Abstract
Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) is expressed on the surface of infected erythrocytes where it plays a central role in both infected erythrocytes cytoadhesion and immune evasion. Switches in clonal expression of PfEMP1 result in antigenic variation that facilitates long-term chronic infection of the host. The var gene family encodes PfEMP1 variants, with transcriptional switching between different var variants providing the molecular basis for antigenic variation. Despite the importance of var transcriptional switching in the evasion of the immune response, little is known about the way in which this process is regulated. Here we report the measurement of transition on and off rates for a series of var gene variants. We find (i) that on and off rates for a given variant are dissimilar, (ii) that these rates vary dramatically among different variants, and (iii) that in isogenic clones expressing the same var gene, both on and off rates are constant and appear to be an intrinsic property of that particular gene. These data would suggest that the information that determines the probability of the activation or silencing of var genes is present in their surrounding DNA. Furthermore, some transitions appear to be disallowed depending on the recent variant antigen expression history of the parasite clone. These findings have important implications for both the underlying molecular mechanisms of antigenic variation and the processes that promote chronicity of infection in vivo.
Antigenic variation has evolved in a number of pathogenic microorganisms to facilitate evasion of the host immune response. In Plasmodium falciparum, the parasite responsible for the most severe form of human malaria, antigenic variation is manifested by a change of variant antigen expression on the infected erythrocyte (IE) surface (1, 2). The major IE surface variant antigen is termed P. falciparum erythrocyte membrane protein 1 (PfEMP1) and consists of a family of 250- to 350-kDa proteins that are important targets of the host-protective antibody response (3–6). In addition to their role in immune evasion, members of the PfEMP1 family are important virulence factors through the ability of individual members to mediate adhesion of IE to single or multiple host receptors on endothelial cells, uninfected red cells (leading to the formation of rosettes), dendritic cells, and the placenta (7–13). This process results in the accumulation of IE in particular organs, sometimes leading to organ dysfunction that may be exacerbated by the presence of rosettes that may further obstruct blood flow. In the case of dendritic cells, it may interfere with the generation of an appropriate immune response. The PfEMP1 family is thus central to both the pathogenesis of disease and the induction of immunity.
PfEMP1 is encoded by the var multigene family that comprises ≈60 members per haploid genome (14). The transcription of var genes appears to be tightly regulated such that only a single full-length copy is transcribed in one parasite at any one time (15, 16). This process results in the mutually exclusive expression of PfEMP1 variants at the IE surface. Transcriptional switching occurs in situ and, in contrast to similar processes in other organisms, does not require specific gene rearrangements or DNA deletion events (16). No changes in the pattern of DNA methylation or DNase I hypersensitivity have been associated with the expressed variant, nor does expression rely on the presence of variant specific promoter sequences (1). These observations have led to the suggestion that the specific transcriptional activation or repression of var genes is under epigenetic control. The molecular basis for these processes, however, remains elusive. Recent data have suggested that the silencing of nontranscribed var genes involves the interaction of promoter and intron sequences; although the manner in which genes become activated is entirely unclear (17, 18). To better understand the molecular basis of var transcriptional switching, we have attempted to measure the transition rates at which different var gene variants become transcriptionally activated or repressed.
Experiments designed to study var gene switching in vitro are complicated by the fact that switching rates away from a defined variant are as high as 2% per generation (19). Thus, long-term cultures in the absence of selective pressure rapidly become extremely heterogeneous with respect to the var genes they express. In an attempt to overcome this problem, we have carried out phenotypic selection in vitro followed by immediate cloning of the cultures. This process results in a high proportion of clones that express a single var gene variant. In this article, we have sought to use this panel of isogenic clones to measure both on and off rates of specific variants. The results show that these rates are constant for any given var gene. They also suggest that var genes may exist in three independent states: active, activatable, and highly silenced. The implications of these results for the in vivo dynamics of antigenic variation and for mathematical models that try to mimic this process are discussed.
Materials and Methods
Plasmodium Culture and Cloning. P. falciparum clones and isolates were cultured in vitro in donated human type O erythrocytes (20). The clone A4 was derived from the Ituxi (IT) isolate, as described in ref. 19 and is genetically indistinguishable from many FCR3 laboratory lines (21). A4 was rosette-selected by differential sedimentation of trophozoite IE over Plasmagel (Bellon, France) and cloned by limiting dilution (22), deriving 38 clones from which clones A–F were analyzed further. The A4 culture was also selected for expression of the type 41 var PfEMP1 molecule by using the specific mouse mAb BC6 (mAbBC6) (19, 22). After selection, >75% of infected erythrocytes bound mAbBC6. Clones G and H were derived from this culture by limiting dilution. During the subsequent 40 cycles of in vitro growth, all parasite cultures were treated identically and maintained at a 2–3% hematocrit and 0.5–4% parasitemia by dilution with uninfected erythrocytes.
Rosetting Assay. The 0.5 ml of trophozoite-infected erythrocytes (2–3% parasitemia) at 5% hematocrit containing 10 μg/ml ethidium bromide was rotated for 30 min at room temperature. The proportion of infected erythrocytes bound to two or more uninfected erythrocytes was determined by microscopy (FITC filter with Nikon Eclipse E600 fluorescence microscope). Data presented represent the mean of three independent counts of 200 infected erythrocytes.
DBL1α Expression Sequence Tag Determination. Ring stage RNA (1 μg), treated with RNase-free DNase (Promega), was reverse-transcribed from random hexamers (Invitrogen). αAF and αBR are universal primers validated for unbiased amplification of the duffy binding-like 1α (DBL1α) domain of PfEMP1 from var genes (23). DBL1α expression sequence tags were amplified from isolated RNA by using these oligonucleotides. RT-PCR products were cloned and between 6 and 10 independent clones were sequenced to confirm the identity of the DBL1α expressed sequence tag. Furthermore, RT-PCR products were radiolabeled and hybridized to a nylon filter on which all available A4 genomic DNA DBL1α sequence tags (European Molecular Biology Laboratory Database AJ319680–AJ319712) had been arrayed. Var variants transcribed in ring stage parasites were determined from these DBL1α arrays after stringent hybridization (65°C) and wash (68°C, 0.1× SSC and 0.1% SDS) conditions. Finally, cloned DBL1α sequence tags were hybridized to Northern blots (see below) to confirm hybridization back to a single predominant var transcript identified by using a conserved VarC probe (21).
Northern Blot Analysis. Total RNA was isolated, size-fractionated, and blotted by using a standard P. falciparum protocol (24). DNA probes were sequentially hybridized to the nylon filters and washed to an appropriate stringency for the probe used (21). Specific var gene probes were derived by PCR amplification of cloned DBL1α sequence tags from A4 genomic DNA. Signal intensities were determined by using a Fuji film FLA-2000 phosphorimager with subsequent image analysis provided by aida (Advanced Image Data Analyzer, version 2) software. Var transcript signal intensity was normalized for each sample by using signals obtained from the P. falciparum EF-1α, 3.8 gene, and proliferating cell nuclear antigen transcripts. The mean of these normalized values relative to their value at the start of the experiment (assumed to be 100%) was plotted over time.
mAbs, Human Serum Samples, and Flow Cytometry. Human serum samples were obtained from anonymous healthy adult donors from either the United Kingdom National Blood Service or the Kenya Medical Research Institute/Wellcome Trust Collaborative Research Programme located within a Kenyan coastal town in a malaria-endemic region. Serum sample kilifi#20 was shown by flow cytometry analysis to specifically recognize intact erythrocytes infected with parasites transcribing var type 6, but not var types 14, 19, 32, 41, 43, or R29var, these other variants being transcribed by rosette or mAbBC6-selected parasite populations (19, 22, 25).
Human IgG bound to P. falciparum trophozoite-IE was detected by using FITC-conjugated anti-human CH2 (DAKO) and ethidium bromide (1 μg/ml). Mouse Ig bound to IE was detected first by using rabbit anti-mouse Ig followed by FITC-conjugated swine anti-rabbit Ig (DAKO). The number of FITC- and ethidium bromide-stained cells as a proportion of the ethidium bromide-stained cells (infected erythrocytes) was determined by flow cytometry (FACSCalibur, Becton Dickinson).
var Transition Rate Determination. The transcript off rate (roff) is defined as roff = 1 – (p1/n), where p is the proportion of population remaining of the original phenotype/transcript after n cycles of growth. The transcript on rate (ron) is determined as follows: after n cycles, the proportion of parasites transcribing a given variant (pn) with on rate r(on) is given by: pn = ron(1) + ron(1 – ron)(2) + ron[1 – ron(1 – ron)](3)... (n). This approximates to pn = nron for situations where nron < 0.1. Accurate simulations were carried out on Microsoft excel spreadsheets iteratively taking into account a range of potential on and off rates. Over the period of 40 cycles, it was found that including the ron rate (where roff rates were being determined) or roff rates (where ron rates were being determined) did not make a significant difference to the final estimate in most cases.
Results and Discussion
The repertoire of var genes expressed by an A4 parasite clone was restricted after in vitro selection for adhesion to uninfected erythrocytes (rosetting) immediately before cloning. Previous rosette selection studies using the A4 clone indicate that a limited subset of PfEMP1 variants are transcribed in these selected populations (22, 25, 26).
Thirty-eight clones were isolated, and sequencing of RT-PCR products derived from the DBL1α domain and Northern blots of extracted RNA were used to identify the var gene(s) transcribed in each clone (Fig. 1). More than half of the clones transcribed a single predominant var gene, with var type 6 by far the most common. Most of the remaining clones transcribe var type 6 in combination with var types 32 and/or 19. Six clones transcribed only type 14 var in a manner that appeared to be mutually exclusive to any other var genes detected. Analysis of rosetting frequency in these clones indicates that this phenotype is principally associated with transcription of var types 14, 19, and 43 (Fig. 1). This finding is consistent with the fact that the DBL1α domain of var 19 is highly homologous to that of the R29var rosetting gene variant, the latter having been shown to mediate adhesion to erythrocytes via complement receptor 1 (25). The R29var clone was not transcribed by any of the progeny clones described because transcription of this variant appears to require truncation of the telomere immediately adjacent to the gene, an event not represented in the clones examined here (data not shown) (26).
Fig. 1.
Var transcription patterns in a panel of isogenic P. falciparum clones. The var variant transcribed by each of the 38 clones derived by cloning of rosette-selected A4 isolate are indicated. Filled black boxes represent a single major transcript, with gray boxes representing a mix of var transcript signals. Clones selected for further analysis are indicated in bold. Two further clones derived from an A4 isolate selected for expression of the type 41 var PfEMP1 product (G and H) are also indicated. Rosetting frequency (R+ Freq.) represents the percentage of infected erythrocytes bound to two or more uninfected erythrocytes. The mean of three independent experiments is shown.
We were intrigued by the high frequency of clones transcribing type 6 var because expression of this gene was not associated with rosetting in this or a previous study in ref. 22. This observation might reflect some inherent probability associated with the transcriptional activation of this particular variant. To test this hypothesis, we decided to measure var transition rates in clones expressing single var gene variants (var 6, 14, 32, and 43; see Fig. 1). Two further clones expressing only the var type 41 variant were also investigated. These were obtained by selection of the A4 clone for adhesion to a mAbBC6 specific for the PfEMP1 encoded by var type 41 (19) immediately before cloning.
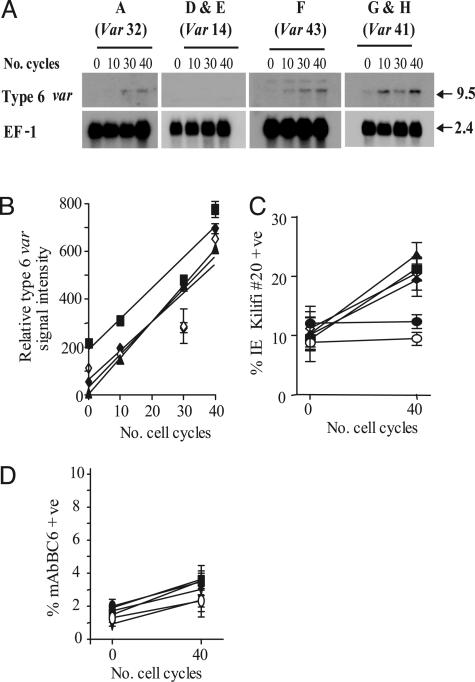
All clones were cultured in vitro for 40 generations over which period RNA was harvested every 10 generations, size-fractionated, and blotted. Transcribed var genes were identified by hybridization with specific probes. We measured the increase of steady-state levels of var transcripts over this time by using several housekeeping transcripts as internal loading controls. In clones that did not already transcribe var type 6 (A and D–H), a readily quantifiable increase in this transcript level with time was observed in all except two clones (D and E; Fig. 2 A and B). Although each of the clones A and F–H start with a different level of var 6 transcript, the rate at which this transcript accumulates in each clone is almost exactly the same (ron = 0.25% per generation, Fig. 2B). Similarly, a small increase in the accumulation of var type 41 (ron = 0.025% per generation, data not shown) was noted in each of clones A–F. Increases in the signal intensity of other var transcripts identified by RT-PCR analysis of these clones were not detected above background.
Fig. 2.
Var transcript on-rates (ron) are fixed in P. falciparum.(A) Steady-state transcript levels of type 6 var in clones A and D–H over 40 cycles of intraerythrocytic growth. The predominant var type transcribed in each of these parasite clones is indicated above their respective panels. Clones B and C are not shown because var type 6 is the predominant transcript in these clones. The size of each transcript is described in kb. The hybridization signal from the control transcript EF-1α is indicated as used for one of several loading and staging controls. (B) Analysis of the relative increase in type 6 var transcript in clones A and F–H over 40 cycles of cell growth. Logarithmic regression of the mean relative signal intensity (±SEM, n = 3) indicates that type 6 var accumulates at the same rate (ron = 0.25% per generation) in all of these clones. Data points for each clone are represented as follows; clone A (▴), clone F (▪), and clones G and H (♦ and ⋄). (C) Flow cytometry analysis of type 6 var-encoded PfEMP1 molecule expression. The proportion of infected erythrocytes (mean ± SEM, n = 3) recognized by var type 6-specific serum (kilifi#20) over 40 cycles of growth is indicated. Whereas the proportion of infected erythrocytes in clones A and F–H recognized by kilifi#20 increases at a mean ron of 0.25% per generation, there is no appreciable increase in kilifi#20 binding to clones D and E (• and ○) over the same period. (D) Flow cytometry analysis of type 41 var-encoded PfEMP1 molecule expression in clones A–F. The proportion of infected erythrocytes (mean ± SEM, n = 3) recognized by mAbBC6 over 40 cycles of growth is indicated. In these six clones, not originally transcribing the type 41 var gene variant, the proportion of parasites recognized by mAbBC6 increases at a mean ron of 0.025% per generation. Symbols used are the same as for above.
To confirm that the increases in the level of the var type 6 and 41 transcripts corresponded to a concomitant increase in the level of PfEMP1 at the erythrocyte surface, we carried out flow cytometry analysis on intact infected red cells. We used mAbBC6 to detect var 41 PfEMP1 and a serum identified from a healthy adult donor (kilifi#20) from a malaria-endemic region that specifically recognizes the var 6 PfEMP1 product but not any other PfEMP1 variant expressed by the rosette and mAbBC6-selected A4 clones (Fig. 2 C and D). Together, these data show that the var variants 6 and 41 accumulate at fixed rates of ≈0.25% and 0.025% per generation, respectively.
We noted above that accumulation of var type 6 was immeasurably low in clones D and E (Fig. 2 A and C), both of which transcribe var type 14, despite the fact that the var 6 locus appeared to be intact by detailed restriction enzyme mapping (data not shown). Taken together with the exclusive nature of var type 14 transcription in the 38 clones (Fig. 1), this observation would suggest that the ability to switch to certain variant types might depend on the antigenic switching history of the parasite, e.g., a switch to var 6 would not normally be permitted in a clone transcribing var type 14.
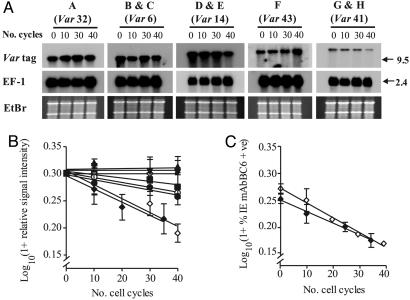
We used the same eight clones and methodology to measure the rate at which individual transcripts were silenced over the 40-generation culture period. The data show that individual var genes exhibit a range of off rates (roff), from >1% per generation to an undetectably low level (Fig. 3 A and B). These off rates were identical within experimental error in different parasite clones expressing the same variant (Fig. 3B). Again we confirmed that levels of transcripts correlated with level of cell surface expressed PfEMP1 for the type 41 var by using fluorescence-activated cell sorting analysis with mAbBC6 (Fig. 3C). The mean off rate of 1.37 ± 0.07% per generation deduced by flow cytometry was indistinguishable from the roff of 1.35 ± 0.12% per generation measured by Northern blot.
Fig. 3.
Var transcript off-rates (roff) are fixed in P. falciparum.(A) Northern blot analysis of decreasing var transcript steady-state levels during 40 cycles of cell growth. Variant-specific probes to each of the predominant var transcribed (indicated by var type) in clones A–H hybridized. The hybridization signal from the control transcript EF-1α is shown as is the ethidium bromide-stained gel to indicate equal loading. The size of each transcript (kb) is indicated. (B) Analysis of the decrease in relative signal intensity for var in clones A–H. Log-transformed mean signal intensity (±SEM, n = 3) for the predominant transcript present in each clone at the start of the experiment is plotted over 40 cycles of cell growth. Clone A (var type 32, ▴, undetectably low roff), clones B and C (var type 6, ▾ and ▿, undetectably low roff), clones D and E (var type 14, • and ○, mean roff = 0.47 ± 0.04% per generation), clone F (var type 43, ▪, roff = 0.30% per generation), and clones G and H (var type 41, ♦ and ⋄, mean roff = 1.35 ± 0.12% per generation). (C) Flow cytometry analysis of type 41 var-encoded PfEMP1 molecule expression in clones G and H. The proportion of infected erythrocytes (mean ± SEM, n = 3) in each clone recognized by the var type 41 PfEMP1-specific mAbBC6 was log-transformed and plotted over 40 cycles of growth (mean type 41 PfEMP1 expression roff in clones G and H is 1.37 ± 0.07% per generation).
The range of var transition rates that we observe could be explained in one of three ways. It could be that all var genes switch at the same rate but have individual growth rates that are slightly different from each other. The data could also be explained entirely by differences in transcriptional switch rates for each variant. Finally, a combination of these mechanisms may operate. We favor the second explanation for the following reasons. First, we have measured the growth rates of two clones with the most divergent transition rates (expressing var types 6 and 41), comparing these to that of the genotypically distinct 3D7 clone. We find no difference in the multiplication rate over two cycles of in vitro growth. We similarly show no difference in the length of the cell cycle for the two A4-derived clones but clearly demonstrate that 3D7 has a cell cycle that is ≈2 hr shorter (data not shown). Unfortunately, although we did detect genotype-specific differences, the experiments are unable to measure differences as small as 1% per generation, a rate that may underlie our observed results. Second, the most direct data supporting the transcriptional switch rate hypothesis comes from results with clones B and C that both transcribe type 6 var. One of these clones has undergone a spontaneous deletion at one end of chromosome 2 that deletes the gene encoding the knob-associated histidine-rich protein as well as all genes telomeric to it. This is a frequent event during in vitro culture and, when it occurs, the resultant knobless parasites overgrow the culture, suggesting that they have an observable growth advantage. We, however, see no difference in the transition rates between these two clones, implying that the switch rate is larger than the growth advantage. Third, PfEMP1 is only present on the cell surface at ≈10,000 molecules per cell and is thus, unlike trypanosome variant surface glycoprotein, a very minor cell protein. Unless additional transcriptional changes accompany a var switch, it is unlikely that transcription of different var genes per se would alter the growth rate. We consider this unlikely because we know that in the case of at least one var gene (type 41), transcriptional activation does not spread beyond the gene itself (22, 26). Finally, given that phenotypic variation results from switches in var transcription, presumably the simplest means of regulating the rate of phenotypic variation is through the regulation of the rate of var transcriptional activation and silencing. In any event, even if differential growth rates were to be part of the explanation, the observed phenotype is unaltered. Parasites expressing different var genes still appear and disappear from the population at the rates that we have observed. From here on we assume that it is differences in switch rates that explain our data.
In other protozoan organisms that use antigenic variation, the variant gene families involved may contain as many as 1,000 variants with transcriptional switches being associated with gene duplication events and genomic rearrangements (27). P. falciparum, however, has only ≈60 var genes per haploid genome (14). It has therefore been difficult to conceive how 60 var variants are enough to maintain chronic infection in humans, particularly because acute-phase parasitemias may contain >1010 parasites. Under these circumstances, even var gene variants with extremely low on rates would be expected to be expressed, thus exhausting the entire variant repertoire early during the infection.
Our data may shed some light on this problem. We envisage three possible transcription states for a var gene (Fig. 4): active (A), inactive but capable of being activated (I), or highly silenced (S). Our data suggests that transitions between the A and I states are intrinsic properties of these variants and appear to be reproducible in different parasite clones. The events that determine the highly silenced state (e.g., var 6 in parasites expressing var 14) are unknown. Although DNA methylation has previously been shown not to be involved in var transcriptional activation/inactivation (1), we examined whether it may play some role in maintenance of a highly silenced state. Restriction analysis with methylation-sensitive endonucleases of the type 6 var locus in parasite clones where this variant is actively transcribed (B and C), inactive but capable of being activated (A and G), or highly silenced (D and E) confirm that DNA methylation was not involved in this silencing mechanism (data not shown).
Fig. 4.
Three transcriptional states are proposed for var genes. The first two are transcriptionally active (A) and transcriptionally inactive (I), although the latter are still capable of being activated. The transition rates between the A and I states appear to be invariant among clones, suggesting they are intrinsic properties of each var gene variant. Var genes appear also to exist in a third highly silenced state (S). Irrespective of the rate of the I to A transition, these S-state var gene variants remain transcriptionally inactive within the limits of detection of this experiment. Data presented herein indicates that var transcriptional switching history may determine the S to I transition. The molecular mechanisms that mediate this highly silenced state remain unknown.
We propose that because A to I transition rates for the same var gene variant are the same in different clones, irrespective of the var variant predominantly transcribed, that these are in effect “hard wired.” This finding presumably reflects some aspect of local or extended primary DNA sequence that acts to modulate their local chromatin environment. Var genes are distributed in three broadly defined locations within the genome: either immediately adjacent to the telomere, close to a telomeric var gene, or in internal clusters (14). These locations are each associated with a specific 5′ UTR termed UpsA, B, or C (14, 28). We determined the Ups-promoter sequence immediately adjacent to each var gene in this study and found no association between var on or off rates and a particular 5′ UTR (data not shown). Thus, neither genomic position per se nor the 5′ UTR sequence determine switch rates, and the relevant DNA sequence must therefore lie elsewhere. Other distinct changes in the extended chromatin environment must presumably exist in genes in the S state to explain their highly silenced phenotype.
In other systems of antigenic variation, particularly in the extensively studied African trypanosome, it has been possible to measure variant specific on rates both in vitro and in vivo (29–31). These data remain controversial and vary over some orders of magnitude, particularly when measured in recently tsetse fly-transmitted pleiomorphic organisms (31). It has proved impossible, however, to measure off rates in vivo, because these are confounded by the immune response, and in vitro, because pleiomorphic trypanosomes cannot be maintained for a sufficient length of time without differentiation. In the case of malaria, similar considerations apply. On rates have been measured in vivo in the mouse malaria model P. chabaudi, and we have previously reported the off rate in vitro for one variant in P. falciparum (19, 32). However, considerations discussed earlier have limited the extent of these studies. Attempts to infer variant transition rates in humans in vivo through mathematical modeling have been made, but these are complicated by the problem of in vivo host selection for particular cytoadherence and hence antigenic types (33, 34).
Our data provide a direct measure of on and off rates (I to A to I) and show that these are highly reproducible for any given var variant. The variation in the rates that we observe among different var genes may be crucial in determining the in vivo dynamics of new variant appearance. Variants with a very high on rate and immeasurably slow off rate are likely to dominate the initial stages of infection, preserving the remainder of the repertoire for maintaining chronicity. The range of on rates that we have observed may also serve to structure the appearance of variants in a statistically preferred order as has been observed in both malaria and other systems (32, 35, 36). The order of var transcript appearance in vivo would thus be determined by a combination of probabilistic events (the intrinsic on rate) and the proportion of genes that have entered the highly silenced (S) state, leading to a situation where subsets of var genes could be turned on in a branched switching pathway. This process might be a useful evolutionary strategy to preserve some high-frequency variants for late in infection. At this stage, parasitemia is extremely low and high on rates would be necessary to achieve an antigenic switching to maintain a chronic infection.
Attempts to provide a mathematical framework that is capable of reproducing the sequential dominance of individual antigenic types seen in vivo in many systems of antigenic variation have been problematic and have had to make assumptions that are not necessarily based on biological observation (37–39). It has recently been shown that the presence of short-lived immune responses to minor antigenic epitopes shared between malarial variant surface antigens can readily achieve sequential dominance (40). We believe that this latter model, in combination with the observations made in this article, may provide a robust framework with which to understand the way in which the appearance of antigenic variants is controlled in vivo.
Acknowledgments
We thank B. Lowe and his colleagues at the Kenya Medical Research Institute/Wellcome Trust Collaborative Research Programme for providing adult serum samples and P. Bull, B. Urban, G. Rudenko, and S. Gupta for comments and discussion during the preparation of the manuscript. A Wellcome Trust program grant supported this work.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DBL1α, duffy binding-like 1α domain; IE, infected erythrocyte; IT, Ituxi; PfEMP1, Plasmodium falciparum erythrocyte membrane 1; roff, var transcript off rate; ron, var transcript on rate.
References
- 1.Kyes, S., Horrocks, P. & Newbold, C. (2001) Annu. Rev. Microbiol. 55, 673–707. [DOI] [PubMed] [Google Scholar]
- 2.Newbold, C. I. (1999) Curr. Opin. Microbiol. 2, 420–425. [DOI] [PubMed] [Google Scholar]
- 3.Bull, P. C., Lowe, B. S., Kortok, M., Molyneux, C. S., Newbold, C. I. & Marsh, K. (1998) Nat. Med. 4, 358–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baruch, D. I., Pasloske, B. L., Singh, H. B., Bi, X., Ma, X. C., Feldman, M., Taraschi, T. F. & Howard, R. J. (1995) Cell 82, 77–87. [DOI] [PubMed] [Google Scholar]
- 5.Smith, J. D., Chitnis, C. E., Craig, A. G., Roberts, D. J., Hudson-Taylor, D. E., Peterson, D. S., Pinches, R., Newbold, C. I. & Miller, L. H. (1995) Cell 82, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su, X. Z., Heatwole, V. M., Wertheimer, S. P., Guinet, F., Herrfeldt, J. A., Peterson, D. S., Ravetch, J. A. & Wellems, T. E. (1995) Cell 82, 89–100. [DOI] [PubMed] [Google Scholar]
- 7.Urban, B. C., Ferguson, D. J., Pain, A., Willcox, N., Plebanski, M., Austyn, J. M. & Roberts, D. J. (1999) Nature 400, 73–77. [DOI] [PubMed] [Google Scholar]
- 8.Ringwald, P., Peyron, F., Lepers, J. P., Rabarison, P., Rakotomalala, C., Razanamparany, M., Rabodonirina, M., Roux, J. & Le Bras, J. (1993) Infect. Immun. 61, 5198–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Udomsangpetch, R., Wahlin, B., Carlson, J., Berzins, K., Torii, M., Aikawa, M., Perlmann, P. & Wahlgren, M. (1989) J. Exp. Med. 169, 1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urban, B. C. & Roberts, D. J. (2003) J. Exp. Med. 197, 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner, C. M. (1997) FEMS Microbiol. Lett. 153, 227–231. [DOI] [PubMed] [Google Scholar]
- 12.Fried, M. & Duffy, P. E. (1996) Science 272, 1502–1504. [DOI] [PubMed] [Google Scholar]
- 13.Rowe, A., Obeiro, J., Newbold, C. I. & Marsh, K. (1995) Infect. Immun. 63, 2323–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner, M. J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R. W., Carlton, J. M., Pain, A., Nelson, K. E., Bowman, S., et al. (2002) Nature 419, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, Q., Fernandez, V., Sundstrom, A., Schlichtherle, M., Datta, S., Hagblom, P. & Wahlgren, M. (1998) Nature 394, 392–395. [DOI] [PubMed] [Google Scholar]
- 16.Scherf, A., Hernandez-Rivas, R., Buffet, P., Bottius, E., Benatar, C., Pouvelle, B., Gysin, J. & Lanzer, M. (1998) EMBO J. 17, 5418–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calderwood, M. S., Gannoun-Zaki, L., Wellems, T. E. & Deitsch, K. W. (2003) J. Biol. Chem. 278, 34125–34132. [DOI] [PubMed] [Google Scholar]
- 18.Deitsch, K. W., Calderwood, M. S. & Wellems, T. E. (2001) Nature 412, 875–876. [DOI] [PubMed] [Google Scholar]
- 19.Roberts, D. J., Craig, A. G., Berendt, A. R., Pinches, R., Nash, G., Marsh, K. & Newbold, C. I. (1992) Nature 357, 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trager, W. & Jensen, J. B. (1976) Science 193, 673–675. [DOI] [PubMed] [Google Scholar]
- 21.Kyes, S. A., Christodoulou, Z., Raza, A., Horrocks, P., Pinches, R., Rowe, J. A. & Newbold, C. I. (2003) Mol. Microbiol. 48, 1339–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horrocks, P., Pinches, R., Kyes, S., Kriek, N., Lee, S., Christodoulou, Z. & Newbold, C. I. (2002) Mol. Microbiol. 45, 1131–1141. [DOI] [PubMed] [Google Scholar]
- 23.Taylor, H. M., Kyes, S. A., Harris, D., Kriek, N. & Newbold, C. I. (2000) Mol. Biochem. Parasitol. 105, 13–23. [DOI] [PubMed] [Google Scholar]
- 24.Kyes, S., Pinches, R. & Newbold, C. (2000) Mol. Biochem. Parasitol. 105, 311–315. [DOI] [PubMed] [Google Scholar]
- 25.Rowe, J. A., Moulds, J. M., Newbold, C. I. & Miller, L. H. (1997) Nature 388, 292–295. [DOI] [PubMed] [Google Scholar]
- 26.Horrocks, P., Kyes, S., Pinches, R., Christodoulou, Z. & Newbold, C. (2004) Mol. Biochem. Parasitol. 134, 193–199. [DOI] [PubMed] [Google Scholar]
- 27.Borst, P. (2002) Cell 109, 5–8. [DOI] [PubMed] [Google Scholar]
- 28.Kraemer, S. M. & Smith, J. D. (2003) Mol. Microbiol. 50, 1527–1538. [DOI] [PubMed] [Google Scholar]
- 29.Cross, G. A. (1990) Annu. Rev. Immunol. 8, 83–110. [DOI] [PubMed] [Google Scholar]
- 30.Cross, G. A. (1996) BioEssays 18, 283–291. [DOI] [PubMed] [Google Scholar]
- 31.Turner, C. M. & Barry, J. D. (1989) Parasitology 99, 67–75. [DOI] [PubMed] [Google Scholar]
- 32.Brannan, L. R., Turner, C. M. & Phillips, R. S. (1994) Proc. R. Soc. London Ser. B 256, 71–75. [DOI] [PubMed] [Google Scholar]
- 33.Molineaux, L., Diebner, H. H., Eichner, M., Collins, W. E., Jeffery, G. M. & Dietz, K. (2001) Parasitology 122, 379–391. [DOI] [PubMed] [Google Scholar]
- 34.Paget-McNicol, S., Gatton, M., Hastings, I. & Saul, A. (2002) Parasitology 124, 225–235. [DOI] [PubMed] [Google Scholar]
- 35.Handunnetti, S. M., Mendis, K. N. & David, P. H. (1987) J. Exp. Med. 165, 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vickerman, K. (1978) Nature 273, 613–617. [DOI] [PubMed] [Google Scholar]
- 37.Agur, Z., Abiri, D. & Van der Ploeg, L. H. (1989) Proc. Natl. Acad. Sci. USA 86, 9626–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank, S. A. (1999) Proc. R. Soc. London Ser. B 266, 1397–1401. [Google Scholar]
- 39.Molineaux, L. & Dietz, K. (1999) Parassitologia 41, 221–231. [PubMed] [Google Scholar]
- 40.Recker, M., Nee, S., Bull, P. C., Kinyanjui, S., Marsh, K., Newbold, C. & Gupta, S. (2004) Nature 429, 555–558. [DOI] [PubMed] [Google Scholar]