Abstract
Although a consensus has emerged that an HIV vaccine should elicit a cytotoxic T lymphocyte (CTL) response, the characteristics of an effective vaccine-induced T lymphocyte response remain unclear. We explored this issue in the simian human immunodeficiency virus/rhesus monkey model in the course of assessing the relative immunogenicity of vaccine regimens that included a cytokine-augmented plasmid DNA prime and a boost with DNA or recombinant pox vectors. Recombinant vaccinia virus, recombinant modified vaccinia Ankara (MVA), and recombinant fowlpox were comparable in their immunogenicity. Moreover, whereas the magnitude of the peak vaccine-elicited T lymphocyte responses in the recombinant pox virus-boosted monkeys was substantially greater than that seen in the monkeys immunized with plasmid DNA alone, the magnitudes of recombinant pox boosted CTL responses decayed rapidly and were comparable to those of the DNA-alone-vaccinated monkeys by the time of viral challenge. Consistent with these comparable memory T cell responses, the clinical protection seen in all groups of experimentally vaccinated monkeys was similar. This study, therefore, indicates that the steady-state memory, rather than the peak effector vaccine-elicited T lymphocyte responses, may be the critical immune correlate of protection for a CTL-based HIV vaccine.
Recent nonhuman primate studies have shown that a vaccine-elicited cytotoxic T lymphocyte (CTL) response does not provide sterilizing immune protection against a simian immunodeficiency virus (SIV) or simian human immunodeficiency virus (SHIV) challenge but can confer protection against disease progression after infection (1–6). Infected monkeys with preexisting vaccine-elicited CTL responses demonstrate lower viral loads and more benign clinical courses than do infected monkeys without vaccine-induced T cell responses. These findings have provided an impetus for the development of HIV vaccines that elicit virus-specific CTL responses.
One of the most active areas of investigation currently being pursued in the development of vaccine strategies for eliciting HIV-specific CTL responses is the use of recombinant pox vectors. A variety of attenuated pox viruses are being examined as vectors for use as either single modality vaccines or as boosting immunogens in association with heterologous priming immunizations (6–14). These pox vectors include a number of avian pox viruses as well as various attenuated vaccinia viruses (10–14). It is, however, unclear whether one of these pox vectors is superior to another for use in this context.
A central unresolved issue in the effort to develop effective CTL-based HIV vaccines is the type of T lymphocyte responses that will confer optimal protection. For example, T lymphocytes can exist as memory or effector cells, and the T lymphocyte subpopulation that will expand most readily after an infection and will mediate the most effective antiviral activity has not been defined (15–17). Whether different vaccine vectors generate antigen-specific T lymphocytes with different functional repertoires remains unknown.
The present study was initiated to evaluate the relative ability of various pox vectors to boost a plasmid DNA-primed CTL response in rhesus monkeys. The results suggest that recombinant vaccinia virus (rVac), recombinant modified vaccinia Ankara (rMVA), and recombinant fowlpox (rFPV) were comparable in boosting CTL responses. Interestingly, the magnitude of the vaccine-elicited memory CTL populations in all groups of recombinant poxvirus boosted monkeys were no greater than those elicited by plasmid DNA alone.
Materials and Methods
Generation of Poxvirus Recombinants Expressing SHIV89.6P env and SIVmac239 gag. The recombinant vaccinia viruses (rVac) expressing SHIV89.6P env and SIVmac239 gag were constructed by inserting these genes in the HindIII M region of TBC-Wy, Therion strain of vaccinia as described (18). rFPV viruses expressing these same genes were constructed by inserting the genes in the BamJHI region of POXVAC-TC (Schering-Plough) strain of FPV, as described (19). rMVA were generated by inserting these genes in the deletion III region of a plaque-purified isolate of the replication-defective strain of vaccinia virus designated MVA (20). The env and gag genes of the recombinant viruses were under the control of the vaccinia virus 40K(H5R) promoter (21). All of the viruses also contained the Escherichia coli lacZ gene under the control of the fowlpox C1 promoter (19) to facilitate their use in a colorimetric screen for recombinant viruses. The genomic structure of these recombinant viruses was determined by PCR amplification and sequencing. Expression of Gag p55 was demonstrated by Western blot assay with an anti-p27 antibody (Advanced Biotechnologies, Columbia, MD), and expression of Env gp160/gp41 was demonstrated by Western blot assay with an anti-gp41 antibody. Purity of the recombinant viruses was assessed by in situ immunostaining by using the same antibodies. Nonrecombinant wild-type vaccinia virus (Wyeth strain) was designated VV-WT, wild-type fowlpox virus was designated FPV-WT, and wild-type modified vaccinia Ankara was designated MVA-WT. These wild-type viruses were used as control vector immunogens.
Selection and Vaccination of Monkeys. A PCR-based assay was used to select adult rhesus monkeys (Macaca mulatta) that expressed the Mamu-A*01 MHC class I allele (1). Monkeys were housed at Advanced BioScience Laboratories. The animals were maintained in accordance with National Institutes of Health and Harvard Medical School guidelines.
Twenty-eight monkeys were vaccinated by separate intramuscular injections of 5 mg of HIV-1 89.6P Env (KB9) DNA and 5 mg of SIV mac239 Gag DNA in sterile saline without adjuvant. Half the dose was delivered to each quadriceps muscle, and each injection was delivered in a 0.5-ml volume by using a needle-free Biojector apparatus and a no. 4 syringe (Bioject, Portland, OR). These monkeys also received 5 mg of IL-2/Ig plasmid on day 2 after DNA vaccination. In week 42 of the study, 7 of the 28 monkeys were vaccinated by both intradermal and intramuscular injections of 109 plaque-forming units (pfu) of rFPV expressing HIV-1 89.6P Env and 109 pfu of the same virus expressing SIV mac239 Gag. Seven monkeys received 109 pfu rMVA-HIV-1 89.6P Env and 109 pfu rMVA-SIVmac239 Gag, and another seven monkeys received 109 pfu rVac-HIV-1 89.6P Env and 109 pfu rVac-SIVmac239 Gag administered both intradermally and intramuscularly. Another 28 monkeys received 10 mg of sham plasmid DNA and 2 × 109 pfu of empty pox vectors.
Tetramer Staining. One microgram of phycoerythrin-labeled tetrameric Mamu-A*01/peptide complex was used in conjunction with FITC-labeled anti-human CD8α (Leu2a; Becton Dickinson), phycocerythrin-Texas red-labeled anti-human CD8αβ (2ST8-5H7; Beckman Coulter), and allophycocyanin-labeled anti-rhesus monkey CD3 (FN18; BioSource International, Camarillo, CA) monoclonal antibodies to stain peptide-specific CD8+ T cells (22–24). One hundred microliters of whole blood from the vaccinated or control monkeys was directly stained with these reagents, lysed, washed, and fixed. Samples were analyzed by four-color flow cytometry with a Becton Dickinson FACS-Calibur system, and gated CD3+ CD8αβ+ T cells were examined for staining with tetrameric Mamu-A*01/peptide complex.
Enzyme-Linked Immunospot (ELISPOT) Assays. Multiscreen 96-well plates were coated overnight with 100 μl per well of 5 μg/ml anti-human IFN-γ (B27; BD Pharmingen) in endotoxin-free Dulbecco's PBS (D-PBS). The plates were then washed three times with D-PBS containing 0.25% Tween 20, blocked for 2 h with D-PBS containing 5% FBS to remove the Tween 20, and incubated with peptide pools and 2 × 105 peripheral blood mononuclear cells (PBMCs) in triplicate in 100-μl reaction volumes. Each peptide pool was comprised of 15-aa peptides overlapping by 11 aa. The pools covered the entire SIVmac239 Gag protein and the HIV-1 89.6P (KB9) Env protein. Each peptide in a pool was present at a 1 μg/ml concentration. After an 18-h incubation at 37°C, the plates were washed nine times with D-PBS containing 0.25% Tween 20 and once with distilled water. The plates were then incubated with 2 μg/ml biotinylated rabbit anti-human IFN-γ (BioSource) for 2 h at room temperature, washed six times with Coulter Wash (Beckman Coulter), and incubated for 2.5 h with a 1:500 dilution of streptavidin-AP (Southern Biotechnology Associates). After five washes with Coulter Wash and one with D-PBS, the plates were developed with NBT/BCIP chromogen (Pierce), stopped by washing with tap water, air dried, and read with an ELISPOT reader (Hitech Instruments, Edgemont, PA) by using image-pro plus image-processing software (Version 4.1) (Media Cybernetics, Des Moines, IA).
Anti-Env Antibody ELISA and Neutralizing Antibody Assays. A direct ELISA was used to measure plasma titers of anti-gp120 antibodies as described (25). Ninety-six-well Maxisorp ELISA plates (Nunc) were coated overnight at 4°C with 100 μl of PBS containing 1 μg/ml recombinant gp120 protein from HIV-1MN (ImmunoDiagnostics, Woburn, MA). After a wash with PBS containing 0.05% Tween 20, the wells were blocked for 2 h with a blocking buffer containing 5% nonfat dry milk, 2% FBS (HyClone) and 0.05% Tween 20 in PBS. Plasma samples were serially diluted in the blocking buffer and added to the ELISA wells. After a 1-h incubation, the plates were washed three times and then incubated with a 1:2,000 dilution of a peroxidase-conjugated anti-human IgG + IgM secondary antibody (The Jackson Laboratory) for 1 h. The plates were washed three times, developed with 3,3′,5,5′-tetramethylbenzidine (Kirkegaard & Perry Laboratories), stopped with 1% HCl, and analyzed at 450 nm with a Dynatech MR5000 ELISA plate reader.
Neutralizing antibodies were measured in TZM-bl cells (National Institutes of Health AIDS Research and Reference Reagent Program), as described (26), except that we added trypsinized cells (10,000 cells per well) to the virus serum dilutions and added indinavir to inhibit progeny virions. Titers of neutralizing antibodies are the reciprocal plasma dilution at which relative luminescence units were reduced 50% compared to virus control wells (no sample). The assay stock of SHIV-89.6P was grown in human PBMC.
CD4+ T Lymphocyte Counts and Viral RNA Levels. CD4+ T lymphocyte counts were determined by multiplying the total lymphocyte count by the percentage of CD3+ CD4+ T cells determined by flow cytometric analysis. Plasma viral RNA levels were measured by an ultrasensitive branched DNA amplification assay with a detection limit of 125 copies per ml (Bayer Diagnostics, Berkeley, CA).
Results
Vaccine Trial Design. Twenty-one Mamu-A*01+ and 35 Mamu-A*01-rhesus monkeys were distributed into four experimental and four control groups, each consisting of seven animals. Three of the seven monkeys in each of the experimental and three of the four control groups were Mamu-A*01+, whereas one of the control groups had no Mamu-A*01+ monkeys. The monkeys in the four experimental groups received priming immunizations with HIV-1 89.6P Env gp140 (KB9) and SIV mac239 Gag DNA expressed by the pV1R plasmid, adjuvanted by the coadministration of a pV1R plasmid expressing IL-2/Ig (25). Five milligrams of the env DNA vaccine and 5 mg of the gag DNA vaccine were administered intramuscularly as separate injections to all of the monkeys in the four experimental groups by using the needleless Biojector apparatus (Bioject) at weeks 0, 4, and 8. Ten milligrams of a sham plasmid were similarly administered to the control monkeys according to the same schedule. The monkeys in the experimental groups received 5 mg of IL-2/Ig plasmid, and the monkeys in the control groups received 5 mg of sham plasmid on day 2 after the week 0 and 4 vaccinations.
At week 42, groups of experimental monkeys were boosted with 109 pfu of recombinant poxvirus vectors expressing HIV-1 89.6P env and SIVmac239 gag administered both by intramuscular and intradermal routes either as rFPV, rMVA, or rVac. The fourth group of experimental monkeys was boosted with 5 mg of the env DNA and 5 mg of the gag DNA vaccines.
Vaccine-Elicited Immune Responses. The cellular immune responses elicited by these vaccine constructs were assessed by using tetramer staining and pooled peptide ELISPOT assays. CTL specific for the Mamu-A*01-restricted immunodominant SIV Gag p11C and subdominant HIV-1 Env p41A epitope (27) were monitored in the 21 Mamu-A*01-positive monkeys by tetramer staining of fresh PBMC. SIV Gag p11C/Mamu-A*01 tetramer-binding CD3+CD8+ cells were detected in freshly isolated PBMC 2 weeks after the second immunization. Two weeks after the third DNA immunization, 0.7 ± 0.2% circulating CD3+CD8+ cells bound the Gag p11C/Mamu-A*01 tetramer and 0.2 ± 0.05% bound the Env p41A/Mamu-A*01 tetramer in the experimentally vaccinated Mamu-A*01+ monkeys (data not shown). Vaccine-elicited PBMC ELISPOT responses to SIV Gag and HIV-1 Env peptides were detected 2 weeks after the first inoculation, reaching 1,337 ± 273 total spot-forming cells (SFC) per million PBMC 2 weeks after the third DNA immunization (Fig. 6, which is published as supporting information on the PNAS web site).
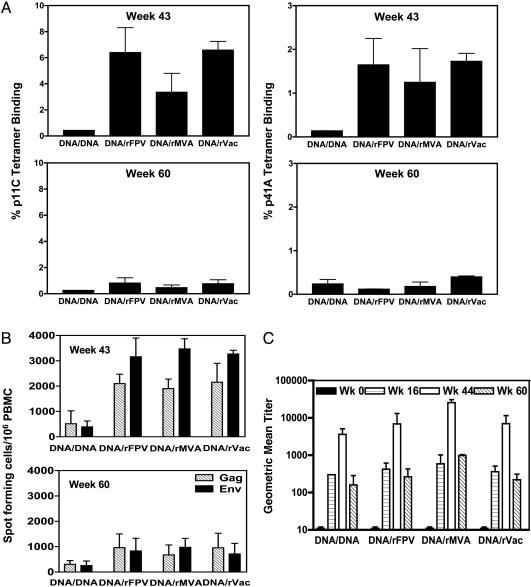
After the boosting immunization at week 42, the Mamu-A*01+ monkeys receiving inoculations of recombinant pox vectors demonstrated dramatic expansions of their p11C- and p41A-specific CD8+ T cell responses (Fig. 1A). Interestingly, the three recombinant pox vectors boosted CTL responses to comparable magnitudes, with peak Gag p11C/Mamu-A*01 tetramer-binding responses of 6.4 ± 3.3% for rFPV, 3.3 ± 2.5% for rMVA, and 6.6 ± 1.1% for rVac vectors at week 43. The HIV-1 Env p41A/Mamu-A*01 tetramer-binding responses were 1.7 ± 0.6% for rFPV, 1.3 ± 0.77% for rMVA, and 1.7 ± 0.18% for rVac-boosted monkeys. The monkeys boosted with plasmid DNA vaccines had ≈10-fold lower peak tetramer-binding cell responses, 0.43 ± 0.02% Gag p11C/Mamu-A*01 tetramerbinding, and 0.22 ± 0.02% Env p41A/Mamu-A*01 tetramer-binding circulating CD8+ T cells. The control vaccinated monkeys demonstrated no detectable tetramer-binding PBMC responses (data not shown).
Fig. 1.
Evolution of the vaccine-elicited cellular and humoral immune responses detected by tetramer staining, PBMC IFN-γ ELISPOT, and anti-Env antibody ELISAs. Tetramer staining and IFN-γ ELISPOT assays are shown for the PBMC samples obtained in week 43 (1 week after the final immunization) and week 60 (day of challenge). (A) Vaccine-elicited CD8+ T cell responses specific for the immunodominant SIV Gag p11C and subdominant HIV-1 Env p41A epitopes were measured by tetramer staining of freshly isolated PBMC. The percent CD3+ CD8+ T cells that bound the Mamu-A*01/peptide-tetramer complexes are shown. The bars represent mean ± SEM for each group. PBMC of the control vaccinated monkeys demonstrated percent tetramer-binding responses of <0.02%. (B) Freshly isolated PBMC were assessed for IFN-γ ELISPOT responses after in vitro exposure to peptide pools spanning the SIVmac239 Gag and HIV-1 Env proteins. The bars represent the mean ± SEM of values of SFC responses to individual viral proteins for seven monkeys in each group. PBMC of the control vaccinated monkeys demonstrated SFC responses of <20. (C) Serum samples from the vaccinated and control monkeys were analyzed for anti-Env antibodies by direct ELISA. The bars represent geometric mean titers ± SE for each group before immunization, postprime, postboost, and day of challenge.
Consistent with the tetramer staining results, the PBMC ELISPOT responses to SIV Gag and HIV-1 Env peptides elicited by each of the three recombinant poxvirus vectors were comparable in magnitude (Fig. 1B). These ELISPOT responses represented both CD4+ and CD8+ T lymphocyte responses, as determined by assays performed using unfractionated and CD8+ T lymphocyte-depleted PBMC from the monkeys (data not shown). PBMC of the control vaccinated monkeys demonstrated SFC responses of <20 per 106 cells (data not shown).
The tetramer-binding CD8+ T cell responses declined in the PBMC of the vaccinated monkeys, reaching a plateau level reflecting memory CD8+ T cell responses 10 weeks after the boost immunizations. Importantly, this contraction was most rapid and dramatic in the groups of animals receiving the recombinant pox vectors. By week 60, tetramer-binding CD8+ T cell responses elicited by the recombinant poxvirus vectors had contracted almost 10-fold from their peak levels (Fig. 1 A). The monkeys boosted with plasmid DNA demonstrated only a 2-fold contraction of the peak p11C-specific CD8+ T cell response to 0.26 ± 0.002% and no contraction in the p41A-specific CD8+ T cells. Consistent with the tetramer staining results, there was a greater contraction of the vaccine-elicited IFN-γ-secreting T cell responses in the groups of monkeys boosted with recombinant pox vectors than in the group of animals boosted with plasmid DNA (Fig. 1B). Thus, the magnitudes of the virus-specific T cell responses as measured by both tetramer and ELISPOT assays were comparable at the time of challenge in the monkeys receiving DNA/pox and those receiving DNA alone immunogens.
To assess the vaccine-elicited humoral immune responses, serum samples from the vaccinated and control monkeys were analyzed for anti-Env antibodies by a direct ELISA by using recombinant gp120 protein (Fig. 1C). Anti-Env antibodies were detected in the vaccinated monkeys after the priming with plasmid DNA. After the boosting immunization at week 42, monkeys in all four vaccination groups had a 10- to 20-fold increase in their anti-Env antibody titers that were comparable in magnitudes in all of the vaccination groups. At week 60, all 4 groups of vaccinated animals showed a decline in antibody titers. In contrast, the sham-vaccinated animals did not develop detectable anti-Env antibodies (data not shown). No HIV-1 89.6P-specific neutralization above background was detected in plasma of the vaccinated monkeys at the time of peak vaccine-elicited immunity or on the day of challenge by using peripheral blood lymphocyte-grown virus in a luciferase reporter gene assay (25) (data not shown).
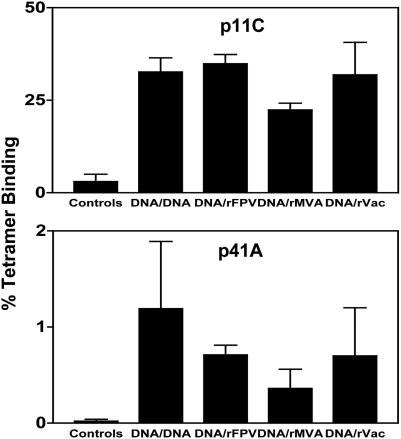
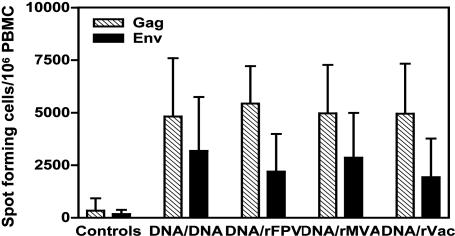
Immune Responses After Viral Challenge. Eighteen weeks after the final immunization, all animals were challenged with 50 MID50 of cell-free SHIV-89.6P by the i.v. route. Because viral replication in these monkeys is likely controlled by anamnestic SHIV-specific CTL populations, we determined the magnitude of their virus-specific T cell responses after challenge. As shown in Fig. 2, the control monkeys developed tetramer-binding CD8+ T lymphocyte responses that were maximal 2 weeks after viral challenge, 3 ± 2% SIV Gag p11C-specific, and no detectable p41A-specific CD8+ T cells. In contrast, all four groups of vaccinated monkeys developed robust secondary p11C-specific CTL responses that were comparable in magnitude. Two weeks after challenge, SIV Gag p11C-specific CD8+ T cell responses were 34.9 ± 2.5%, 22.4 ± 1.8%, and 31.9 ± 8.8% in the groups of monkeys boosted with rFPV, rMVA, and rVac, respectively. Importantly, the animals boosted with plasmid DNA had Gag p11C-specific CD8+ T cell responses of 32.7 ± 3.8%, similar in magnitude to that seen in the recombinant pox virus-boosted animals (Fig. 2). The p41A-specific responses also were ≈30-fold lower than the p11C-specific responses but were comparable in magnitude between the groups (Fig. 2). Consistent with the results of the tetramer-binding assays, the magnitudes of the postchallenge IFN-γ ELISPOT responses to both vaccine antigens were comparable in all four experimentally vaccinated groups of monkeys (Fig. 3). Therefore, although the prechallenge peak vaccine-elicited immune responses were greater in the groups of monkeys boosted with recombinant pox vectors, the prechallenge plateau and postchallenge peak secondary responses were equivalent in magnitude in all four experimental groups of animals.
Fig. 2.
Peak SHIV-89.6P-specific CTL responses after viral challenge. Monkeys were challenged with SHIV-89.6P and CD8+ T cell responses specific for the SIV Gag p11C, and HIV-1 Env p41A epitopes were determined by tetramer staining of freshly isolated PBMC. The percent CD3+ CD8+ T cells that bound the Mamu-A*01/peptide-tetramer complexes 2 weeks after viral challenge are shown.
Fig. 3.
PBMC IFN-γ ELISPOT responses 2 weeks after viral challenge. Freshly isolated PBMC were assessed for IFN-γ ELISPOT responses after in vitro exposure to peptide pools spanning the SIVmac239 Gag and HIV-1 89.6P Env proteins. The bars represent the mean ± SEM of values of SFC responses to individual viral proteins for 7 monkeys in each group of experimentally vaccinated animals and for 28 monkeys in the group of control vaccinated animals.
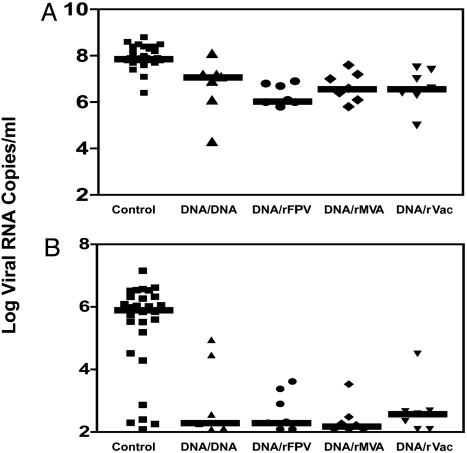
Plasma Viral RNA Levels and CD4 T Cell Counts. Viral replication in the SHIV-89.6P-challenged monkeys was assessed by quantitating viral RNA levels in their plasma by using an ultrasensitive branched DNA assay with a detection limit of 125 copies per ml (Bayer Reference Testing Laboratory, Berkeley, CA). At the time of peak viremia, the control monkeys had higher plasma viremia than the vaccinated monkeys with a median value of eight log copies of viral RNA per milliliter of plasma. The median values of the peak plasma viral RNA levels in the four groups of experimentally vaccinated monkeys were 7.1 (plasmid DNA), 6.1 (rFPV), 6.6 (rMVA), and 6.6 (rVac) log copies (Fig. 4A). Thus, the experimentally vaccinated monkeys had significantly lower peak viral loads than did the control vaccinees (Wilcoxon rank sum tests, P < 0.002). However, comparisons among the four groups of experimentally vaccinated monkeys did not show significant differences in their peak viral loads.
Fig. 4.
Postchallenge plasma viral RNA levels at (A) peak viremia and (B) set point. These values were determined by an ultrasensitive branched DNA amplification assay with a detection limit of 125 copies per ml. Peak viremia values represent plasma viral RNA levels on day 10–14 after challenge. Set-point values represent median viral RNA levels for each animal between days 120 and 180 postchallenge.
Because the steady-state or set-point plasma viral RNA levels have proven robust predictors of the clinical sequelae of chronic HIV/SIV/SHIV infection, we also assessed these levels in the challenged monkeys. The set-point plasma viral RNA values used for evaluation were the median of four data points determined between days 120 and 180 postchallenge. Using these set-point values, 24 of the 28 control monkeys had high viral loads, with a median value of 5.9 log copies per ml. The medians of plasma viral RNA levels at set point in the groups of experimentally vaccinated monkeys were 2.29 log copies per ml for plasmid DNA, 2.33 log copies per ml for rFPV, 2.23 log copies per ml for rMVA, and 2.6 log copies per ml for rVac boosted animals (Fig. 4B). Therefore, at set point, the vaccinated animals had almost 3–3.5 logs lower plasma viral RNA levels than the control animals, as our laboratory and others had previously reported. These values were significantly lower than that of the controls, with P = 0.007 (plasmid DNA), P = 0.007 (rFPV), P = 0.002 (rMVA), and P = 0.007 (rVac), as determined by Wilcoxon rank-sum tests. However, the four groups of experimentally vaccinated monkeys did not have statistically significant differences in their set-point viral loads (see also Fig. 7, which is published as supporting information on the PNAS web site).
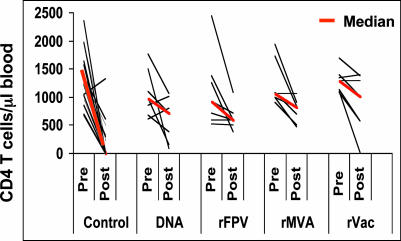
Twenty-three of the 28 control monkeys had a profound loss of their CD4+ T cells between days 14 and 35 after challenge. In contrast, 27 of the 28 vaccinated animals maintained their CD4+ T cells (Fig. 8, which is published as supporting information on the PNAS web site). To maintain consistency with the determination of the set-point plasma viral RNA levels, postchallenge CD4+ T cell counts for each animal were expressed as the median of the CD4+ T cell counts determined between days 120 and 180 postchallenge. Using these set-point values, a highly significant difference in peripheral blood CD4+ T cell counts was evident between the control monkeys and the monkeys that received experimental vaccines. The control vaccinated monkeys had a median CD4+ T cell count of 6, and the vaccinated animals had median CD4+ T cell counts of 723 (plasmid DNA), 602 (rFPV), 824 (rMVA), and 1,015 (rVac) (Fig. 5). These values were significantly higher than that of the control monkeys, with P = 0.004 (plasmid DNA), P = 0.002 (rFPV), P = 0.002 (rMVA), and P = 0.002 (rVac), as determined by Wilcoxon rank-sum tests. Consistent with the plasma viral RNA levels and CD4+ T lymphocyte counts in the infected monkeys, 21 of the 28 control but none of the experimentally vaccinated monkeys were killed because of advanced disease by day 300 postchallenge. However, no significant difference in CD4+ T cell loss was observed between groups of experimentally vaccinated monkeys.
Fig. 5.
Decline in CD4+ T lymphocyte counts after SHIV-89.6P challenge. These values were determined by multiplying percentage of CD3+CD4+ lymphocytes by the total lymphocyte counts. Postchallenge CD4+ T lymphocyte counts for each animal are the median CD4+ T lymphocyte count between days 120 and 180 postchallenge.
Discussion
The present study showed that three different pox vectors were comparable in their ability to boost DNA vaccine-primed CTL responses in monkeys. A number of explanations might account for the comparable immunogenicity of the rFPV, rMVA, and rVac constructs. These different constructs may simply elicit CTL populations of similar magnitude and might also do so even as stand-alone vaccines. Alternatively, these vaccines may differ in their immunogenicity in naïve animals. A number of studies have suggested that considerably less antigen expression is required to expand an already primed population of memory CTL than to elicit such a population of cells in a naïve host (28–30). It is possible that the antigen expression from even the weakest of the recombinant pox immunogens was sufficient to boost optimally the population of memory T lymphocytes elicited by the plasmid DNA vaccination. Higher levels of antigen expression from the other recombinant pox viruses might therefore provide no incremental immunogenicity in these monkeys.
Although the various DNA prime/recombinant pox boost vaccine strategies elicited higher-frequency peak CTL responses than did the single modality plasmid DNA vaccination, all of these vaccine approaches generated comparable plateau memory responses. Thus, there was a rapid and dramatic contraction of the vaccine-elicited CTL populations in the recombinant pox virus-boosted monkeys, and no detectable differences in the magnitudes of the virus-specific T cell responses in the various groups of vaccinated monkeys were observed at the time of challenge. The apparent equivalence of these vaccine-elicited memory CTL responses was confirmed by their comparable expansions postchallenge. The equivalence of the postchallenge viral loads and CD4+ T lymphocyte counts is consistent with the comparable postchallenge immune responses of these groups of monkeys. However, it is possible that the rhesus monkey/SHIV-89.6P model is not sensitive enough to detect differences in the degree of clinical protection conferred by these vaccine-elicited T cell responses.
The findings in the present study have a number of important implications for plans to advance specific HIV vaccine candidates forward into efficacy trials. First, there appears to be no evidence in preclinical studies that any one of these pox vectors will outperform the others. Second, and perhaps of greater significance, judging the relative potential of one vaccine over another on the basis of the magnitude of a peak vaccine-elicited T lymphocyte response may be misleading. Rather, the best immune correlate of clinical protection may be the magnitude of the vaccine-elicited memory CTL population.
Supplementary Material
Acknowledgments
We acknowledge Nancy Miller, Shawn Jackson, Karen Hershberger, Kristin Beaudry, Nicole Hall, Sharon Orndorff, Jim Treece, and Deborah Weiss for generous advice and assistance; and Gail Mazzara, Michael Wyand, Linda Gritz, and Alicia Gomez-Yafal for reagents (all from Therion Biologics, Cambridge, MA). This work was supported by National Institutes of Health Grants AI-15430, AI-26507, and AI-30033.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CTL, cytotoxic T lymphocyte; ELISPOT, enzyme-linked immunospot; SIV, simian immunodeficiency virus; SHIV, simian human immunodeficiency virus; MVA, modified vaccinia Ankara; rMVA, recombinant MVA; rVac, recombinant vaccinia virus; rFPV, recombinant fowlpox; pfu, plaque-forming units; D-PBS, Dulbecco's PBS; PBMC, peripheral blood mononuclear cells; SFC, spot-forming cells.
References
- 1.Barouch, D. H., Santra, S., Schmitz, J. E., Kuroda, M. J., Fu, T.-M., Wagner, W., Bilska, M., Craiu, A., Zheng, X. X., Krivulka, G. R., et al. (2000) Science 290, 486–492. [DOI] [PubMed] [Google Scholar]
- 2.Egan, M. A., Charini, W. A., Kuroda, M. J., Schmitz, J. E., Racz, P., Tenner-Racz, K., Manson, K., Wyand, M., Lifton, M. A., Nickerson, C. E., et al. (2000) J. Virol. 74, 7485–7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amara, R. R., Villinger, F., Altman, J. D., Lydy, S. L., O'Neil, S. P., Staprans, S. I., Montefiori, D. C., Xu, Y., Herndon, J. G., Wyatt, L. S., et al. (2001) Science 292, 69–74. [DOI] [PubMed] [Google Scholar]
- 4.Rose, N. F., Marx, P. A., Luckay, A., Nixon, D. F., Moretto, W. J., Donahoe, S. M., Montefiori, D., Roberts, A., Buonocore, L. & Rose, J. K. (2001) Cell 106, 539–549. [DOI] [PubMed] [Google Scholar]
- 5.Shiver, J. W., Fu, T.-M., Chen, L., Casimiro, D. R., Davies, M. E., Evans, R. K., Zhang, Z. Q., Simon, A. J., Trigona, W. L., Dubey, S. A., et al. (2002) Nature 415, 331–335. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch, V. M., Fuerst, T. R., Sutter, G., Carroll, M. W., Yang, L. C., Goldstein, S., Piatak, M., Elkins, W., Alvord, W. G., Montefiori, D. C., et al. (1996) J. Virol. 70, 3741–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belyakov, I. M., Wyatt, L. S., Ahlers, J. D., Earl, P., Pendleton, C. D., Kelsall, B. L., Strober, W., Moss, B. & Berzofsky, J. A. (1998) J. Virol. 72, 8264–8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ourmanov, I., Brown, C. R., Moss, B., Carroll, M., Wyatt, L., Pletneva, L., Goldstein, S., Venzon, D. & Hirsch, V. M. (2000) J. Virol. 74, 2740–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egan, M. A., Pavlat, W. A., Tartaglia, J., Paoletti, E., Weinhold, K. J., Clements, M. L. & Siliciano, R. F. (1995) J. Infect. Dis. 171, 1623–1627. [DOI] [PubMed] [Google Scholar]
- 10.Seth, A., Ourmanov, I., Kuroda, M. J., Schmitz, J. E., Carroll, M. W., Wyatt, L. S., Moss, B., Forman, M. A., Hirsch, V. M. & Letvin, N. L. (1998) Proc. Natl. Acad. Sci. USA 95, 10112–10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent, S. J., Zhao, A., Best, S. J., Chandler, J. D., Boyle, D. B. & Ramshaw, I. A. (1998) J. Virol. 72, 10180–10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson, H. L., Montefiori, D. C., Johnson, R. P., Manson, K. H., Kalish, M. L., Lifson, J. D., Rizvi, T. A., Lu, S., Hu, S.-L., Mazzara, G. P., et al. (1999) Nat. Med. 5, 526–534. [DOI] [PubMed] [Google Scholar]
- 13.Santra, S., Schmitz, J. E., Kuroda, M. J., Lifton, M. A., Nickerson, C. E., Lord, C. I., Pal, R., Franchini, G. & Letvin, N. L. (2002) J. Immunol. 168, 1847–1853. [DOI] [PubMed] [Google Scholar]
- 14.Allen, T. M., Vogel, T. U., Fuller, D. H., Mothe, B. R., Steffen, S., Boyson, J. E., Shipley, T., Fuller, J., Hanke, T., Sette, A., et al. (2000) J. Immunol. 164, 4968–4978. [DOI] [PubMed] [Google Scholar]
- 15.Seder, R. A. & Ahmed, R. (2003) Nat. Immunol. 4, 835–842. [DOI] [PubMed] [Google Scholar]
- 16.Sallusto, F., Lenig, D., Forster, R., Lipp, M. & Lanzavecchia, A. (1999) Nature 401, 708–712. [DOI] [PubMed] [Google Scholar]
- 17.Wherry, E. J., Teichgraber, V., Becker, T. C., Masopust, D., Kaech, S. M., Antia, R., von Andrian, U. H. & Ahmed, R. (2003) Nat. Immunol. 4, 225–234. [DOI] [PubMed] [Google Scholar]
- 18.Mazzara, G. P., Destree, A. & Mahr, A. (1993) Methods Enzymol. 217, 557–581. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins, S., Gritz, L., Fedor, C. H., O'Neill, E. M., Cohen, L. K. & Panicali, D. L. (1991) AIDS Res. Hum. Retroviruses 7, 991–998. [DOI] [PubMed] [Google Scholar]
- 20.Mayr, A., Hochstein-Mintzel, V. & Stickl, H. (1975) Infection 3, 6–14. [Google Scholar]
- 21.Gritz, L., Destree, A., Cormier, N., Day, E., Stallard, V., Caiazzo, T., Mazzara, G. & Panicali, D. (1990) J. Virol. 64, 5948–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altman, J. D., Moss, P. A. H., Goulder, P. J. R., Barouch, D. H., McHeyzer-Williams, M. G., Bell, J. I., McMichael, A. J. & Davis, M. M. (1996) Science 274, 94–96. [DOI] [PubMed] [Google Scholar]
- 23.Kuroda, M. J., Schmitz, J. E., Barouch, D. H., Craiu, A., Allen, T. M., Sette, A., Watkins, D. I., Forman, M. A. & Letvin, N. L. (1998) J. Exp. Med. 187, 1373–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroda, M. J., Schmitz, J. E., Charini, W. A., Nickerson, C. E., Lord, C. I., Forman, M. A. & Letvin, N. L. (1999) J. Virol. 73, 1573–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barouch, D. H., Craiu, A., Kuroda, M. J., Schmitz, J. E., Zheng, X. X., Santra, S., Frost, J. D., Krivulka, G. R., Lifton, M. A., Crabbs, C. L., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 4192–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei, X., Decker, J. M., Wang, S., Hui, H., Kappes, J. C., Wu, X., Salazar-Gonzalez, J. F., Salazar, M. G., Kilby, J. M., Saag, M. S., et al. (2003) Nature 422, 307–312. [DOI] [PubMed] [Google Scholar]
- 27.Egan, M. A., Kuroda, M. J., Voss, G., Schmitz, J. E., Charini, W. A., Lord, C. I., Forman, M. A. & Letvin, N. L. (1999) J. Virol. 73, 5466–5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Stipdonk, M. J., Lemmens, E. E. & Schoenberger, S. P. (2001) Nat. Immunol. 2, 423–429. [DOI] [PubMed] [Google Scholar]
- 29.Kaech, S. M. & Ahmed, R. (2001) Nat. Immunol. 2, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong, P. & Pamer, E. G. (2001) J. Immunol. 166, 5864–5868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.