Abstract
Reovirus is a benign human virus that was recently found to have oncolytic properties and is currently in clinical trials as a potential cancer therapy. We have previously demonstrated that activation of Ras signaling, a common event in cancer, renders cells susceptible to reovirus oncolysis. In this study, we investigate which elements downstream of Ras are important in reovirus infection. By using a panel of NIH 3T3 cells transformed with activated Ras mutated in the effector-binding domain, we found that only the RasV12G37 mutant, which was unable to signal to Raf or phosphatidylinositol 3-kinase but retained signaling capability to guanine nucleotide-exchange factors (GEFs) for the small G protein, Ral (known as RalGEFs), was permissive to reovirus. Expression of the activated mutant of the RalGEF, Rlf, also allowed reovirus replication. Specific inhibition of the Ral pathway by using dominant-negative RalA rendered normally permissive H-Ras cells (cells expressing activated Ras) resistant to reovirus. To further identify elements downstream of RalGEF that promote reovirus infection, we used chemical inhibitors of the downstream signaling elements p38 and JNK. We found that reovirus infection was blocked in the presence of the p38 inhibitor but not the JNK inhibitor. Together, these results implicate a Ras/RalGEF/p38 pathway in the regulation of reovirus replication and oncolysis.
Keywords: Ras signaling pathway, reovirus cancer therapy
Mammalian reovirus is a small, nonenveloped icosahedral virus with a segmented double-stranded RNA genome (reviewed in ref. 1). Found ubiquitously in the environment, most humans have been infected with reovirus, although infections are usually subclinical and go unnoticed. Despite its benign nature in humans, reovirus has served well as a tool for the study of many viral phenomena, including reassortment of viral genes, viral tropism in mice, receptor binding and entry, and induction of apoptosis in host cells.
An exciting characteristic of reovirus is that it is naturally oncolytic. Hashiro et al. (2) and Duncan et al. (3) have reported that reovirus can replicate in transformed and cancer cells. More recently, we demonstrated that cells that are nonpermissive to reovirus can be rendered permissive by overexpression or constitutive activation of the epidermal growth factor receptor (EGFR), whose signaling induces transformation (4, 5). Subsequent studies have revealed that activation of elements downstream of the EGFR, namely, the guanine nucleotide-exchange factor (GEF) Sos and the small G protein Ras, also renders cells permissive to reovirus by a mechanism that involves promotion of viral protein synthesis (6).
Ras proteins are known for their versatile signaling capabilities in various organisms, communicating extracellular cues to elicit such outcomes as differentiation, proliferation, and motility, yielding significant physiological consequences. Constitutively active (e.g., the V12 mutant) Ras is predominantly GTP-bound, because of lower intrinsic GTPase activity and its resistance to the GTPase-stimulating activity of RasGAPs (7, 8). Activated Ras is a potent inducer of cellular transformation, mediated by the cooperative activation of several downstream effectors (reviewed in ref. 9). Ras–GTP stimulates >18 effectors, with the most well known elements being the Raf kinases, phosphatidylinositol 3-kinase (PI3-kinase), and GEFs for the small G protein Ral. Together, signaling through these and other effectors cooperates to contribute to tumorigenesis in many ways. For example, oncogenic Ras promotes metastasis, angiogenesis, and loss of growth control. Activating mutations in ras genes have been found in >30% of cancers, and constitutive Ras pathway signaling brought about by oncogenic changes in upstream and downstream elements arises in an even greater proportion of human tumors. Thus, aberrant Ras signaling has major clinical implications.
Given the prevalence of Ras activation in human tumors and the propensity of reovirus to replicate in ras-transformed cells, several preclinical studies (10–18) have demonstrated the potential usefulness of reovirus as a Ras pathway-directed cancer therapy. Reovirus therapy efficacy has been demonstrated in both immunocompromised and immunocompetent animal models of cancer, with reovirus administered by direct, i.v., and i.p. injection, as well as intracerebral delivery. Virus replication has also been documented in ex vivo human tumor surgical specimens. Altogether, reovirus oncolysis has been demonstrated in many cancer types, including human breast, colon, ovarian, neurological, hematological, pancreatic, and bladder neoplasms. Ultimately, these studies have led to human clinical trials testing the effectiveness of reovirus therapy of cancer.
Although much preclinical work has been carried out establishing that reovirus could serve as a cancer therapy, the mechanisms governing the permissiveness of transformed cells to reovirus infection remain to be fully characterized. Because reovirus replicates in cancers of very diverse origin, it is likely that the virus exploits cellular signals that occur often in transformation and tumorigenesis. Thus, delineating this usurpation should in turn shed light on signaling that is common among various cancers (and perhaps necessary for tumorigenesis), and it may reveal novel therapeutic targets. In this study, we focus on signaling downstream of activated Ras that confers host cell permissiveness to reovirus. By using various mutants of Ras and downstream elements, as well as signaling inhibitors, we show that reovirus exploits an activated Ras/RalGEF/p38 pathway in the host cell for infection.
Materials and Methods
Cell Lines and Virus. L-929 and NIH 3T3 cells were obtained from the American Type Culture Collection. Rlf–CAAX, Rlf–CAAX (inact.), and parental A14 cells (a variant of NIH 3T3 expressing the human insulin receptor) were kindly provided by J. Bos (Utrecht University Medical Center, Utrecht, The Netherlands) (19). Ras effector mutant cell lines were a kind gift from C. Webb and G. Vande Woude (Van Andel Research Institute, Grand Rapids, MI). H-Ras cells (cells expressing activated Ras) were generously provided by D. Faller (Boston University School of Medicine, Boston). L-929 cells were maintained in Joklik's modified Eagle's medium (JMEM), containing 5% FBS (Cansera, Etobicoke, Ontario, Canada) and antibiotics, and all other cells were maintained in high-glucose DMEM (Invitrogen) with 10% FBS and antibiotics. The Dearing strain of reovirus serotype 3 was propagated in L-929 cells that were grown in suspension, and it was purified according to the protocol of Smith et al. (20).
Molecular Constructs and Reagents. Activated Ras and Ras effector mutant constructs in pBabepuro were generously provided by C. Der and A. Cox (University of North Carolina, Chapel Hill). dnRalA A26 in pBabepuro was a kind gift from A. Chan (Mount Sinai School of Medicine, New York) (21). The identity of all constructs was verified by sequencing.
Retroviral Infection. Retroviral DNA was transfected by using Superfect (Qiagen, Valencia, CA) into the Bosc 23 packaging cell line according to the manufacturer's specifications, and retrovirus-containing supernatant was harvested and filtered at 48 h after transfection, flash-frozen, and stored at –70°C until use. NIH 3T3 cells were infected with retrovirus in the presence of polybrene (8 μg/ml), and at 48 h after infection, cells were subjected to selection in medium containing 2 μg/ml puromycin. All experiments were carried out on pools of puromycin-resistant cells within 2–3 weeks of retroviral infection, and experiments were performed in the absence of puromycin.
Reovirus Infection, Metabolic Radiolabeling, and Immunoprecipitation. For infection with reovirus, cells were plated in six-well plates (3 × 105 cells per well) at 24 h before infection. Cells were then infected at a multiplicity of infection of 10–100 plaque-forming units per cell. For drug treatment, cells were incubated with the indicated inhibitor for the duration of the experiment. The following inhibitors were used: LY294002, SP600125, U0126, PP2, and SB203580 (Calbiochem). At designated time points after infection, the medium was replaced with methionine and cysteine-free DMEM with 10% dialyzed FBS (Invitrogen) and [35S]methionine (50 μCi/ml; 1 Ci = 37 GBq; Amersham Biosciences). After 5 h, medium was removed and cells were washed in ice-cold PBS and lysed in PBS containing 1% Triton X-100, 0.5% sodium deoxycholate, 1 mM EDTA, and 10 μg/ml aprotinin. Nuclei were removed by centrifugation and supernatants were stored at –70°C until use.
Polyclonal rabbit antireovirus serotype 3 serum prebound to protein A/acrylic beads [prepared with 30 μl of serum per 1 ml of beads in TBS (20 mM Tris/137 mM NaCl, pH 7.3)] was used for immunoprecipitation of [35S]methionine-labeled reovirus proteins from cell lysates. Cell lysates were diluted 1:5 in TBS/1% Triton X-100, and 50 μl of Ab/beads was added. Immunoprecipitation reactions were incubated for1 h, and bead complexes were washed three times in TBS/Triton X-100 and then boiled in 50 μl of 2× protein sample buffer. Samples were subjected to 10% SDS/PAGE, as described by Laemmli (22), followed by autoradiography.
RT-PCR. Cells were infected with reovirus, and at various times after infection, RNA was harvested by extraction with TRIzol (Invitrogen) according to the manufacturer's directions. RNA was analyzed by RT-PCR, as described by Strong et al. (6).
Small GTPase Assays and Western Blot Analysis. Ras–GTP and Ral–GTP levels were measured by using Ras and Ral activation assay kits (Upstate Biotechnology, Lake Placid, NY). Subconfluent cells in 15-cm dishes were washed two times in ice-cold TBS and lysed by scraping in 500 μl of Mg2+ lysis buffer (MLB, Upstate Biotechnology). We then incubated 600 μg of precleared lysate with 30 μl of GST–Raf Ras binding domain or GST–RalBP1 Ral binding domain bound to glutathione agarose for 45 min at 4°C. Beads were then washed three times in MLB and boiled in 40 μl of 2× protein sample buffer with 2 μl of 1M DTT. We then subjected 20 μl of pull-down sample and 40 μg of total lysate to 15% SDS/PAGE and transferred them to nitrocellulose membrane. Blots were probed overnight with 1,000-fold-diluted RAS10 mAb (Upstate Biotechnology) or RalA mAb (Transduction Laboratories, Lexington, KY) and then washed three times for 5 min in TBS/0.1% Tween-20, followed by incubation in goat anti-mouse-HRP Ab (1:2500, Santa Cruz Biotechnology). After three more washes, blots were processed and signals were visualized by using enhanced chemiluminescence (Amersham Biosciences).
For detection of activated AKT, extracellular signal-regulated kinase (ERK), and mitogen-activated protein kinase kinase (MEK) cells were serum-starved for 24 h in 0.5% FBS, washed in ice-cold TBS, and lysed in standard radioimmunoprecipitation assay buffer with phosphatase inhibitors (TBS/0.1% SDS/0.5% sodium deoxycholate/1% Triton X-100/10 mM sodium pyrophosphate/25 mM β-glycerophosphate/1 mM sodium orthovanadate/25 mM sodium fluoride). We analyzed 40 μg of precleared lysate by Western blotting using the following Abs obtained from Cell Signaling Technology (Beverly, MA): anti-phospho-AKT, anti-total AKT, anti-phospho-ERK1/2, anti-total ERK 1/2, anti-phospho-MEK1/2, and anti-total MEK1/2.
Results
Reovirus Replication Does Not Depend on Raf or PI3-Kinase Signaling Downstream of Ras. Previously, we found that cellular transformation by oncogenic Ras or activated upstream signaling elements rendered NIH 3T3 cells acutely susceptible to reovirus infection through promotion of viral protein synthesis. Because Ras itself has a multitude of effectors, initial studies were aimed at narrowing the field of potential downstream elements needed for reovirus infection. To this end, we used NIH 3T3 cell lines expressing effector binding domain mutants of activated Ras. Each mutant is an active form (V12) of Ras harboring an additional point mutation in the effector-binding domain, which impairs interaction (and signaling) with some, but not all effectors (23, 24). For example, RasV12C40 is an activated form of Ras that has lost its ability to bind Raf kinase and RalGEFs but retains signaling capability to PI3-kinase. RasV12G37 activates RalGEFs but not Raf or PI3-kinase, and the RasV12S35 mutant cannot activate RalGEFs or PI3-kinase but does stimulate Raf activity. In assessing the relative capacity of these mutants to support reovirus protein synthesis, it would be possible to determine which effectors are not essential for Ras-abetted reovirus oncolysis.
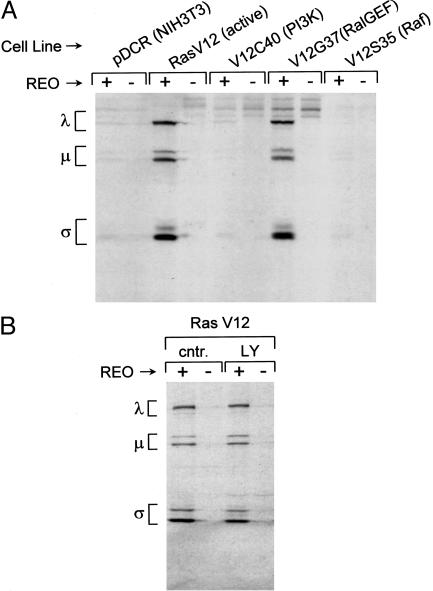
To assess reovirus protein synthesis, infected or mock-infected cells were radiolabeled with [35S]methionine at 60 h after infection and then lysed. Reovirus proteins were immunoprecipitated from the lysate and subjected to SDS/PAGE. As expected, we found that RasV12-expressing cells were much more permissive to reovirus than the empty vector-containing NIH 3T3 cells (pDCR) (Fig. 1A). Of the three effector mutants, RasV12C40 and RasV12S35 were much less permissive for reovirus protein synthesis than the RasV12G37-expressing cells. This preference was confirmed in another set of NIH 3T3 cells in which Ras mutants were expressed by retroviral vectors (data not shown). The V12G37 mutant is unable to stimulate Raf or PI3-kinase efficiently, suggesting that signaling via these pathways is not involved in Ras promotion of reovirus protein synthesis.
Fig. 1.
Involvement of Ras effector pathways in reovirus susceptibility. (A) Infection of Ras effector binding domain mutant-expressing NIH 3T3 cells. NIH 3T3 cells expressing empty vector (pDCR), activated Ras (V12), or activated Ras with a point mutation in the effector binding domain (V12C40, V12G37, and V12S35) were infected with reovirus at a multiplicity of infection of 40 plaque-forming units per cell and pulse-labeled with [35S]methionine-containing medium for 5 h at 60–65 h after infection. The cells were then lysed, and reovirus proteins were immunoprecipitated from part of the lysate by using rabbit polyclonal antireovirus Ab. Immunoprecipitated proteins were resolved by 10% SDS/PAGE, followed by autoradiography. Migration of the three size classes of reovirus proteins (λ, μ, and σ) is indicated on the left. (B) Effect of the PI3-kinase inhibitor, LY294002, on reovirus infection of Ras-transformed NIH 3T3 cells. NIH 3T3 cells expressing activated Ras were infected with reovirus at a multiplicity of infection of 40 plaque-forming units per cell and treated with 20 μM LY294002 (LY) or left untreated (cntr.) for the duration of the experiment. Cells were metabolically radiolabeled with [35S]methionine at 43–48 h after infection, and lysates were prepared and analyzed as described in A. Reovirus protein migration (λ, μ, and σ) is indicated on the left.
To further confirm these results, we treated cells expressing RasV12 with the PI3-kinase inhibitor LY294002 (Fig. 1B). We found that this inhibitor did not suppress signaling downstream of Ras that is necessary for reovirus infection. Moreover, whereas reovirus protein synthesis was largely unaffected, total cellular protein synthesis was partially suppressed by LY294002, suggesting that viral and cellular translation are differently regulated (data not shown). Consistent with our previous demonstration that inhibition of MEK with PD98059 has no effect (6), we found that another MEK inhibitor, U0126, also does not block reovirus infection (data not shown). Together with the Ras effector domain mutant data, these results suggest that oncogenic Ras stimulation of Raf and PI3-kinase is not involved in promotion of reovirus protein synthesis.
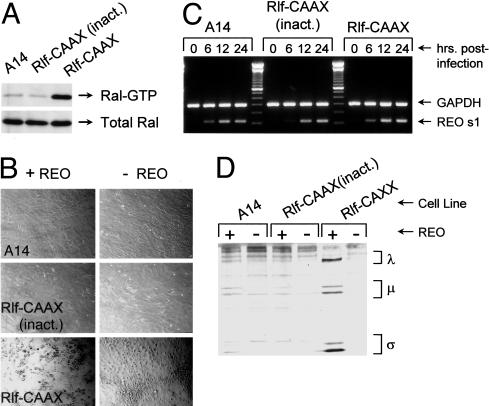
Activation of RalGEF Signaling Renders Cells Permissive to Reovirus. Although the V12G37 mutant does not signal to Raf or PI3-kinase, it does retain signaling capability to members of the GEF family for the small G protein Ral. To determine whether RalGEF signaling can render cells permissive to reovirus, we tested NIH 3T3 cells expressing an active mutant of the RalGEF, Rlf. Because Ras promotes effector activity through their targeting to the plasma membrane, effectors are commonly constitutively activated through attachment of the CAAX farnesylation signal, which helps to anchor the protein to membrane (25). Fig. 2A shows that cells expressing Rlf–CAAX manifested enhanced Ral activity relative to parental A14 cells (control). Upon infection with reovirus, the Rlf–CAAX cells were indeed permissive to reovirus, as demonstrated by the presence of cytopathic effects by 72 h after infection (Fig. 2B); parental A14 cells were nonpermissive to the virus. Furthermore, because Rlf has been found to interact with the kinase PDK1 in a non-GEF activity-dependent manner, we sought to determine whether Rlf protein expression alone could promote permissiveness. Accordingly, we tested infectibility of cells expressing Rlf–CAAX mutated in the GEF catalytic domain (inact.), which is incapable of stimulating Ral (25) (Fig. 2 A). As seen with the parental cells, Rlf–CAAX (inact.)-expressing cells could not support virus replication (Fig. 2B), indicating that RalGEF activity is necessary to render cells permissive to reovirus.
Fig. 2.
Reovirus infection of cells with activated RalGEF. (A) Ral activity in A14 cells expressing empty vector, active Rlf–CAAX, or Rlf–CAAX with a point mutation in the catalytic domain (inact.). Cells were lysed and Ral–GTP as well as total Ral levels were assessed, as described in Materials and Methods. (B) Cytopathic effects induced by reovirus. Cells were infected with reovirus at a multiplicity of infection of 40 plaque-forming units per cell or mock-infected, and photomicrographs were taken at 72 h after infection. (C) Reovirus S1 RNA synthesis in infected A14, Rlf–CAAX (inact.), and Rlf–CAAX cells. Cells were infected with reovirus, and at various times after infection RNA was extracted and subjected to RT-PCR for reovirus S1 RNA and cellular GAPDH RNA (control). Reactions were resolved by 2% agarose gel electrophoresis and visualized by ethidium bromide staining. (D) Reovirus protein synthesis in A14, Rlf–CAAX (inact.), and Rlf-CAXX-expressing cells. Infected cells were pulse-labeled with [35S]methionine-containing medium for 5 h at 67–72 h after infection. After labeling, cells were lysed and reovirus proteins were analyzed by immunoprecipitation and 10% SDS/PAGE/autoradiography, as described in Fig. 1. The three size classes of reovirus proteins (λ, μ, and σ) are indicated on the right.
Previous work has demonstrated that reovirus can successfully enter both permissive and nonpermissive cells (e.g., NIH 3T3 and H-Ras) to produce reovirus transcripts yet can synthesize protein only in permissive cells. Therefore, we next tested whether A14, Rlf–CAAX (inact.) and Rlf–CAAX cells all have the capacity to allow viral transcription. Cells were first infected with reovirus, and at various times after infection, RNA was extracted for RT-PCR to assess the presence of reovirus transcripts (Fig. 2C). As found previously, both resistant and permissive cell lines supported production of the reovirus S1 transcript. Examination of other reovirus transcripts also revealed no differences in the ability of nonpermissive and permissive cells to allow transcription (data not shown), suggesting that Rlf promotes after posttranscriptional events in the reovirus replication cycle.
To determine whether Rlf, like Ras, promotes reovirus protein synthesis, cells were infected with reovirus, and at 67 h after infection, proteins were metabolically labeled with [35S]methionine (Fig. 2D). SDS/PAGE analysis of reovirus protein imumuoprecipitates indicated that parental control cells and mutant Rlf–CAAX-expressing cells were relatively incapable of supporting reovirus protein synthesis. Active Rlf–CAAX-expressing cells, however, allowed effective synthesis of viral proteins. It thus appears that active Rlf acts similarly to Ras to promote reovirus infection at the level of translation.
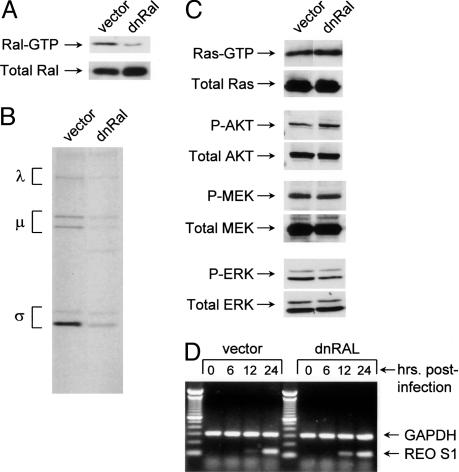
Down-Regulation of RalGEF Activity Impairs Reovirus Replication in H-Ras Cells. To further support the notion that RalGEF signaling mediates reovirus permissiveness, we tested the effect of Ral-GEF inhibition in H-Ras cells. H-Ras cells were infected with pBabepuro retrovirus (vector control) or a dominant-negative Ral (dnRal)-expressing retrovirus and put under selection in puromycin-containing medium, and surviving cell pools were tested for inhibition of Ral activation. Fig. 3A shows that Ral activation was suppressed in cells expressing dnRal compared with control cells. Correspondingly, these cells also showed impaired ability to support reovirus protein synthesis (Fig. 3B). This impairment was not due to nonspecific inhibition of Ras or Raf/PI3-kinase pathway signaling because there was no detectable decrease in MEK, ERK, AKT, or Ras activation (Fig. 3C). These results further qualify RalGEF as the Ras effector necessary for reovirus protein synthesis.
Fig. 3.
Reovirus infection of H-Ras cells expressing dnRal. (A) Down-regulation of Ral activity in H-Ras cells expressing dnRal. H-Ras cells were infected with pBabepuro (vector) retrovirus or retrovirus expressing dnRalA A26 (dnRal). The levels of Ral–GTP and total Ral were assessed as described in Materials and Methods. (B) H-Ras cells harboring vector alone or expressing dnRal were infected with reovirus. Infected cells were pulse-labeled with [35S]methionine-containing medium for 5 h at 67–72 h after infection, and reovirus protein immunoprecipitates were subjected to 10% SDS/PAGE, followed by autoradiography. Reovirus protein migration is indicated on the left. (C) Ras, AKT, MEK, and ERK activity in control (vector) H-Ras and dnRal/H-Ras cells. For Ras activity, serum-starved cell lysates were subjected to pull-down reactions by using GST–Raf Ras binding domain conjugated to glutathione beads, followed by Western blot analysis for Ras. To assess downstream signaling, cells were serum-starved overnight and cell lysates were examined by 10% SDS/PAGE and Western blot analysis for the phosphorylated form of each kinase. (D) Reovirus transcription in control (vector) H-Ras and dnRal/H-Ras cells. Cells were infected with reovirus, and at various times after infection, RNA was extracted and examined by RT-PCR for reovirus S1 RNA and cellular GAPDH RNA control.
Because of its role in vesicular trafficking and endocytosis, it is possible that inhibition of Ral signaling artifactually inhibited early steps in replication, namely, viral entry and transcription. To test this possibility, reovirus transcripts were examined in empty vector- and dnRal-expressing H-Ras cells. Fig. 3D shows that viral transcription (exemplified by the S1 transcript) was unimpaired in the dnRal cells. Again, similar results were obtained for other reovirus transcripts (data not shown). Together, these data indicate that down-regulation of RalGEF activity reinstates a posttranscriptional antiviral mechanism that is normally inhibited in H-Ras cells.
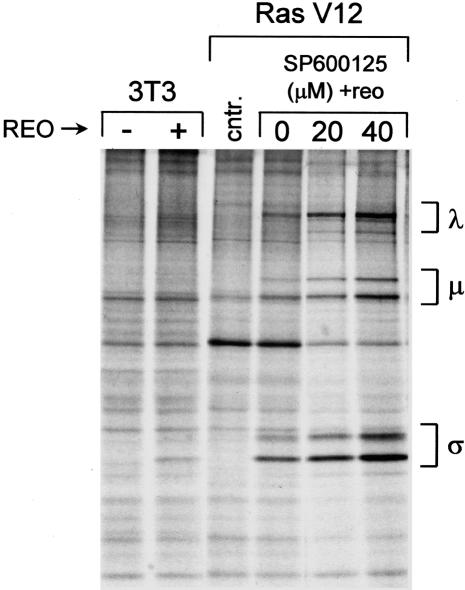
Reovirus Requires p38 kinase, but Not JNK Activity, for Replication. Ras and RalGEFs have been implicated together in many cellular phenomena, including transformation, NF-κB and cyclin D activation, and activation of the stress-activated protein kinases JNK and p38 (9, 26–29). Because cellular stresses promote reovirus protein synthesis in nonpermissive NIH 3T3 cells (K.L.N., M. C. Coffey, and P.W.K.L., unpublished data), we examined JNK and p38 for their involvement in reovirus infection. First, a novel inhibitor of JNK, SP600125, was used to assess the contribution of this pathway in Ras-promoted reovirus protein synthesis. Specifically, Ras-transformed cells were infected with reovirus and treated with 0, 20, or 40 μM SP600125 for the duration of the experiment. At 48 h after infection, cells were radiolabeled and reovirus proteins were analyzed by SDS/PAGE. The result (Fig. 4) shows that, although JNK activity has been implicated in reovirus-induced apoptosis (30), it is not involved in Ras promotion of viral protein synthesis in our system. Indeed, viral protein synthesis appeared to be somewhat enhanced, rather than suppressed, at higher concentrations of SP600125. The reason for this enhancement is unclear at present.
Fig. 4.
Effect of the JNK inhibitor SP600125 on reovirus infection of H-Ras cells. NIH 3T3 cells and NIH 3T3 cells expressing activated Ras (Ras V12) were infected with reovirus in the presence of increasing concentrations of the JNK inhibitor SP600125. Cells were radiolabeled as described in Fig. 1 for 5 h at 43–48 h after infection, cell lysates were prepared, and immunoprecipitates were subjected to 10% SDS/PAGE and autoradiography. Migration of reovirus proteins is indicated on the right.
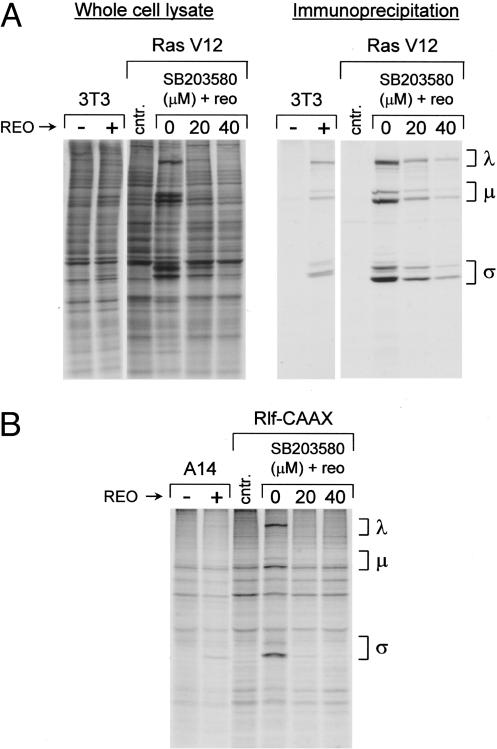
Likewise, p38 activity was inhibited with SB203580 in Ras-transformed cells and reovirus protein synthesis examined at 48 h after infection. Interestingly, we found that unlike the JNK inhibitor, SB203580 potently inhibited reovirus in a dose-dependent manner without affecting host protein synthesis (Fig. 5A). These results were confirmed with another p38 inhibitor, SB202190 (data not shown). Because SB203580 can also inhibit Src family kinases (31), the Src family inhibitor PP2 was used to assess whether these kinases were involved in Ras promotion of reovirus protein synthesis. We found that inhibition of Src family kinases in Ras-transformed cells did not hinder reovirus protein synthesis (data not shown). These data indicate that p38 may function in Ras-transformed cells to regulate viral protein synthesis.
Fig. 5.
Effect of the p38 inhibitor SB203580 on reovirus infecton of H-Ras cells and Rlf–CAAX cells. (A) NIH 3T3 cells and NIH 3T3 cells expressing activated Ras (Ras V12) were infected with reovirus or mock-infected in the presence of increasing concentrations of the p38 inhibitor SB203580. Cells were radiolabeled as described in Fig. 1 for 5 h at 43–48 h after infection, cell lysates were prepared, and whole-cell lysate (Left) or immunoprecipitates (Right) were subjected to 10% SDS/PAGE and autoradiography. cntr., Control uninfected/untreated Ras cells. Migration of reovirus proteins (λ, μ, and σ) is indicated on the right. (B) Control A14 cells and A14 cells expressing activated Rlf–CAAX were infected with reovirus in the presence of increasing concentrations of the p38 inhibitor SB203580. Cells were radiolabeled as described above, cell lysates were prepared, and immunoprecipitates were subjected to 10% SDS/PAGE and autoradiography. Migration of reovirus proteins is indicated on the right.
As mentioned above, activated Rlf can also stimulate p38 activity (28). Therefore, SB203580 was used to determine whether p38 activity is also necessary for reovirus replication in Rlf–CAAX-expressing cells. As in Ras-transformed cells, SB203580 treatment significantly decreased the ability of Rlf–CAAX cells to support reovirus protein synthesis and had little effect on cellular protein synthesis (Fig. 5B and data not shown). Together, our data suggest that Ras promotes reovirus protein synthesis through a RalGEF/p38 pathway.
Discussion
The discovery of the oncolytic properties of reovirus has allowed for the initiation of clinical trials to measure its efficacy in cancer therapy. The aim of the current study was to better characterize the molecular signaling events downstream of Ras that dictate host cell permissiveness to reovirus. This knowledge was sought in the hope of improving our understanding of reovirus oncolysis and perhaps gaining insight into cellular signaling mechanisms regulating viral replication and transformation.
We have found that reovirus replication in Ras-transformed cells depends primarily on RalGEF signaling. The other two most well known signaling pathways downstream of Ras, namely, the Raf and the PI3-kinase pathways, do not seem to be involved in this process. Support for this notion has come from the use of Ras effector domain mutants, which reveals a distinct preference of reovirus for the RasV12G37 (non-Raf, non-PI3-kinase signaling) mutant-expressing cell line over the RasV12C40 and RasV12S35 (both non-RalGEF signaling) cell lines. The use of the PI3-kinase inhibitor, LY294002, and MEK inhibitors further confirms that permissiveness to reovirus does not depend highly on Raf or PI3-kinase signaling.
The knowledge that it is an activated RalGEF pathway that is exploited by reovirus for infection comes from two additional observations. First, constitutive activation of the RalGEF Rlf renders cells permissive to reovirus. Second, dnRal reverses the permissiveness of a stable H-Ras (V12)-expressing cell line to reovirus. Interestingly, mutational activation of RalGTPase itself did not render NIH 3T3 cells permissive to reovirus (data not shown). This observation is not surprising because phenotypic effects from RasV12G37 and Rlf–CAAX but not from active Ral are commonly seen (25, 32–35). Nonetheless, it is apparent that RalGEF signaling plays a major role in dictating the outcome of reovirus infection.
Ras and RalGEFs have been implicated together in many cellular phenomena, including transformation, NF-κB and cyclin D activation, as well as activation of the stress-activated protein kinases, JNK and p38 (9, 26–29). Given that we have observed promotion of reovirus replication in NIH 3T3 cells under stressful conditions, such as UV and osmotic shock (K.L.N., M. C. Coffey, and P.W.K.L., unpublished data), these kinases were logical contenders for regulating reovirus replication (36). We found that treatment of susceptible cells with the JNK inhibitor SP600125 had no effect on reovirus protein synthesis, whereas inhibition of p38 with SB203580 potently blocked reovirus. From these data, p38 emerges as a promising candidate downstream molecule in Ras promotion of viral replication. Although precisely how RalGEF activation is linked to p38 activation is unclear at present, the observation that SB203580 blocks reovirus infection in Rlf–CAAX-expressing cells supports previous findings that RalGEF is likely to be upstream of p38.
Exactly how activated Ras and RalGEFs exert their effects to promote reovirus replication merits further study. It has been proposed that Ras may up-regulate a step before reoviral translation, namely, viral entry (37). So far, our data are inconsistent with this scenario, because reovirus transcripts are synthesized in both permissive and nonpermissive cells, implying that entry is successful in both cell types (Figs. 2C and 3D; K.L.N., A. D. Yang, and P.W.K.L., unpublished data) (6). It has also been found (6, 38, 39) that activated Ras inhibits the antiviral, double-stranded RNA-activated protein kinase (PKR). Moreover, genetic and chemical inhibition of PKR promotes reovirus protein synthesis (6). Thus, it is conceivable that Ras suppression of PKR activity promotes viral protein synthesis. However, the tripartite relationship between PKR, Ras, and reovirus awaits confirmation by means of the identification of intermediary players. Thus far, we have been unable to establish a definitive link between Rlf and PKR (K.L.N. and P.W.K.L., unpublished data). Finally, it deserves mention that recent studies have shown that p38 can also regulate protein synthesis (40). Further study is underway to examine the possibility of the involvement of PKR and other cooperating factors in Ras and Ral pathway-dependent reovirus oncolysis.
The demonstration in this study of the importance of RalGEFs for reovirus replication is relevant to its use as a cancer therapy, as substantial evidence mounts in the literature that RalGEF signaling is significant in human tumorigenesis and metastasis (reviewed in ref. 41). Activation of p38 has also been reported in a number of human cancers, including non-small-cell lung carcinomas and colon polyps, as well as in the progression from follicular to diffuse large B cell lymphoma (42–44). In light of our findings, the fact that these signaling pathways are commonly activated in cancer is promising for reovirus use as a cancer therapy. Future work could possibly focus on the identification of additional pathway elements and the precise molecular mechanism by which they promote reovirus infection and oncolysis in human tumors.
Acknowledgments
We thank Drs. Johannes Bos, Andrew Chan, Steve Robbins, Channing Der, Adrienne Cox, Craig Webb, and George Vande Woude for their generous gifts of constructs and cell lines. We also thank Megan Patrick and Ann Hornby-Smith for technical assistance. K.L.N. was supported by studentships from the Natural Sciences and Engineering Research Council of Canada and the Alberta Heritage Foundation for Medical Research. K.H. was a recipient of a Canadian Institutes for Health Research and an Alberta Heritage Foundation for Medical Research Fellowship. This work was supported by grants (to P.W.K.L.) from the Canadian Institutes for Health Research, the Canadian Breast Cancer Foundation (Alberta Chapter), and the Cancer Research Society.
Abbreviations: GEF, guanine nucleotide-exchange factor; PI3-kinase, phosphatidylinositol 3-kinase; dnRal, dominant-negative Ral; ERK, extracellular signal-regulated kinase; MEK, mitogen-activated protein kinase kinase; PKR, double-stranded RNA-activated protein kinase.
References
- 1.Nibert, M. & Schiff, L. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott–Raven, Philadelphia).
- 2.Hashiro, G., Loh, P. C. & Yau, J. T. (1977) Arch. Virol. 54, 307–315. [DOI] [PubMed] [Google Scholar]
- 3.Duncan, M. R., Stanish, S. M. & Cox, D. C. (1978) J. Virol. 28, 444–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strong, J. E., Tang, D. & Lee, P. W. K. (1993) Virology 197, 405–411. [DOI] [PubMed] [Google Scholar]
- 5.Strong, J. E. & Lee, P. W. K. (1996) J. Virol. 70, 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strong, J. E., Coffey, M. C., Tang, D., Sabinin, P. & Lee, P. W. K. (1998) EMBO J. 17, 3351–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheffzek, K., Ahmadian, M. R., Kabsch, W., Wiesmuller, L., Lautwein, A., Schmitz, F. & Wittinghofer, A. (1997) Science 277, 333–338. [DOI] [PubMed] [Google Scholar]
- 8.Krengel, U., Schlichting, L., Scherer, A., Schumann, R., Frech, M., John, J., Kabsch, W., Pai, E. F. & Wittinghofer, A. (1990) Cell 62, 539–548. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, S. L., Khosravi-Far, R., Rossman, K. L., Clark, G. J. & Der, C. J. (1998) Oncogene 17, 1395–1413. [DOI] [PubMed] [Google Scholar]
- 10.Coffey, M. C., Strong, J. E., Forsyth, P. A. & Lee, P. W. K. (1998) Science 282, 1332–1334. [DOI] [PubMed] [Google Scholar]
- 11.Norman, K. L., Coffey, M. C., Hirasawa, K., Demetrick, D. J., Nishikawa, S. G., DiFrancesco, L. M., Strong, J. E. & Lee, P. W. K. (2002) Hum. Gene Ther. 13, 641–652. [DOI] [PubMed] [Google Scholar]
- 12.Hirasawa, K., Nishikawa, S. G., Norman, K. L., Alain, T., Kossakowska, A. & Lee, P. W. K. (2002) Cancer Res. 62, 1696–1701. [PubMed] [Google Scholar]
- 13.Yang, W. Q., Senger, D., Muzik, H., Shi, Z. Q., Johnson, D., Brasher, P. M., Rewcastle, N. B., Hamilton, M., Rutka, J., Wolff, J., et al. (2003) Cancer Res. 63, 3162–3172. [PubMed] [Google Scholar]
- 14.Wilcox, M. E., Yang, W., Senger, D., Rewcastle, N. B., Morris, D. G., Brasher, P. M., Shi, Z. Q., Johnston, R. N., Nishikawa, S., Lee, P. W., et al. (2001) J. Natl. Cancer Inst. 93, 903–912. [DOI] [PubMed] [Google Scholar]
- 15.Alain, T., Hirasawa, K., Pon, K. J., Nishikawa, S. G., Urbanski, S. J., Auer, Y., Luider, J., Martin, A., Johnston, R. N., Janowska-Wieczorek, A., et al. (2002) Blood 100, 4146–4153. [DOI] [PubMed] [Google Scholar]
- 16.Thirukkumaran, C. M., Luider, J. M., Stewart, D. A., Cheng, T., Lupichuk, S. M., Nodwell, M. J., Russell, J. A., Auer, I. A. & Morris, D. G. (2003) Blood 102, 377–387. [DOI] [PubMed] [Google Scholar]
- 17.Etoh, T., Himeno, Y., Matsumoto, T., Aramaki, M., Kawano, K., Nishizono, A. & Kitano, S. (2003) Clin. Cancer Res. 9, 1218–1223. [PubMed] [Google Scholar]
- 18.Kilani, R. T., Tamimi, Y., Hanel, E. G., Wong, K. K., Karmali, S., Lee, P. W. & Moore, R. B. (2003) Virus Res. 93, 1–12. [DOI] [PubMed] [Google Scholar]
- 19.Burgering, B. M., Medema, R. H., Maassen, J. A., van de Wetering, M. L., van der Eb, A. J., McCormick, F. & Bos, J. L. (1991) EMBO J. 10, 1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, R. E., Zweerink, H. J. & Joklik, W. K. (1969) Virology 39, 791–810. [DOI] [PubMed] [Google Scholar]
- 21.Osada, M., Tolkacheva, T., Li, W., Chan, T. O., Tsichlis, P. N., Saez, R., Kimmelman, A. C. & Chan, A. M. (1999) Mol. Cell. Biol. 19, 6333–6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. (1970) Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 23.Webb, C. P., van Aelst, L., Wigler, M. H. & Woude, G. F. (1998) Proc. Natl. Acad. Sci. USA 95, 8773–8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White, M. A., Nicolette, C., Minden, A., Polverino, A., van Aelst, L., Karin, M. & Wigler, M. H. (1995) Cell 80, 533–411. [DOI] [PubMed] [Google Scholar]
- 25.Wolthuis, R. M., de Ruiter, N. D., Cool, R. H. & Bos, J. L. (1997) EMBO J. 16, 6748–6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henry, D. O., Moskalenko, S. A., Kaur, K. J., Fu, M., Pestell, R. G., Camonis, J. H. & White, M. A. (2000) Mol. Cell. Biol. 20, 8084–8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Ruiter, N. D., Wolthuis, R. M., van Dam, H., Burgering, B. M. & Bos, J. L. (2000) Mol. Cell. Biol. 20, 8480–8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ouwens, D. M., de Ruiter, N. D., van der Zon, G. C., Carter, A. P., Schouten, J., van der, B. C., Kooistra, K., Bos, J. L., Maassen, J. A. & van Dam, H. (2002) EMBO J. 21, 3782–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bulavin, D. V., Demidov, O. N., Saito, S., Kauraniemi, P., Phillips, C., Amundson, S. A., Ambrosino, C., Sauter, G., Nebreda, A. R., Anderson, C. W., et al. (2002) Nat. Genet. 31, 210–215. [DOI] [PubMed] [Google Scholar]
- 30.Clarke, P., Meintzer, S. M., Widmann, C., Johnson, G. L. & Tyler, K. L. (2001) J. Virol. 75, 11275–11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eyers, P. A., Craxton, M., Morrice, N., Cohen, P. & Goedert, M. (1998) Chem. Biol. 5, 321–328. [DOI] [PubMed] [Google Scholar]
- 32.White, M. A., Vale, T., Camonis, J. H., Schaefer, E. & Wigler, M. H. (1996) J. Biol. Chem. 271, 16439–16442. [DOI] [PubMed] [Google Scholar]
- 33.Goi, T., Rusanescu, G., Urano, T. & Feig, L. A. (1999) Mol. Cell. Biol. 19, 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamad, N. M., Elconin, J. H., Karnoub, A. E., Bai, W., Rich, J. N., Abraham, R. T., Der, C. J. & Counter, C. M. (2002) Genes Dev. 16, 2045–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, L., Frankel, P., Jackson, D., Rotunda, T., Boshans, R. L., D'Souza-Schorey, C. & Foster, D. A. (2003) Mol. Cell. Biol. 23, 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karin, M. (1998) Ann. N.Y. Acad. Sci. 851, 139–146. [DOI] [PubMed] [Google Scholar]
- 37.Golden, J. W., Linke, J., Schmechel, S., Thoemke, K. & Schiff, L. A. (2002) J. Virol. 76, 7430–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mundschau, L. J. & Faller, D. V. (1992) J. Biol. Chem. 267, 23092–23098. [PubMed] [Google Scholar]
- 39.Mundschau, L. J. & Faller, D. V. (1994) Biochimie 76, 792–800. [DOI] [PubMed] [Google Scholar]
- 40.Waskiewicz, A. J., Flynn, A., Proud, C. G. & Cooper, J. A. (1997) EMBO J. 16, 1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feig, L. A. (2003) Trends Cell Biol. 13, 419–425. [DOI] [PubMed] [Google Scholar]
- 42.Elenitoba-Johnson, K. S., Jenson, S. D., Abbott, R. T., Palais, R. A., Bohling, S. D., Lin, Z., Tripp, S., Shami, P. J., Wang, L. Y., Coupland, R. W., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 7259–7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenberg, A. K., Basu, S., Hu, J., Yie, T. A., Tchou-Wong, K. M., Rom, W. N. & Lee, T. C. (2002) Am. J. Respir. Cell Mol. Biol. 26, 558–564. [DOI] [PubMed] [Google Scholar]
- 44.Hardwick, J. C., van den Brink, G. R., Offerhaus, G. J., van Deventer, S. J. & Peppelenbosch, M. P. (2001) Oncogene 20, 819–827. [DOI] [PubMed] [Google Scholar]