted in a higher RhoGAP activity in cells. We thus analyzed RhoA activation using a GST-Rhotekin pulldown assay in p190AWT- or p190AΔPLS-transfected cells (Fig. 5 B). The expression of both constructs resulted in the inhibition of RhoA, with a more important effect observed for the p190AΔPLS mutant, arguing for an enhanced GAP activity in cells. Hence, deletion of the PLS increases the protein capacity to inhibit cellular RhoA and remodel actin cytoskeleton. The strong effect of ΔPLS may result from increased intrinsic GAP catalytic activity toward RhoA. To analyze the level of activation of p190AWT and -ΔPLS, we performed an affinity precipitation assay with the activated form of RhoA (RhoAQ63L) that binds with high affinity to activated GAPs and effectors in the cell (García-Mata et al., 2006). Recombinant GST-RhoAQ63L was incubated with protein extracts of Huh7 cells expressing either HA-p190AWT or HA-p190AΔPLS. Interestingly, p190AΔPLS exhibited a higher affinity for active RhoA than p190AWT (Fig. 5 C), indicating that the GAP domain is more inclined to interact with active RhoA in the ΔPLS version of the protein. As cortactin is a partner of the PLS responsible for p190A localization, we then tested whether cortactin interaction affects the binding capacity of p190A to active RhoA. Surprisingly, we found that overexpression of cortactin decreases the ability of p190A to bind active RhoA (Fig. 5 D). Overall, our results show that besides its role in targeting p190A to cell protrusions, the PLS domain is important for the regulation of p190A interaction with active RhoA. Moreover, these data suggest that the PLS is committed in an autoinhibited conformation of p190A that is relieved upon PLS deletion and promoted by cortactin interaction.
Figure 5.
Regulation of p190A GAP activity. (A) Huh7 cells expressing p190AWT or p190AΔPLS were plated for 3 h on fibronectin and stained for HA (green), F-actin (red, phalloidin), and nuclei (blue, Hoechst). # indicates a cell without stress fibers. (B) Huh7 cells transfected with p190AWT or p190AΔPLS were used for a RhoA GTPase pulldown assay using GST-Rhotekin. RhoA is detected using an anti-RhoA antibody. Quantification of RhoA activity is represented as a histogram. (C) Huh7 cells were transfected with p190AWT or p190AΔPLS. Affinity purification of Huh7 cell lysate was performed with GST-RhoAQ63L and analyzed by immunoblot using anti-HA antibodies. Quantification of the affinity between p190A and active RhoA is represented as a histogram. (D) Huh7 cells were transfected or not (NT) with mcherry-cortactin. Affinity purification on Huh7 cell lysate was performed with GST-RhoAQ63L and analyzed with anti-p190A antibodies. Quantification of the affinity between endogenous p190A and active RhoA is represented as a histogram. On graphs, values were calculated by measuring the band intensity of pulldown/input and are represented as the mean ± SEM of three independent experiments. P-values from the ANOVA (B) or the unpaired t test (C and D) are indicated. *, P < 0.05; **, P < 0.01.
Figure 6.
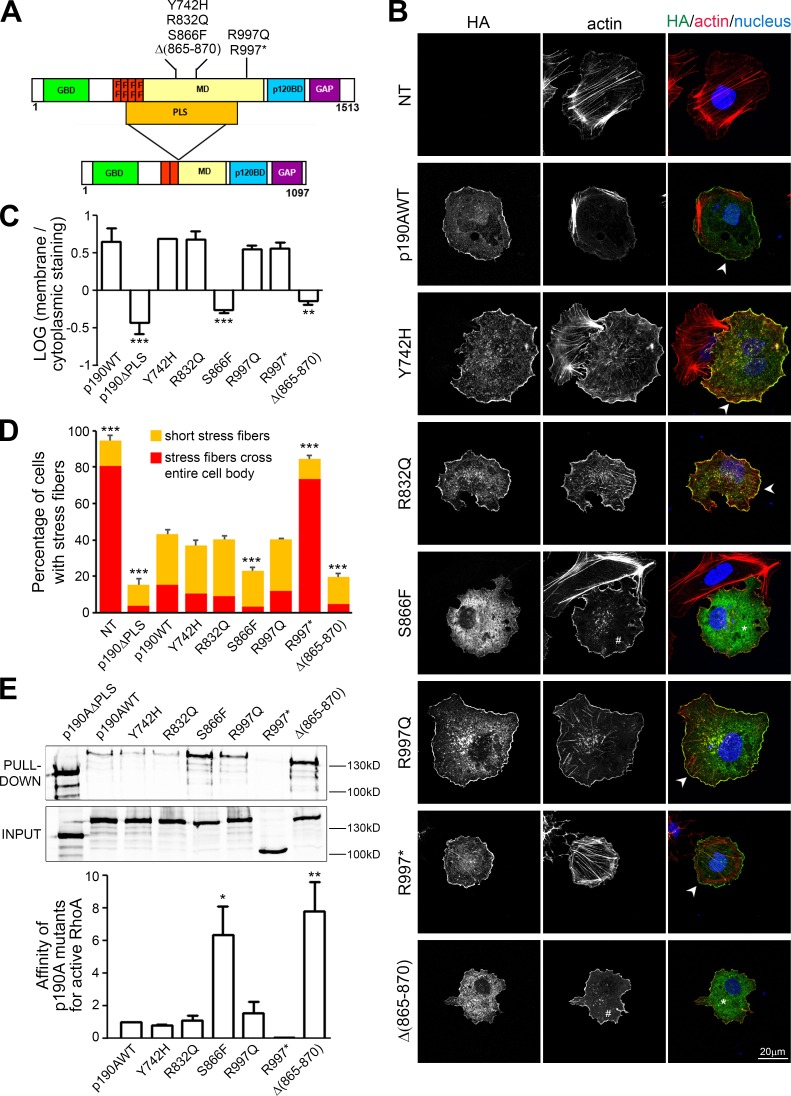
Analysis of p190A mutations found in tumors. (A) Schematic representation of the different p190A mutants. Point mutations, deletion, and the PLS domain are indicated on the protein. (B) Huh7 cells transfected with p190AWT or p190A mutants were plated for 3 h on fibronectin, fixed, and stained for HA (green), F-actin (red), and nuclei (blue). NT, nontransfected cells. Arrowheads show localization of mutants at cell protrusions; * shows cytoplasmic localization of mutants; and # points out absence of stress fibers. (C) Localization of p190A mutants is analyzed by quantification of membrane staining intensity/cytoplasmic staining intensity ratio. The graph presents the mean ± SEM of three independent experiments (n = 20 cells per condition). **, P < 0.01; ***, P < 0.001. (D) Quantification of cells bearing stress fibers in the experiment described in B. Statistical significance was calculated relative to the control (p190WT) condition. ***, P < 0.001. (E) Huh7 cells were transfected with indicated constructs or mutants, and their affinity for active RhoA was tested by pulldown using recombinant GST-RhoAQ63L and revealed by immunoblot with anti-HA antibodies. Quantification of the affinity between p190A mutants and active RhoA is represented as a histogram in which values were calculated by measuring the band intensity of pulldown/input and are represented as the mean ± SEM of three independent experiments. *, P < 0.05; **, P < 0.01.
Cancer-associated mutations present in the PLS impact on p190A properties
In a recent study (Lawrence et al., 2014), the somatic point mutations in exome sequence from 4,742 tumor-normal pairs across 21 cancer types were analyzed. This large-scale genomic analysis identified the p190RhoGAP-encoding gene ARHGAP35 as highly significantly mutated in 2% of global cancers and more specifically in 15% of endometrial tumors (data available at http://www.tumorportal.org/). Among the mutations found in tumors, a high frequency of nonsense mutations such as the recurrent R997* were detected, arguing for a loss of function. In light of our study, we focused on mutations located in the PLS domain of p190A (i.e., three point mutations: Y742H, R832Q, and S866F) and the intriguing in-frame deletion Δ(865–870), all found in endometrial tumors (Fig. 6 A and Table S1). In addition, we also selected the R997* nonsense mutation (lacking the GAP domain) and the R997Q point mutation, both localized in the MD of p190A outside the PLS, but, respectively, found in six endometrial tumors and in colon cancer (Fig. 6 A and Table S1). HA-tagged constructs of p190A bearing these mutations were generated by site-directed mutagenesis, and their consequences on p190A functionality were analyzed in Huh7 cells. We first studied the localization of the mutated proteins in cells plated on fibronectin. Interestingly, whereas most mutants localized to actin protrusions similar to the WT protein, p190A-S866F and Δ(865–870) failed to reach the cell edge and displayed mainly a cytoplasmic staining (Fig. 6, B and C). As described earlier, actin remodeling (i.e., alteration of stress fibers) is a consequence of the RhoGAP activity of p190A expression (Fig. S1 and Table S3). We then classified stress fibers into two phenotypes: stress fibers crossing the entire cell body and short stress fibers (Fig. S5 A and Fig. 6 D). Whereas p190A-R997* expression showed no impact on stress fibers, confirming its lack of GAP activity, p190A-Y742H, R832Q, and R997Q expression decreased the number of stress fibers as observed upon expression of p190AWT (Fig. 6, B and D). Of interest, cells expressing p190A-S866F and Δ(865–870) showed very few stress fibers, suggesting a stronger RhoGAP activity of these mutants. We further measured the affinity of the mutant proteins toward active RhoA and demonstrated that S866F and Δ(865–870) mutations increase the affinity of p190A for active RhoA (Fig. 6 E), as previously described for p190AΔPLS. p190A-Y742H, R832Q, and R997Q showed similar affinity for active RhoA to p190AWT, and the absence of binding of R997* to active RhoA is explained by its lack of GAP domain. These results suggest that S866F and Δ(865–870) mutations mimic the effect of PLS deletion on both p190A GAP activity and cellular localization. We then addressed whether these p190A mutants present a defective interaction with cortactin. Surprisingly, unlike p190AΔPLS, the cancer-associated mutants of p190A remain able to interact with cortactin (Fig. S5 B), demonstrating that these amino acids are not involved in cortactin–p190A interaction. As their leading edge localization is impaired, this result also indicates that cortactin binding is necessary but not sufficient to target p190A to actin-rich protrusions.
To get more insights into the function of cancer-associated p190A mutations, we then used mouse embryonic fibroblasts (MEFs), which display a spindle shape with a ruffling cell process easier to identify and measure than that of Huh7 cells. Moreover, tension applied by stress fibers induces focal adhesion formation in this model. HA-tagged versions of p190A were expressed in p190A knockout MEFs to avoid additive effect of endogenous p190A, and the number of focal adhesions was evaluated. A decreased focal adhesion area was observed in p190A−/− MEFs transfected with HA-p190AWT compared with nontransfected cells after overnight plating on fibronectin (Fig. 7, A and B). Expression of the mutants with predicted high GAP activity led to an even more dramatic decrease of focal adhesion area and a marginal decrease of the number of focal adhesions per cell. Focal adhesions regulate ruffles and lamellipodia stability (Schwarz and Gardel, 2012). Because persistent membrane ruffling favors cell process growth, we hypothesized that p190A mutants may affect cell process length. To test this, we measured the distance between the nucleus and ruffle edges of spindle shape MEFs plated overnight on fibronectin (Fig. 7 C). MEFs transfected with p190AWT displayed a cell process length similar to nontransfected cells, arguing that overexpression of p190AWT does not disturb cell process stability in MEFs. However, p190AΔPLS, S866F, and Δ(865–870) significantly decreased the cell process length, indicating that their higher GAP activity reduces lamellipodial persistence, a mechanism involving cortactin. As a consequence, we found that p190AΔPLS, S866F, and Δ(865–870) expression in MEFs increase random cell migration in chemokinesis assays when compared with untransfected or p190AWT-expressing cells (Fig. 7 D).
Figure 7.
Mutations in PLS affect cell process growth. (A) p190A−/− MEFs expressing the indicated HA-tagged p190A constructs were plated overnight on fibronectin, fixed, and stained for nucleus (Hoechst, blue), cortactin (red), focal adhesions (vinculin, gray), and HA (green). Arrowheads show cortactin-rich edges, and arrows show focal adhesions. (B) Quantification of focal adhesion area in cells transfected as described in A (n = 15 cells per condition). Inset represents the number of focal adhesions (FA) per cell in the same experiment. (C) Quantification of ruffling cell process length. Graph presents the mean distance between nucleus and ruffle edges in cells transfected as described in A (n = 15 cells per condition). (D) Transfilter chemokinesis of cells transfected as described in A. Results are expressed as a percentage of basal migration without chemoattractant (n > 100 cells). For all graphs, values are expressed as the mean ± SEM of three (B and C) or five (D) independent experiments. Statistical significance was calculated relative to p190AWT condition. *, P < 0.05; **, P < 0.01; ***, P < 0.001. NT, nontransfected cells.
Finally, we tested the impact of these p190A mutations on cancer cell migration using MDA-MB-231 breast carcinoma cells plated on gelatin-coated dishes. Time-lapse microscopy experiments were performed after cotransfection of the cells with p190A- and EGFP-expressing constructs (Videos 1–5). Analysis of the transfected cell paths revealed that the directionality of the cells expressing p190A mutants is altered compared with their WT counterpart (Fig. 8, A and B), whereas the speed of migration remains unaffected (Fig. 8 C). Thus, this result demonstrates that the S866F and Δ(865–870) cancer-associated mutations may favor turning behavior, which reflects exploration of cancerous cells.
Figure 8.
Mutations in PLS affect MDA-MB-231 directed cell migration. MDA-MB-231 cells were cotransfected with p190A- and EGFP-expressing constructs, spread on gelatin coating, and visualized by epifluorescence and contrast time-lapse microscopy for 16 h. Frames were collected every 10 min. (A) Representative tracings of paths traveled over 16 h by MDA-MB-231 cells transfected with p190A mutants. (B) Quantification of movement directionality represented as a ratio of the distance between the start and end point over the total path traveled. (C) Cell velocity is calculated by length of the path traveled divided by time. Values are expressed as the mean ± SEM (n = 20 cells per condition). Statistical significance was calculated relative to nontransfected (NT) condition. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s, not significant.
Discussion
To timely and spatially regulate RhoA, the localization of p190A is itself tightly controlled. In the present study, we investigated the mechanisms by which p190A is recruited to cell edges. We demonstrated that the region comprising aa 380–971 corresponds to its PLS. The PLS is the minimal domain necessary and sufficient for p190A localization to ruffles and lamellipodia in migrating cells. This PLS-based localization was also observed in fibronectin-, C8N6-BPA–, or V12Rac1-induced lamellipodia. Because lamellipodia induced by constitutively active Rac1 are devoid of active RhoA (Pertz et al., 2006), it is unlikely that membrane-bound RhoA mediates p190A recruitment to protrusion. This is further emphasized by the fact that the PLS does not contain the RhoA-binding GAP domain of p190A. In addition, the PLS lacks the p120BD, which was described to mediate p190A recruitment to the plasma membrane upon stimulation by integrins during cell attachment and subsequent tyrosine phosphorylation of p190A by Src or Arg (Hu and Settleman, 1997). It was previously shown that p190A has to be released from p120RasGAP to efficiently access and inactivate RhoA at the cell periphery. This mechanism occurs through a competition between the phosphorylated forms of p190A and paxillin for p120RasGAP binding during cell adhesion (Tsubouchi et al., 2002). We thus describe PLS as a novel functional domain involved in the targeting of p190A.
It is striking that the identified PLS almost corresponds to the region (aa 382–1,007) defined as the Rnd3-binding domain of p190B isoform (Wennerberg et al., 2003; Bustos et al., 2008). However, silencing of Rnd3 does not alter p190A targeting to membrane protrusions in Huh7 cells (unpublished data), suggesting that Rnd3 is not involved in p190A localization. The Rnd3-binding domain was further restricted to an aa 382–607 fragment sufficient to promote the interaction between activated Rac1 and p190B and subsequent p190B membrane recruitment (Bustos et al., 2008). However, authors failed to detect a GTP-dependent association between activated Rac1 and p190A, suggesting a different targeting mechanism between both isoforms.
By affinity purification assays, we identified cortactin as a binding partner of the PLS. siRNA-mediated silencing of cortactin revealed that cortactin is necessary for p190A targeting to membrane ruffles and lamellipodia. Cortactin is an F-actin regulating factor that allows directional migration by stabilizing lamellipodia (Krause and Gautreau, 2014). Although not essential for the formation of the lamellipodia, cortactin regulates migration by increasing lamellipodial persistence through the binding of the Arp2/3 complex and F-actin (Bryce et al., 2005). We documented the p190A/cortactin interaction through four types of experiments: GST-pulldown, coimmunoprecipitation, indirect immunostaining, and PLA. Although a strong colocalization between p190A and cortactin was found by indirect immunostaining, only few dots could be seen by PLA. This difference may be because of a technical caveat of the PLA. Indeed, considering that p190A and cortactin are relatively large proteins, the Fc part of the antibodies used may be often too far to allow annealing. Another possible explanation may be that the proteins are located at the same place but only interact in a stimuli-dependent (i.e., regulation of RhoA activity or F-actin elongation) and tightly regulated manner. This hypothesis is supported by the fact that an increased interaction between p190A and cortactin inhibits p190A GAP accessibility (Fig. 5 D). Hence, a dynamic regulation of p190A activity by cortactin likely occurs at the leading edge, implying transient binding/dissociation events while the proteins remain in proximity. Cortactin redistribution from the cytoplasm to the membrane ruffles and lamellipodia is controlled by Rac1 activation (Weed et al., 1998). Beyond its role in F-actin polymerization, cortactin controls the cell shape by interacting with GTPase regulators such as Fgd1 and Dock4, respectively, GEFs of Cdc42 and Rac1 (Hou et al., 2003; Ueda et al., 2013). Cortactin-mediated control of lamellipodial persistence could therefore be driven by both the stimulation of Rac1 and Cdc42 activity by GEFs and the inhibition of RhoA activity by p190A. Furthermore, Rac1-mediated recruitment of the cortactin–p190A complex to the lamellipodia and subsequent RhoGAP domain activation could account for the negative regulation of RhoA by Rac1 observed in various cell lines (Kozma et al., 1997; Leeuwen et al., 1997; Sander et al., 1999; Zondag et al., 2000; Nimnual et al., 2003). Recent spatiotemporal analyses of lamellipodia formation revealed that RhoA is activated directly at the leading edge, but is switched off to allow Rac1 and Cdc42 activation during the protrusion phase (Machacek et al., 2009). Therefore, as suggested for p190B, p190RhoGAP proteins may be involved in the local antagonistic cross talk between these GTPases. We believe that the PLS of p190A is functionally implicated in this mechanism.
Accordingly, in addition to the involvement of the PLS in p190A localization, we found that the same sequence is required to inhibit p190A RhoGAP activity. Indeed, RhoA activity decrease is more pronounced in cells expressing p190AΔPLS than p190AWT. However, we were not able to demonstrate a modification of Rac1 activity upon expression of either p190AWT or p190AΔPLS in Huh7 cells (unpublished data). It has to be noted that the PLS excludes the poly basic region identified in p190A as crucial for its RacGAP activity (Lévay et al., 2013). The lowest RhoA activity observed with p190AΔPLS has phenotypic consequences (e.g., loss of stress fibers, a process controlled by RhoA; Chrzanowska-Wodnicka and Burridge, 1996). The nonspecific localization of the mutant may provide more RhoA substrate and explain this discrepancy. However, affinity purification of p190A with the active form of RhoA (RhoAQ63L), a method already used to detect p190A RhoGAP activity upon cadherin engagement (Noren et al., 2003), demonstrated that the strong effect of p190AΔPLS arises from a drastic increase of its affinity toward active RhoA, in accordance with an enhanced GAP activity in cells. Even if we cannot exclude the possibility that this stronger binding titrates out active RhoA, making the latter unavailable for effector binding and resulting in the same phenotype (i.e., loss of stress fibers), these results account for a PLS-mediated cis-inhibition of the GAP domain. This hypothesis is consistent with previous studies showing that p190B lacking the Rac1-binding sequence, described above, also exhibited enhanced RhoGAP activity (Bustos et al., 2008). Collectively, these data indicate that the same region in both p190 isoforms accounts for a similar autoinhibitory function. Autoinhibition due to protein folding that controls the GAP activity has been described for several GAPs such as p50RhoGAP, oligophrenin, Abr, and chimerins (Ligeti et al., 2012). Thus, this reinforces the model proposed earlier by S.H. Hansen and colleagues (Wennerberg et al., 2003; Bustos et al., 2008), in which p190 is present in a “closed” conformation in the cytoplasm that may be opened upon stimuli, leading to the translocation and full activation of p190 restricted to membrane ruffles and lamellipodia, thus promoting cell protrusion and migration. Whereas for p190B it was proposed that Rac1 or Rnd3 could relieve its autoinhibition, the mechanism for p190A remains to be established. We found that cortactin is not involved in this mechanism and in contrast favors the closed conformation of p190AWT. Thus, another additive signal may be necessary to locally activate p190A by competing with cortactin antagonism. Interestingly, our data pointed out serine 866 as an important amino acid for p190A regulation. Indeed, the cancer-associated S866F missense mutation and the Δ865–870 deletion phenocopy the absence of PLS, affecting both p190A localization and GAP activity regulation. Because phosphorylation is frequently involved in functional regulation, it is tempting to hypothesize that phosphorylation of serine 866 keeps p190A in its closed conformation and that its dephosphorylation activates the protein.
Our study also highlights that mutations found in tumors may have opposite effects on the activity of the protein. Indeed, whereas p190A-R997* mutant has no GAP activity, p190A-S866F and Δ865–870 showed a high RhoGAP activity. Our data further demonstrated that p190AΔPLS, S866F, and Δ(865–870) significantly decreased cell process length, indicating that their higher GAP activity reduces lamellipodia persistence. Thus, as demonstrated earlier by the polarized cell migration defect of p190A−/− cells (Jiang et al., 2008), the RhoGAP activity of p190A must be tightly regulated to allow for proper cell migration. As a consequence, we found that gain-of-function mutations such as S866F and Δ865–870 favor random cell migration and decrease the ability of cancer cells to migrate in a directed manner. Hence, acquisition of p190A mutations may result into an enhanced exploratory behavior of tumor cells. Given the high prevalence of ARHGAP35 mutations in tumors, these data open up a new path of investigations with clinical implications.
Materials and methods
Cell lines
Huh7 and MDA-MB-231 cell lines were purchased from American Type Culture Collection. p190−/− MEFs were a gift from A. Koleske (Yale University, New Haven, CT). Huh7 cells and MEFs were cultured in DMEM containing Glutamax and 4.5 g/l glucose (Gibco), supplemented with 10% FCS, 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen). MDA-MB-231 cells were maintained in L-15 medium and Glutamax-I (Invitrogen) supplemented with 10% FCS and penicillin–streptomycin. PAE expressing EE-tagged V12Rac1 under the control of an IPTG-inducible promoter were established previously (Welch et al., 1998). They were cultured in Ham F12 medium supplemented with 10% heat-inactivated FCS, penicillin-streptomycin, 100 µM hygromycin B, and 500 nM puromycin. V12Rac1 expression was achieved by adding 1 mM IPTG to the culture for 15 h. All cells were maintained at 37°C in a 5% CO2 humidified atmosphere.
Antibodies and reagents
Monoclonal mouse anti-p190A (clone 30) and anti-VASP (clone 43) antibodies were purchased from BD; mouse anti-p190A (clone D2D6), mouse anti-vinculin (clone hVIN1) antibodies, and IgG1 isotype control from mouse myeloma were purchased from Sigma-Aldrich, mouse anti-Rac1 (clone 23A8) and anti-cortactin (clone 4F11) were purchased from Millipore; and mouse anti-GFP and anti-RhoA were purchased from Santa Cruz Biotechnology, Inc. Monoclonal rat anti-HA (clone 3F10) and mouse anti-HA (clone 12CA5) were obtained from Roche. Polyclonal rabbit anti-cortactin (H222) antibodies were obtained from Cell Signaling Technology. Alexa Fluor 546–phalloidin and Alexa Fluor 488–labeled secondary antibodies were purchased from Molecular Probes. IPTG, puromycin, and hygromycin B were obtained from EMD Millipore.
Transfection
DNA transfections in Huh7, MEFs, and PAE cells were realized with promofectin-hepatocyte transfection reagent (Promocell) according to the manufacturer’s protocol. MDA-MB-231 cells were transfected using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s protocol. siRNA oligos were purchased from Eurofins Genomics and transfected into Huh7 cells with Lipofectamine RNAiMax Reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. siCortactin1 targets human cortactin mRNA at 5′-CCCAGAAAGACUAUGUGAAAGGGUU-3′ and siCortactin2 targets at 5′-GGAGAAGCACGAGUCACAGAGAGAU-3′ (Jia et al., 2008). The nonsilencing siRNA siControl 5′-AATTCTCCGAACGTGTCACGT-3′ was used as a control.
Plasmid constructs
All of the truncated p190A constructs were engineered using PCR with the rat full-length p190RhoGAP (GenBank under accession no. M94721) as matrix and subcloned into pKH3 expression vector using BamHI and EcoRI restriction sites. The ΔPLS mutant was constructed as followed: two PCR fragments corresponding to N-terminal part (aa 1–380) and C-terminal part (aa 800–1,513) were amplified from pKH3-p190WT and subcloned into pGEM-T easy vectors. Both fragments were then purified and ligated. This ΔPLS fragment was finally inserted into the BamHI-EcoRI sites of pKH3 vector. All primers used for these constructs are listed in Table S2. All constructs were verified by DNA sequencing. pKH3-p190AWT was a gift from I. Macara (Vanderbilt University, Nashville, TN). pGEX-RhoAQ63L was a gift from K. Burridge (University of North Carolina at Chapel Hill, Chapel Hill, NC). Cortactin-pmCherryC1 was a gift from C. Merrifield (Institut de Biologie Intégrative de la Cellule, Gif-sur-Yvette, France; plasmid 27676; Addgene; Taylor et al., 2011). α-Actinin–GFP construct was a gift from G. Giannone (Interdiscipliniary Institute for Neuroscience, Bordeaux, France; Giannone et al., 2004). pGEX-2T-Rgd1 and pGEX-PH–PLC-γ1 were gifts from F. Doignon (Bordeaux University, Bordeaux, France) and M. Lemmon (University of Pennsylvania, Philadelphia, PA), respectively. pEGFP-C1 was purchased from Takara Bio Inc.
Western blot analysis, immunoprecipitation, and pulldown assay
Cells were scraped off on ice and homogenized in Tris-HCl lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.6% NP-40) with protease and phosphatase inhibitors. Cell lysates were cleared of cellular debris and nuclei by a 10,000 g centrifugation step for 10 min. Lysates were denatured with Laemmli loading buffer containing 2.5% 2-β-mercaptoethanol, analyzed by SDS-PAGE, and blotted onto nitrocellulose membranes. Blots were incubated overnight at 4°C with primary antibodies, and then incubated with infrared fluorescent dye-conjugated secondary antibodies (LI-COR Biosciences), and activity was visualized with the Odyssey infrared imaging system (LI-COR Biosciences).
Immunoprecipitation was performed with anti-HA affinity matrix beads (Roche) according to the manufacturer's instructions. In brief, cells were extracted with Tris-HCl lysis buffer and centrifuged for 10 min at 10,000 g. The supernatant was incubated for 30 min with the beads. Bead pellets were washed three times with Wash Buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% NP-40), resuspended in loading buffer, and analyzed by SDS-PAGE. Recombinant GST-proteins were expressed in bacteria, purified on GSH-coated sepharose beads (Sigma-Aldrich), and incubated with protein extracts of Huh7 cells. Bacteria lysis was performed with Tris-HCl lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100, and 1 mM DTT) with protease and phosphatase inhibitors. GST-RhoAQ63L pulldown was performed as previously described (García-Mata et al., 2006). pGEX-2T construct containing Rhotekin-Rho binding domain was provided by Martin Schwartz (University of Virginia, Charlottesville, VA). Transfected Huh7 cells were lysed, and protein extracts were used for RhoA pulldown assays as previously described (Ren et al., 1999). The lysis buffer for RhoA GTPase pulldown assay consists of 50 mM Tris, pH 7.2, 500 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 10 mM MgCl2, 1 mM PMSF, and a mixture of protease inhibitors. In brief, cleared cell lysates were incubated with Rhotekin-RBD beads (Cytoskeleton, Inc.), and then the beads were washed and suspended in 5× Laemmli loading buffer. The whole-cell lysate was also used to examine the total amount of Rho GTPase in the cells. Samples were analyzed by Western blotting with anti-RhoA antibody. Immunoprecipitation of endogenous proteins was performed with anti-p190A (clone D2D6) and anti-cortactin (clone 4F11). In brief, cells were extracted with Tris-HCl lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.6% NP-40, and 0.2% Triton X-100) and centrifuged for 10 min at 10,000 g. The supernatant was incubated overnight with control IgG and specific antibodies for 3 h with Dynabeads Protein G (Invitrogen). Bead pellets were washed three times with Wash Buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% NP-40), resuspended in loading buffer, and analyzed by SDS-PAGE.
Immunofluorescence and confocal imaging
Glass coverslip–plated cells were prepared for immunofluorescence microscopy as previously described (Moreau et al., 2003). Coverslips were mounted on slides with Fluoromount G mounting medium. Cells were imaged using an SP5 confocal microscope (Leica Biosystems) using a 63×/NA 1.4 Plan Neofluor objective lens and the LAS-AF-Lite 2.4.1 acquisition software (Leica Biosystems). To prevent contamination among fluorochromes, each channel was imaged sequentially using the multitrack recording module before merging. Images were processed using LAS-AF-Lite 2.4.1 (Leica Biosystems), Gimp 2.0, or ImageJ software (National Institutes of Health). Quantification of protrusion localization was performed in a double-blinded manner by comparing actin and HA signal at the plasma membrane on 30–200 transfected cells per condition.
PLA (Sigma-Aldrich) is an immunofluorescence-based method allowing the visualization of protein interaction with a distance <16 nm. Antigens were immunolabeled with two primary antibodies from different species and then specific secondary antibodies conjugated to complementary oligonucleotides. Close proximity of the antibodies allows the ligation, amplification, and hybridization of fluorescent probes appearing as distinct puncta. Quantification of dots was performed in ImageJ (National Institutes of Health) with the “analyze particles” command. The staining intensities at the membrane were measured inside a 2 × 0.28-µm rectangle, of which the longest dimension follows the membrane. 60 measurements were made for each condition. Cytoplasmic staining intensities were measured on the same surface parallel to the rectangle used for membrane measurement 6 µm behind the membrane. Cell process length measurement was performed in three independent experiments. For each experiment, mean size was calculated from 15 cells with spindle shape. The measurement was done from a line starting from the nucleus edge on the side of the ruffling process and ending at the most distant part of the ruffling edge. Focal adhesion area was measured as previously described (Horzum et al., 2014). In brief, using ImageJ software (National Institutes of Health), we sequentially subtracted background, enhanced contrast, minimized background, and adjusted brightness and contrast automatically, ran LOG3D filter, thresholded the image, and then executed the “analyze particles” command.
Cell migration assays
Migration of MEFs was performed with Boyden chambers (8-µm pore size filter; BD). 105 cells were seeded per chamber on top of the filter. Chemokinesis assays were performed with 10% serum on both sides of the filter. Migration was performed for 6 h, and membranes were then fixed with 4% PFA. Nonmigrated cells were removed from the top compartment with a cotton swab. Cells that had migrated to the lower side of the filter were stained with Hoechst 33258 and counted. Data are expressed as a percentage of basal migration without serum. Time-lapse microscopy experiments were performed with transfected MDA-MB-231 cells plated on gelatin-coated six-well dishes. The cells were placed in a 37°C chamber equilibrated with humidified air containing 5% CO2 throughout the experiment. Time-lapse microscopy was performed using a microscope composed of an inverted DMI 6000 microscope (Leica Biosystems) equipped with a resolutive HQ2 camera (Photometrics). The objective used was an HCX PL Fluotar 10× dry 0.3 NA PH1. The 37°C atmosphere was created with an incubator box and an air heating system (Life Imaging Services). This system was controlled by MetaMorph software (Molecular Devices). Images were taken every 10 min during 16 h. The videos were created from the time-lapse series using ImageJ (National Institutes of Health). Movements of cells expressing the green EGFP plasmid reporter were tracked in ImageJ with MtrackJ plugin.
Phospholipid binding assays
GST-PLS, GST-Rgd1, and GST expressions were induced in BL21 bacterial strains with 0.1 mM IPTG overnight at 20°C. The proteins were eluted from Glutathione Sepharose beads (Sigma-Aldrich) just before the experiments and quantified by SDS-PAGE. The PIP Strip membranes (Echelon Biosciences) were used according to the manufacturer’s instructions. GST-PLC-γ1-PH and GST-Rgd1 were used as positive controls (Prouzet-Mauléon et al., 2008). Each GST fusion protein was incubated overnight at 4°C with the PIP Strip membranes at 1 µg/ml and revealed by a rabbit anti-GST antibody (EMD Millipore) used at 1:2,500.
Site-directed mutagenesis
All p190A point mutations and the Δ(865–870) deletion were generated in the parental vector pKH3-p190A using site-directed mutagenesis with the QuikChange II XL kit according to the manufacturer’s protocol (Agilent Technologies). All primers used for these constructs are listed in Table S2. Point mutations and deletion were verified by sequencing. Note that the aa R997 found mutated in human tumors corresponds to the amino acid R998 in the rat p190A sequence.
Statistical analysis
Statistical analysis was performed with Prism software (GraphPad Software). Data are presented as mean ± SEM of at least three independent experiments. Comparisons between groups were analyzed by t test or one-way analysis of variance (ANOVA) followed by Bonferroni’s posttest when the number of conditions was higher than two, Mann–Whitney test to compare two conditions, and Spearman test when specified. Significance was accepted for values where *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material
Fig. S1 shows the localization of all truncated versions of p190A. Fig. S2 shows the localization of the different truncated versions of the 2F-MD construct. Fig. S3 demonstrates that PLS is necessary and sufficient for Rac1-induced lamellipodia targeting in PAE cells. Fig. S4 reveals that PLS does not interact with phospholipids in vitro, but interacts with cortactin. Fig. S5 shows the interaction of p190A cancer-associated mutations with cortactin. Table S3 summarizes the localization and impact on the actin cytoskeleton of the truncated versions of p190A. Table S1 describes the cancer-associated mutations in p190A from patients. Table S2 contains the list of primers used to generate p190A constructs and mutants. Video 1 shows MDA-MB-231 cells transfected with an EGFP-expressing construct. Videos 2, 3, 4, and 5 show MDA-MB-231 cells cotransfected with an EGFP-expressing construct and with, respectively, WTp190A, p190AΔPLS, p190AS866F, and p190AΔ(865–870) constructs.
Supplementary Material
Acknowledgments
We thank Drs. K. Burridge, A. Koleske, I. G. Macara, M. Schwartz, C. Merrifield, G. Giannone, and F. Doignon for DNA constructs, Dr. S. Fribourg for structural advice, and Drs. E. Chevet and G. Giannone for helpful discussions. Microscopy was done in the Bordeaux Imaging Center, a service unit of the Centre National de la Recherche Scientifique-Institut National de la Santé et de la Recherche Médicale and Bordeaux University, member of the national infrastructure France BioImaging, with the help of Dr. F. Cordelières.
F. Binamé was supported by an Idex Postdoctoral fellowship from the Université de Bordeaux. A. Bidaud-Meynard was supported by a PhD scholarship from the French Ministry of Research and an end-of-thesis short-term fellowship from the Fondation pour la Recherche Médicale, comité Aquitaine. L. Piquet is supported by a PhD fellowship from the SIRIC BRIO. This work was supported by grants from La Ligue Contre le Cancer (Comité de la Dordogne), La Ligue Contre le Cancer “Equipe labellisée,” and Fondation ARC pour la Recherche sur le Cancer.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ANOVA
- analysis of variance
- Arp2
- actin-related protein 2
- BPA
- branched polyamine
- GAP
- GTPase-activating proteins
- GBD
- GTP-binding segment
- GEF
- guanine nucleotide exchange factor
- MD
- middle domain
- MEF
- mouse embryonic fibroblast
- p120BD
- p120RasGAP binding domain
- p190A
- p190RhoGAP
- PAE
- porcine aortic endothelial
- PLA
- proximity ligation assay
- PLS
- protrusion localization sequence
- VASP
- vasodilator-stimulated phosphoprotein
- WT
- wild-type
References
- Arthur W.T., Petch L.A., and Burridge K.. 2000. Integrin engagement suppresses RhoA activity via a c-Src-dependent mechanism. Curr. Biol. 10:719–722. 10.1016/S0960-9822(00)00537-6 [DOI] [PubMed] [Google Scholar]
- Bass M.D., Morgan M.R., Roach K.A., Settleman J., Goryachev A.B., and Humphries M.J.. 2008. p190RhoGAP is the convergence point of adhesion signals from alpha 5 beta 1 integrin and syndecan-4. J. Cell Biol. 181:1013–1026. 10.1083/jcb.200711129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binamé F., Pawlak G., Roux P., and Hibner U.. 2010. What makes cells move: requirements and obstacles for spontaneous cell motility. Mol. Biosyst. 6:648–661. 10.1039/b915591k [DOI] [PubMed] [Google Scholar]
- Bradley W.D., Hernández S.E., Settleman J., and Koleske A.J.. 2006. Integrin signaling through Arg activates p190RhoGAP by promoting its binding to p120RasGAP and recruitment to the membrane. Mol. Biol. Cell. 17:4827–4836. 10.1091/mbc.E06-02-0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns M.R., Matheson S.F., Hu K.Q., Delalle I., Caviness V.S., Silver J., Bronson R.T., and Settleman J.. 2000. The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development. 127:4891–4903. [DOI] [PubMed] [Google Scholar]
- Brouns M.R., Matheson S.F., and Settleman J.. 2001. p190 RhoGAP is the principal Src substrate in brain and regulates axon outgrowth, guidance and fasciculation. Nat. Cell Biol. 3:361–367. 10.1038/35070042 [DOI] [PubMed] [Google Scholar]
- Bryant S.S., Briggs S., Smithgall T.E., Martin G.A., McCormick F., Chang J.H., Parsons S.J., and Jove R.. 1995. Two SH2 domains of p120 Ras GTPase-activating protein bind synergistically to tyrosine phosphorylated p190 Rho GTPase-activating protein. J. Biol. Chem. 270:17947–17952. 10.1074/jbc.270.30.17947 [DOI] [PubMed] [Google Scholar]
- Bryce N.S., Clark E.S., Leysath J.L., Currie J.D., Webb D.J., and Weaver A.M.. 2005. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr. Biol. 15:1276–1285. 10.1016/j.cub.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Bustos R.I., Forget M.A., Settleman J.E., and Hansen S.H.. 2008. Coordination of Rho and Rac GTPase function via p190B RhoGAP. Curr. Biol. 18:1606–1611. 10.1016/j.cub.2008.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M., and Burridge K.. 1996. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 133:1403–1415. 10.1083/jcb.133.6.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C., Moran M., McCormick F., and Pawson T.. 1990. Phosphorylation of GAP and GAP-associated proteins by transforming and mitogenic tyrosine kinases. Nature. 343:377–381. 10.1038/343377a0 [DOI] [PubMed] [Google Scholar]
- García-Mata R., Wennerberg K., Arthur W.T., Noren N.K., Ellerbroek S.M., and Burridge K.. 2006. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 406:425–437. 10.1016/S0076-6879(06)06031-9 [DOI] [PubMed] [Google Scholar]
- Giannone G., Dubin-Thaler B.J., Döbereiner H.G., Kieffer N., Bresnick A.R., and Sheetz M.P.. 2004. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 116:431–443. 10.1016/S0092-8674(04)00058-3 [DOI] [PubMed] [Google Scholar]
- Guegan F., Tatin F., Leste-Lasserre T., Drutel G., Genot E., and Moreau V.. 2008. p190B RhoGAP regulates endothelial-cell-associated proteolysis through MT1-MMP and MMP2. J. Cell Sci. 121:2054–2061. 10.1242/jcs.025817 [DOI] [PubMed] [Google Scholar]
- Haskell M.D., Nickles A.L., Agati J.M., Su L., Dukes B.D., and Parsons S.J.. 2001. Phosphorylation of p190 on Tyr1105 by c-Src is necessary but not sufficient for EGF-induced actin disassembly in C3H10T1/2 fibroblasts. J. Cell Sci. 114:1699–1708. [DOI] [PubMed] [Google Scholar]
- Herbrand U., and Ahmadian M.R.. 2006. p190-RhoGAP as an integral component of the Tiam1/Rac1-induced downregulation of Rho. Biol. Chem. 387:311–317. 10.1515/BC.2006.041 [DOI] [PubMed] [Google Scholar]
- Horzum U., Ozdil B., and Pesen-Okvur D.. 2014. Step-by-step quantitative analysis of focal adhesions. MethodsX. 1:56–59. 10.1016/j.mex.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P., Estrada L., Kinley A.W., Parsons J.T., Vojtek A.B., and Gorski J.L.. 2003. Fgd1, the Cdc42 GEF responsible for Faciogenital Dysplasia, directly interacts with cortactin and mAbp1 to modulate cell shape. Hum. Mol. Genet. 12:1981–1993. 10.1093/hmg/ddg209 [DOI] [PubMed] [Google Scholar]
- Hu K.Q., and Settleman J.. 1997. Tandem SH2 binding sites mediate the RasGAP-RhoGAP interaction: a conformational mechanism for SH3 domain regulation. EMBO J. 16:473–483. 10.1093/emboj/16.3.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Uekita T., and Sakai R.. 2008. Hyperphosphorylated cortactin in cancer cells plays an inhibitory role in cell motility. Mol. Cancer Res. 6:654–662. 10.1158/1541-7786.MCR-07-0220 [DOI] [PubMed] [Google Scholar]
- Jiang W., Betson M., Mulloy R., Foster R., Lévay M., Ligeti E., and Settleman J.. 2008. p190A RhoGAP is a glycogen synthase kinase-3-β substrate required for polarized cell migration. J. Biol. Chem. 283:20978–20988. 10.1074/jbc.M802588200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight B., Laukaitis C., Akhtar N., Hotchin N.A., Edlund M., and Horwitz A.R.. 2000. Visualizing muscle cell migration in situ. Curr. Biol. 10:576–585. 10.1016/S0960-9822(00)00486-3 [DOI] [PubMed] [Google Scholar]
- Kozma R., Sarner S., Ahmed S., and Lim L.. 1997. Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol. Cell. Biol. 17:1201–1211. 10.1128/MCB.17.3.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., and Gautreau A.. 2014. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 15:577–590. 10.1038/nrm3861 [DOI] [PubMed] [Google Scholar]
- Kshitiz J., Afzal J., Kim D.H., and Levchenko A.. 2014. Concise review: Mechanotransduction via p190RhoGAP regulates a switch between cardiomyogenic and endothelial lineages in adult cardiac progenitors. Stem Cells. 32:1999–2007. 10.1002/stem.1700 [DOI] [PubMed] [Google Scholar]
- Kusama T., Mukai M., Endo H., Ishikawa O., Tatsuta M., Nakamura H., and Inoue M.. 2006. Inactivation of Rho GTPases by p190 RhoGAP reduces human pancreatic cancer cell invasion and metastasis. Cancer Sci. 97:848–853. 10.1111/j.1349-7006.2006.00242.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langanger G., de Mey J., Moeremans M., Daneels G., de Brabander M., and Small J.V.. 1984. Ultrastructural localization of alpha-actinin and filamin in cultured cells with the immunogold staining (IGS) method. J. Cell Biol. 99:1324–1334. 10.1083/jcb.99.4.1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., and Getz G.. 2014. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 505:495–501. 10.1038/nature12912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuwen F.N., Kain H.E., Kammen R.A., Michiels F., Kranenburg O.W., and Collard J.G.. 1997. The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J. Cell Biol. 139:797–807. 10.1083/jcb.139.3.797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévay M., Settleman J., and Ligeti E.. 2009. Regulation of the substrate preference of p190RhoGAP by protein kinase C-mediated phosphorylation of a phospholipid binding site. Biochemistry. 48:8615–8623. 10.1021/bi900667y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévay M., Bartos B., and Ligeti E.. 2013. p190RhoGAP has cellular RacGAP activity regulated by a polybasic region. Cell. Signal. 25:1388–1394. 10.1016/j.cellsig.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Ligeti E., Welti S., and Scheffzek K.. 2012. Inhibition and termination of physiological responses by GTPase activating proteins. Physiol. Rev. 92:237–272. 10.1152/physrev.00045.2010 [DOI] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G.L., Hahn K.M., and Danuser G.. 2009. Coordination of Rho GTPase activities during cell protrusion. Nature. 461:99–103. 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlade J., Brunkhorst B., Anderson D., Mbamalu G., Settleman J., Dedhar S., Rozakis-Adcock M., Chen L.B., and Pawson T.. 1993. The N-terminal region of GAP regulates cytoskeletal structure and cell adhesion. EMBO J. 12:3073–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V., Tatin F., Varon C., and Génot E.. 2003. Actin can reorganize into podosomes in aortic endothelial cells, a process controlled by Cdc42 and RhoA. Mol. Cell. Biol. 23:6809–6822. 10.1128/MCB.23.19.6809-6822.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Mueller S.C., Nomizu M., Yamada Y., Yeh Y., and Chen W.T.. 1998. Activation of β1 integrin signaling stimulates tyrosine phosphorylation of p190RhoGAP and membrane-protrusive activities at invadopodia. J. Biol. Chem. 273:9–12. 10.1074/jbc.273.1.9 [DOI] [PubMed] [Google Scholar]
- Nedeva I., Koripelly G., Caballero D., Chièze L., Guichard B., Romain B., Pencreach E., Lehn J.M., Carlier M.F., and Riveline D.. 2013. Synthetic polyamines promote rapid lamellipodial growth by regulating actin dynamics. Nat. Commun. 4:2165 10.1038/ncomms3165 [DOI] [PubMed] [Google Scholar]
- Nimnual A.S., Taylor L.J., and Bar-Sagi D.. 2003. Redox-dependent downregulation of Rho by Rac. Nat. Cell Biol. 5:236–241. 10.1038/ncb938 [DOI] [PubMed] [Google Scholar]
- Noren N.K., Arthur W.T., and Burridge K.. 2003. Cadherin engagement inhibits RhoA via p190RhoGAP. J. Biol. Chem. 278:13615–13618. 10.1074/jbc.C200657200 [DOI] [PubMed] [Google Scholar]
- Notsuda H., Sakurada A., Endo C., Okada Y., Horii A., Shima H., and Kondo T.. 2013. p190A RhoGAP is involved in EGFR pathways and promotes proliferation, invasion and migration in lung adenocarcinoma cells. Int. J. Oncol. 43:1569–1577. 10.3892/ijo.2013.2096 [DOI] [PubMed] [Google Scholar]
- Pertz O., Hodgson L., Klemke R.L., and Hahn K.M.. 2006. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 440:1069–1072. 10.1038/nature04665 [DOI] [PubMed] [Google Scholar]
- Prouzet-Mauléon V., Lefebvre F., Thoraval D., Crouzet M., and Doignon F.. 2008. Phosphoinositides affect both the cellular distribution and activity of the F-BAR-containing RhoGAP Rgd1p in yeast. J. Biol. Chem. 283:33249–33257. 10.1074/jbc.M805161200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X.D., Kiosses W.B., and Schwartz M.A.. 1999. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18:578–585. 10.1093/emboj/18.3.578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J. 2011. Life at the leading edge. Cell. 145:1012–1022. 10.1016/j.cell.2011.06.010 [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Self A.J., Kasmi F., Paterson H.F., Hall A., Marshall C.J., and Ellis C.. 1993. rho family GTPase activating proteins p190, bcr and rhoGAP show distinct specificities in vitro and in vivo. EMBO J. 12:5151–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof R.W., Haskell M.D., Dukes B.D., Sherman N., Kinter M., and Parsons S.J.. 1998. Phosphotyrosine (p-Tyr)-dependent and -independent mechanisms of p190 RhoGAP-p120 RasGAP interaction: Tyr 1105 of p190, a substrate for c-Src, is the sole p-Tyr mediator of complex formation. Mol. Cell. Biol. 18:7052–7063. 10.1128/MCB.18.12.7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander E.E., ten Klooster J.P., van Delft S., van der Kammen R.A., and Collard J.G.. 1999. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147:1009–1022. 10.1083/jcb.147.5.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz U.S., and Gardel M.L.. 2012. United we stand: integrating the actin cytoskeleton and cell-matrix adhesions in cellular mechanotransduction. J. Cell Sci. 125:3051–3060. 10.1242/jcs.093716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settleman J., Albright C.F., Foster L.C., and Weinberg R.A.. 1992. Association between GTPase activators for Rho and Ras families. Nature. 359:153–154. 10.1038/359153a0 [DOI] [PubMed] [Google Scholar]
- Shen C.H., Chen H.Y., Lin M.S., Li F.Y., Chang C.C., Kuo M.L., Settleman J., and Chen R.H.. 2008. Breast tumor kinase phosphorylates p190RhoGAP to regulate rho and ras and promote breast carcinoma growth, migration, and invasion. Cancer Res. 68:7779–7787. 10.1158/0008-5472.CAN-08-0997 [DOI] [PubMed] [Google Scholar]
- Tatsis N., Lannigan D.A., and Macara I.G.. 1998. The function of the p190 Rho GTPase-activating protein is controlled by its N-terminal GTP binding domain. J. Biol. Chem. 273:34631–34638. 10.1074/jbc.273.51.34631 [DOI] [PubMed] [Google Scholar]
- Taylor M.J., Perrais D., and Merrifield C.J.. 2011. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9:e1000604 10.1371/journal.pbio.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi A., Sakakura J., Yagi R., Mazaki Y., Schaefer E., Yano H., and Sabe H.. 2002. Localized suppression of RhoA activity by Tyr31/118-phosphorylated paxillin in cell adhesion and migration. J. Cell Biol. 159:673–683. 10.1083/jcb.200202117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda S., Negishi M., and Katoh H.. 2013. Rac GEF Dock4 interacts with cortactin to regulate dendritic spine formation. Mol. Biol. Cell. 24:1602–1613. 10.1091/mbc.E12-11-0782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.Z., Nur-E-Kamal M.S., Tikoo A., Montague W., and Maruta H.. 1997. The GTPase and Rho GAP domains of p190, a tumor suppressor protein that binds the M(r) 120,000 Ras GAP, independently function as anti-Ras tumor suppressors. Cancer Res. 57:2478–2484. [PubMed] [Google Scholar]
- Weaver A.M., Heuser J.E., Karginov A.V., Lee W.L., Parsons J.T., and Cooper J.A.. 2002. Interaction of cortactin and N-WASp with Arp2/3 complex. Curr. Biol. 12:1270–1278. 10.1016/S0960-9822(02)01035-7 [DOI] [PubMed] [Google Scholar]
- Weed S.A., Du Y., and Parsons J.T.. 1998. Translocation of cortactin to the cell periphery is mediated by the small GTPase Rac1. J. Cell Sci. 111:2433–2443. [DOI] [PubMed] [Google Scholar]
- Welch H., Eguinoa A., Stephens L.R., and Hawkins P.T.. 1998. Protein kinase B and Rac are activated in parallel within a phosphatidylinositide 3OH-kinase-controlled signaling pathway. J. Biol. Chem. 273:11248–11256. 10.1074/jbc.273.18.11248 [DOI] [PubMed] [Google Scholar]
- Wennerberg K., Forget M.A., Ellerbroek S.M., Arthur W.T., Burridge K., Settleman J., Der C.J., and Hansen S.H.. 2003. Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr. Biol. 13:1106–1115. 10.1016/S0960-9822(03)00418-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf R.M., Draghi N., Liang X., Dai C., Uhrbom L., Eklöf C., Westermark B., Holland E.C., and Resh M.D.. 2003. p190RhoGAP can act to inhibit PDGF-induced gliomas in mice: a putative tumor suppressor encoded on human chromosome 19q13.3. Genes Dev. 17:476–487. 10.1101/gad.1040003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondag G.C., Evers E.E., ten Klooster J.P., Janssen L., van der Kammen R.A., and Collard J.G.. 2000. Oncogenic Ras downregulates Rac activity, which leads to increased Rho activity and epithelial-mesenchymal transition. J. Cell Biol. 149:775–782. 10.1083/jcb.149.4.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.