Abstract
Integrated hepadnaviral DNA in livers and tumors of chronic hepatitis B patients has been reported for many years. In this study, we investigated whether hepatitis B virus DNA integration occurs preferentially at sites of cell DNA damage. A single I-SceI homing endonuclease recognition site was introduced into the DNA of the chicken hepatoma cell line LMH by stable DNA transfection, and double-strand breaks were induced by transient expression of I-SceI after transfection of an I-SceI expression vector. Alteration of the target cleavage site by imprecise nonhomologous end joining occurred at a frequency of ≈10–3 per transfected cell. When replication of an avian hepadnavirus, duck hepatitis B virus, occurred at the time of double-strand break repair, we observed integration of viral DNA at the site of the break with a frequency of ≈10–4 per transfected cell. Integration depended on the production of viral double-stranded linear DNA and the expression of I-SceI, and integrated DNA was stable through at least 17 cell divisions. Integration appeared to occur through nonhomologous end joining between the viral linear DNA ends and the I-SceI-induced break, because small deletions or insertions were observed at the sites of end joining. The results suggest that integration of hepadnaviral DNA in infected livers occurs at sites of DNA damage and may indicate the presence of more widespread genetic changes caused by viral DNA integration itself.
Hepadnaviridae are a family of viruses containing a circular, partially double-stranded DNA of ≈3 kbp that primarily infect the liver. The family prototype, human hepatitis B virus (HBV), can cause chronic hepatitis and hepatocellular carcinomas (HCCs). HBV-induced HCC is one of the most frequently occurring cancers, although typically decades elapse between viral infection and cancer detection (1–3). Spontaneous integration of viral DNA into host chromosomes occurs in both chronic and acute infections, but in contrast to retroviruses, viral integration does not play a role in hepadnavirus replication (4–8). In an animal model of HBV, the woodchuck hepatitis virus, viral DNA found integrated in HCC commonly activates members of the myc family of protooncogenes (9–11). No corresponding association has been shown in human HCC (12), although ≈85% of HCCs contain integrated HBV sequences (13, 14). The possible roles and significance of hepadnavirus DNA integration in chronic liver disease and HCC have been subjects of investigation for many years.
The mechanism(s) of HBV integration into the host genome are unknown. Indirect evidence suggests that at least two types of linear double-strand viral DNAs are substrates for integration. Despite covalent blockage of the 5′ ends of both strands by protein or RNA, the ends of these molecules have been shown to undergo efficient intra- and intermolecular ligation by nonhomologous end joining (NHEJ). We previously suggested that integration into cellular chromosomes may occur by NHEJ at double-strand breaks in cellular DNA (15, 16). In this study, we tested directly whether sites of cellular DNA damage, namely double-strand breaks, are specific targets for viral DNA integration.
Accordingly, we stably inserted a single I-SceI recognition site into the genome of host LMH cells. After transfection of an I-SceI expression plasmid, we observed evidence of cleavage and repair by imprecise NHEJ at the expected site. Replication of duck HBV (DHBV) in the cells undergoing imprecise repair of the induced double-strand break resulted in integration of viral DNA at the repaired site 7–14% of the time. In situ primed linear DHBV was the preferential substrate for such integration. Integrated viral DNA was maintained in the cell population through multiple transfers even though unintegrated replicating viral DNA was rapidly lost. We concluded that the repair of double-strand breaks by imprecise NHEJ is sometimes accompanied by insertion of viral sequences, implying that the amount of integrated viral DNA in the liver may reflect the degree of overall genetic damage sustained by the liver during a course of chronic hepatitis.
Methods
Plasmids. Construction of pUC119CMVDHBV expression plasmid (1165A plasmid) has been described (15, 17). The 1165A mutation introduces a stop codon in the pre-S coding region of the envelope gene, causing a high level of viral DNA accumulation in the nucleus. The 1165A/DR1-13 plasmid containing a single-base change (C to G) on the plus strand at nucleotide position 2547 (18) was a generous gift from Dan Loeb (University of Wisconsin, Madison). The 1165A/DR1-13 plasmid is defective in plus-strand primer translocation, resulting in an ≈1:1 ratio of linear to circular DNA compared with an ≈1:10 ratio for the 1165A plasmid (15, 19, 20). To introduce a unique I-SceI restriction site into cells, we constructed a plasmid, pEGFP/I-SceI, that contained a single 18-bp I-SceI recognition sequence inserted into the enhanced green fluorescent protein (EGFP) gene, rendering the EGFP gene inactive, and a hygromycin-resistance gene to allow for selection of the integrated substrate. The starting plasmid for construction was pCM-VEGFP2 I-SceI(XhoI), a gift from Perry Kim (Queens University, Kingston, ON, Canada) and Jac Nickoloff (University of New Mexico). This plasmid contained two EGFP genes from pEGFP (Clontech) inserted into pCMV-Script (Stratagene). The cytomegalovirus (CMV)-EGFP gene contained an 18-bp I-SceI recognition sequence that had been inserted at an engineered XhoI site. Both EGFP genes were excised from pCM-VEGFP2 I-SceI(XhoI) by BamHI/HindIII digestion and subcloned into pcDNA3.1Hygro(–) (Invitrogen). The wild-type EGFP gene was deleted from this construct by digestion with HpaI/HindIII, filling in of the 5′ overhang and ligating to create EGFP/I-SceI. Plasmid p1929, expressing EGFP, was obtained from Dan Loeb, and mRFP1, expressing monomeric red fluorescent protein, was a gift from Roger Tsien (University of California at San Diego, La Jolla).
Cell Culture and Transfections. Chicken hepatoma LMH cells were routinely maintained in DMEM/F-12 (1:1) supplemented with 10% FBS. To establish a stably integrated cell line containing an I-SceI substrate, pEGFP/I-SceI vector was linearized with SspI and transfected into LMH cells by electroporation. Briefly, 4 × 106 cells in 0.75 ml of PBS containing 1 μg of DNA were transferred to a cuvette with a 0.4-cm electrode gap and shocked with 300 V at 960 μF. Individual clones were selected and grown in medium containing 400 μg/ml hygromycin. Southern blot analysis was performed on isolated genomic DNA to identify a clone (LMH 3.2) containing a single-copy integrant of EGFP/I-SceI. Fifty micrograms of the I-SceI expression vector pCMV3xnlsI-SceI (21) plus 10 μg of 1165A/DR1-13 plasmid or a 1:1 mixture of 1165A and 1165A/DR1-13 were cotransfected into 4 × 106 LMH 3.2 cells by electroporation. Cells were plated and incubated for 3 days before the cell DNA was extracted for an assay of integrations.
In a reconstruction experiment to determine the relative efficiencies of transfection and cotransfection, EGFP- and mRFP1-expressing plasmids were cotransfected by electroporation at the same amounts as the experimental plasmids. The fraction of mRFP1-transfected (21%), EGFP-transfected (9%), and cotransfected (7%) cells was determined by fluorescence microscopy using red and green filters on an epifluorescent microscope (Nikon) by counting a minimum of 103 cells per transfection. These values were used to normalize the frequencies of imprecise NHEJ and DHBV integration to the number of cells transfected.
DNA Extraction. Cellular DNA was prepared from transfected cells by lysis of the cell layer of a 60-mm dish with 0.4 ml of SDS lysis buffer (10 mM Tris·HCl/10 mM Na-EDTA/0.5% SDS, pH 8.0) containing 0.5 mg/ml Pronase. After 1 h at 37°C, the lysate was extracted with an equal volume of phenol and recovered by ethanol precipitation. The nucleic acid pellet was dissolved in 0.1 ml of TE (10 mM Tris·HCl/1 mM Na-EDTA), adjusted to 1 μg/ml RNase A, and incubated for 10 min at 37°C. DNA was recovered by phenol extraction and ethanol precipitation. The final DNA concentration was determined by the optical density at 260 nm.
PCR Amplification. Individual left EGFP/DHBV junctions (see Fig. 1d) were detected by amplifying sequential dilutions of cellular DNA by nested PCR such that products were detected in only a fraction of the individual reactions. Nested PCR was performed in microplates by using 200 nM each of primers 1A and 1B (Table 1) and 25 units/ml AmpliTaq Gold (Applied Biosystems) in buffer containing 3 mM MgCl2 and 200 μM of each dNTP in a total volume of 10 μl. After an initial incubation at 95°C for 3 min, 40 cycles of PCR were performed by using a denaturation step of 95°C for 15 sec, annealing at 58°C for 15 sec, and extension at 72°C for 30 sec. Approximately 0.1 μl from each well was transferred to a replica microplate containing 10 μl of PCR mixture with 200 nM each of primers 2A and 2B and amplified for an additional 40 cycles. PCRs were electrophoresed through 1.3% agarose gels and stained with ethidium bromide. Right EGFP/DHBV junctions were amplified by using a similar nested-PCR strategy. Primer sets 3A and 3B followed by primers 4A and 4B were used to amplify individual right EGFP/DHBV junctions. To measure imprecise NHEJ of double-strand breaks (Fig. 1c), genomic DNA was digested for 12 h at 37°C with I-SceI (New England Biolabs) to enrich for cleavage-resistant sites. Nested PCR then was performed at limiting template dilutions using primer sets 1A and 3B followed by a second round of PCR using the primers 2A and 4B. The amplified products were digested with I-SceI to identify the nuclease-resistant products of imprecise NHEJ and detected by gel electrophoresis as described above. For quantification of replicative intermediates, known amounts of total DNA were amplified by real-time PCR using an iCycler (Bio-Rad). Forty cycles of amplification were performed as described above by using 1× iQ Sybr green supermix (Bio-Rad) containing 200 nM each of primers 5A and 1B (Table 1).
Fig. 1.
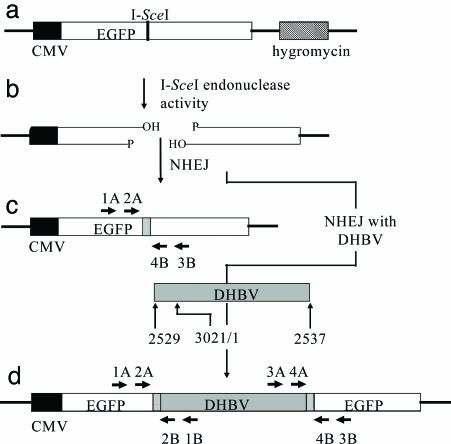
Substrate and potential products of NHEJ. (a) Integration target site present in LMH 3.2 cells, consisting of an I-SceI 18-bp recognition sequence inserted into an EGFP gene and the hygromycin-resistance gene for selection. (b) A double-strand break formed by I-SceI endonuclease activity. (c) Product formed by NHEJ of a double-strand break. Precise joining would recreate the I-SceI site, whereas imprecise NHEJ can result in deletions or insertions with concomitant loss of the recognition sequence (gray box). EGFP-specific primer sets 1A/3B and 2A/4B were used to amplify products (see Fig. 2). (d) Product formed by NHEJ, resulting in the integration of DHBV at the double-strand break. The hypothetical DHBV integration substrate shown represents the larger-than-genome size, in situ primed linear DNA, which is the major form of linear DHBV. Integration can be associated with small deletions or insertions of sequence (gray boxes). Left EGFP/DHBV junctions were amplified by nested PCR of genomic DNA by using the primer pairs 1A/1B followed by 2A/2B. Right EGFP/DHBV junctions were amplified similarly from the same genomic DNA by using primers 3A/3B and then 4A/4B.
Table 1. PCR primers.
| Designation | Sequence | Nucleotides* |
|---|---|---|
| 1A | 5′-GGCCACAAGTTCAGCGTGTC | 73-92 |
| 1B | 5′-TGTGTAGTCTGCCAGAAGTCTTC | 2840-2818 |
| 2A | 5′-TGCAGTGCTTCAGCCGCTAC | 206-225 |
| 2B | 5′-AATGAGATCCACAAAGTGAGTTGC | 2817-2794 |
| 3A | 5′-TGTCCCGAGCAAATATAATCC | 2407-2427 |
| 3B | 5′-GGACCATGTGATCGCGCTTC | 661-642 |
| 4A | 5′-TATAATCCTGCTGACGGCCCA | 2420-2440 |
| 4B | 5′-GTGTTCTGCTGGTAGTGGTC | 560-541 |
| 5A | 5′-TTCGGAGCTGCTTGCCAAGGTATC | 2548-2571 |
| 6A | 5′-CCTTAGCCAATGTGTATGATCTACCA | 2669-2694 |
EGFP nucleotides are based on position 1 starting at the pEGFP start codon (Clontech). DHBV nucleotides are numbered according to ref. 18.
Calculation for Frequencies of NHEJ, Integration, and Replicative Intermediates. NHEJ and integration frequencies were calculated by dividing the total number of individual PCR products obtained from a known number of cell genome equivalents of DNA (3 pg per cell), corrected for the number of transfected cells (21% for NHEJ assays and 7% for integration assays). The copy number of replicative intermediates was calculated from a standard curve generated from serial dilutions of known quantities of BamHI-digested pSPDHBV5.1(2X) using iCycler software. The results were normalized to transfected cell genomes (9% of the total cells).
Southern Blot Analysis of Replicative Intermediates. Procedures used in the analysis of viral DNA replicative intermediates, agarose gel electrophoresis, and Southern blot hybridization have been published (22).
Sequencing. PCR-amplified DNA was excised from agarose gels and purified by using a QIAEX-II gel-extraction kit (Qiagen, Valencia, CA) and sequenced by the DNA Services Core Facility at the University of New Mexico. The EGFP-specific primers 2A and 4B were used to sequence the left and right EGFP/DHBV junctions, respectively. To determine the genotype of replicative intermediates in a mixed transfection, total DNA extracted from transfected cells was amplified by using primers 6A and 1B, and the products were purified by using a Qiagen spin column and sequenced directly by using primer 1B.
Results
Detection of Imprecise NHEJ After Induction of Double-Strand Breaks. Experiments were designed to produce a restriction cut site at a known genomic locus to test the hypothesis that double-strand breaks are targets for the integration of DHBV (Fig. 1). To induce double-strand break formation at the target site, we transfected an I-SceI expression plasmid into LMH 3.2 cells containing the 18-bp restriction recognition sequence, and to allow for integration to occur, we cotransfected the 1165A/DR1-13 plasmid. The 1165A/DR1-13 plasmid was used because indirect evidence based on sequence analysis of viral-cell junctions or subcloning of single cells containing integrated DHBV suggested that linear DHBV DNA was the most likely integration substrate (16, 23). In addition, the excess accumulation of nuclear DNA caused by the 1165A mutation was expected to maximize the frequency at which integration would occur. After transfection, cells were allowed to incubate for 3 days to permit expression of the restriction enzyme, double-strand break formation, repair, and integration.
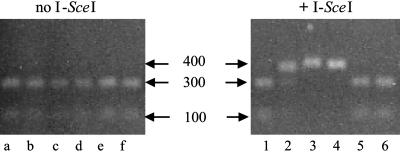
Initially, we looked for evidence of double-stranded breaks having occurred at the I-SceI site. Presumably, the majority of double-strand breaks would undergo precise NHEJ (24), and such products could not be distinguished from sites that failed to cut; however, imprecise NHEJ of double-strand breaks could be measured by the loss of the I-SceI site. Genomic DNA was digested with I-SceI to enrich for altered sites, and then diluted and individual uncut sites were amplified by nested PCR. Because of incomplete digestion of genomic DNA, a fraction of I-SceI sites were not cleaved and were consequently amplified. These products, false-positives, were identified by digesting the PCR-amplified DNA with I-SceI (Fig. 2, lanes a–f, 1, 5, and 6), whereas amplified sites that had lost the I-SceI recognition sequence were no longer digested by the enzyme, resulting in a single band (Fig. 2, lanes 3 and 4). Some sequence degeneracy is tolerated within the I-SceI recognition sequence, and thus single-base changes do not necessarily abolish cleavage but reduce its efficiency to variable extents, resulting in partial digests (lane 2). EGFP sequence analysis of five excised single bands showed four deletions and one insertion within the I-SceI recognition site, consistent with repair by NHEJ of a double-strand break (data not shown). The number of uncut or partially cut PCR bands was determined, and the average frequency of imprecise joining by NHEJ per transfected cell was calculated to be 1.4 × 10–3 (Table 2). These data represent the overall frequency of misjoining of the DNA ends only; because we do not know the rate of site cleavage and rejoining, we were unable to evaluate how frequently cut sites were repaired by NHEJ. These experiments indicate that transfection of an I-SceI expression vector into LMH 3.2 cells resulted in double-strand break formation at the recognition site that could be repaired by imprecise NHEJ.
Fig. 2.
Examples of products formed after imprecise joining of double-strand break and repair by NHEJ. An I-SceI expression vector was electroporated into LMH 3.2 cells (+I-SceI) or untransfected (no I-SceI) and incubated for 3 days. Genomic DNA was isolated, digested with I-SceI restriction enzyme to enrich in altered recognition sites, and amplified by PCR using EGFP-specific primer sets 1A/3B and 2A/4B (see Fig. 1 and Table 1). The PCR product was incubated with I-SceI to cleave wild-type products, electrophoresed through a 1.3% agarose gel and stained with ethidium bromide. Two bands indicate that the PCR product still contained the I-SceI recognition sequence (lanes a–f, 1, 5, and 6), and one band indicates loss of this sequence (lanes 3 and 4). Single base changes within the recognition sequence can result in partial digests (lane 2). The sizes of the fragments are given in base pairs.
Table 2. Imprecise NHEJ frequencies per transfected cell.
| Experiment* | NHEJ frequency |
|---|---|
| Untransfected | <1.4 × 10-4 (0)† |
| 1 | 1.2 × 10-3 (10) |
| 2 | 1.6 × 10-3 (13) |
| Mean | 1.4 × 10-3 (23) |
I-SceI expression and 1165A/DR1-13 plasmids (50 and 10 μg, respectively) were electroporated into 4 × 106 LMH3.2 cells. After 3 days in culture, genomic DNA was purified and assayed for imprecise NHEJ at the target cleavage sites as described in Methods.
Number of NHEJ products observed.
Imprecise NHEJ Results in the Capture of DHBV. Next, we determined whether imprecise NHEJ at the target site was associated with capture of DHBV. There are primarily two forms of linear DNA produced during DHBV replication. The dominant form is in situ primed linear DNA, which is a minor product of abortive replication caused by failure of plus-strand priming to generate a circular genome (20). In addition, cohesive-end linear DNA, a form that is probably derived from denaturation of the cohesive 5′ ends of circular viral DNA and elongation of the resultant recessed 3′ ends (25), has been postulated to be a minor integration substrate (6, 7, 15, 26). After cotransfection of the 1165A/DR1-13 plasmid and I-SceI expression vector, cells were incubated for 3 days to allow for expression of I-SceI, replication of DHBV, and capture of linear substrates. Purified genomic DNA was diluted and amplified by nested PCR of individual left and/or right EGFP/DHBV junctions. We detected no integrations for either left or right EGFP/DHBV junctions without I-SceI expression, which was an expected result considering the small target size of the recognition site (frequency <1.1 × 10–6; Table 3). However, when I-SceI was expressed, left EGFP/DHBV junctions were found at an average frequency of 9.9 × 10–5 per transfected cell with a comparable frequency (4.6 × 10–5) for right junctions (Table 3). Thus, left EGFP/DHBV junctions at cleavage sites were detected at least 90-fold more frequently than spontaneous integrations. Assuming that the orientation of integrated viral sequences was random, then the measured frequency of junctions was twice that observed, or ≈1–2 × 10–4 per transfected cell. Again, the true frequency of integration per double-strand break could not be estimated, because the actual number of site-specific cleavages could not be determined. However, imprecise NHEJ (Table 2) without DHBV integration occurred at a 7- to 14-fold greater frequency than that of DHBV integration (Table 3) when both repair outcomes were possible.
Table 3. Frequency of left and right viral-cell junctions per transfected cell.
| Experiment* | Frequency of left junctions | Frequency of right junctions |
|---|---|---|
| Control† | <1.1 × 10-6 (0)‡ | <6 × 10-6 (0) |
| 1 | 7.8 × 10-5 (27) | 4.6 × 10-5 (8) |
| 2 | 1.2 × 10-4 (41) | ND |
ND, not determined.
I-SceI expression and 1165A/DR1-13 plasmids (50 and 10 μg, respectively) were electroporated into 4 × 106 LMH 3.2 cells. After 3 days in culture, genomic DNA was purified and assayed for viral-cell junctions at the target cleavage sites as described in Methods.
Control transfections electroporated with DHBV vector alone.
Number of viral-cell junctions observed.
Sequence Analysis of EGFP/DHBV Junctions. Capture of DHBV at the I-SceI site is likely to occur via NHEJ. Because this pathway is typically associated with deletions and sometimes insertions of sequences at the termini, we investigated the sequences at the left and right EGFP/DHBV junctions. More samples were analyzed for left junctions, because the position of the left viral-cell junction can distinguish between integration of in situ primed linear and cohesive-end linear DNA. Viral-cell junctions from three separate experiments were excised from agarose gels, DNA-purified, and sequenced (Table 4). Assuming that in situ primed linear DNA was the DHBV substrate, 80% of left junctions had small deletions of sequence (2–58 bp) from the target genome and/or DHBV. Thirteen percent of left junctions had small insertions (5–18 bp) or a single large insert (374 bp) of unknown origin in addition to DHBV. In addition, 7% of left junctions contained some sequence from the 40 nucleotides upstream of the in situ primed linear DNA, consistent with a cohesive-end linear DNA substrate. Although 7 of 12 right junctions were associated with deletion of viral and/or cell DNA sequences, 5 of 12 products had no apparent loss of sequence. Overall, every product (75 of 75 for left junctions and 12 of 12 for right junctions) was consistent with capture of the postulated linear DHBV substrate by an NHEJ mechanism.
Table 4. Sequence summary of left and right viral-cell junctions 3 days posttransfection.
| Junction | No. with no loss of sequence | No. of deletions | No. of insertions | No. containing cohesive-end linear DNA |
|---|---|---|---|---|
| Left | 0 | 60 | 10 | 5 |
| Right | 5 | 7 | 0 | NA* |
Not applicable because both linear DNAs have equivalent right ends.
Linear DNA Is the Preferential Substrate for Integration. Although the results suggest a linear DHBV substrate for integration, we wished to confirm this substrate specificity. Therefore, we used two plasmid vectors: wild-type 1165A, which carries out a normal DHBV replication pathway and produces circular to linear DNA at an ≈10:1 ratio, and mutant 1165A/DR1-13, which produces circular to linear DNA at a 1:1 ratio (15, 20). Cotransfection of equal amounts of these two plasmids with the I-SceI expression vector should result in enrichment of the mutant among the integrated genomes, whereas an enrichment of wild-type genomes would be predicted if circular DNA were the preferred substrate. To confirm replication of both genotypes, replicative intermediates were isolated at 1, 3, and 6 days posttransfection from the cells that were analyzed for integration (Table 5, experiment 1). Southern blot analysis of the replicative intermediates showed an enrichment over time in the amounts of relaxed circular DNA being produced, suggesting an enrichment of 1165A DNA over time (Fig. 3). This result was confirmed by sequence analysis of amplified DHBV intermediates (results shown in Fig. 3). Despite the preferential replication of 1165A genomes, all viral-cell junctions detected in two independent transfections were derived from the 1165A/DR1-13 mutant (Table 5), as determined by sequencing the individual amplified products. The results are consistent with a strong bias for the linear DNA over relaxed circular DNA as integration substrates. DHBV Is Stably Integrated at Double-Strand Breaks. Although we have shown that one end of DHBV is attached at each genomic terminus at the I-SceI-induced double-strand break, these events were assayed independently, and thus it is possible that DHBV is transiently attached to the genomic DNA ends and not stably integrated. Therefore, we determined the stability of integrated DHBV within the EGFP locus by cotransfecting 1165A/DR1-13 plasmid with the I-SceI expression vector into LMH 3.2 cells as done before and determining the integration frequency during the subsequent five transfers of the cells, compared with the frequency of total viral DNA replicative intermediates. This analysis revealed no significant change in frequency of viral-cell junctions during five 1:10 cell transfers spanning 27 days post-transfection (Table 6), indicating that, once formed, viral-cell junctions were stably maintained. In contrast, unintegrated replicative intermediates failed to be maintained, decreasing by ≈1 order of magnitude with each transfer.
Table 5. Sequence distribution of integrated wild-type 1165A and mutant 1165A/DR1-13 DHBV.
| Experiment* | No. analyzed | Wild type, % | Mutant, % |
|---|---|---|---|
| 1 | 8 | 0 | 100 |
| 2 | 37 | 0 | 100 |
| Total | 45 | 0 | 100 |
I-SceI expression and a 1:1 mixture of 1165A and 1165A/DR1-13 plasmids (50 and 10 μg, respectively) were electroporated into 4 × 106 LMH3.2 cells. After 3 days in culture, genomic DNA was purified and assayed for viral-cell junctions at the target cleavage sites as described in Methods. Genotypes were determined by direct sequencing of the PCR products.
Fig. 3.
Southern blot and genotype analyses of DHBV replicative intermediates. I-SceI expression vector, wild-type 1165A, and 1165A/DR1-13 mutant plasmids were cotransfected into LMH 3.2 cells. After 1, 3, and 6 days of incubation, replicative intermediates were isolated, electrophoresed through a 1.3% agarose gel, transferred to a nylon membrane, and detected by hybridization with a riboprobe specific for detection of the minus strand. The three major forms of replicative intermediates are indicated: rc, relaxed circular DNA; lin, linear double-stranded DNA; ss, single-stranded DNA. The ratios of relaxed circular and linear double-strand DNA (rc/lin) were determined by phosphorimaging. In addition, viral sequences (nucleotides 2669–2840) were amplified by PCR and directly sequenced to determine the ratios of 1165A/1165A-DR1-13 (wt/mut) at five different sites of single-nucleotide polymorphism between the two strains at nucleotide positions 2736, 2742, 2751, 2762, and 2790.
Table 6. Frequency of viral-cell junctions and replicative intermediates per transfected cell in serial transfers after transfection.
| Transfers (1:10) | Frequency of left junctions | Frequency of right junctions | Frequency of replicative intermediates |
|---|---|---|---|
| 0 | 2.4 × 10-4 (46)* | 7.8 × 10-5 (8) | 2.3 × 102 |
| 1 | 8.1 × 10-5 (7) | ND | 5.0 × 100 |
| 2 | 1.4 × 10-4 (12) | ND | 5.5 × 10-1 |
| 3 | 1.8 × 10-4 (15) | ND | 1.0 × 10-1 |
| 5 | 3.2 × 10-4 (106) | 1.8 × 10-4 (19) | 4.9 × 10-3 |
Transfected cells were assayed for viral-cell junctions and for viral replicative intermediates after the indicated number of transfers and outgrowth of cells. One-tenth of the total cells were subcultured at each transfer. Frequencies are expressed as copies per transfected cell. ND, not determined.
Number of viral-cell junctions observed
Agarose gels of left EGFP/DHBV junctions from the original population and after five transfers (Fig. 4) showed that size variation caused by deletions/insertions at the sites of DNA capture seen initially were lost by the fifth transfer. Sequence analysis of a sample of nine of these EGFP/DHBV junctions confirmed this loss of complexity such that all analyzed had the same junction sequence. We attribute this loss of complexity to the random recovery and loss of clones during transfer (see Supporting Text, which is published on the PNAS web site). On the other hand, expansion of one clone during the transfers directly confirms the stability of that integrated genome.
Fig. 4.
Agarose gel analysis of left EGFP/DHBV junctions after multiple cell transfers. LMH 3.2 cells were transfected with I-SceI expression vector and 1165A/DR1-13 plasmid and incubated for 3 days (transfer 0) or split ≈1:10 to keep cells in logarithmic growth until 27 days (transfer 5) posttransfection. Genomic DNA was isolated and nested PCR performed as described in the Fig. 1 legend. PCR products were electrophoresed through a 1.3% agarose gel and visualized by ethidium-bromide staining. Molecular marker lanes (m) are included.
Discussion
Using DHBV as a model for the Hepadnaviridae family and host chicken LMH cells, we have shown that the presence of a double-strand break in cellular DNA stimulates viral integration at that site >90-fold (Table 3). Cellular double-strand breaks can result from endogenous metabolism (27) or from exogenous damage (28). Repair of such double-strand breaks is important to the survival of eukaryotic cells, because a single unrepaired cellular double-strand break can result in cell death. Double-strand breaks can be repaired by homology- and nonhomology-dependent mechanisms, although repair of double-strand breaks in higher eukaryotes in the absence of any significant homology occurs through NHEJ (28–30). In our model system, an I-SceI-induced double-strand break within the target gene was repaired by either precise (no biological consequences, not measured) or imprecise (Fig. 2) NHEJ. Imprecise NHEJ resulted in mutations (small deletions or insertions) at the site of joining.
When DHBV linear DNA produced by virus replication was present during repair of the double-strand break, ligation of either end of the break to linear viral DNA by NHEJ could be observed, whereas no such ligations were detected in the absence of induced double-strand breaks. Linear DNA was highly preferred over relaxed circular DNA as the ligation substrate. The viral-cell junctions produced at the sites of double-strand breaks were stable through a 105-fold expansion of the population of cells (five transfers of 1:10), or ≈17 cell divisions (105 ≈ 216.6), and therefore, they represented stable insertions of viral sequences rather than transient ligation products. The fact that almost all viral-cell junctions that we have characterized previously in transient and chronic hepadnavirus infections in vivo bear the typical features of NHEJ, i.e., small deletions of viral sequences at the site of joining, suggests that double-strand breaks are the primary source of integrations in infected hepatocytes.
Double-strand breaks in cells can be produced by genotoxic agents (ionizing radiation, oxidative damage, chemical agents), often through conversion of single-strand lesions into double-strand breaks during DNA replication in growing cells. Double-strand breaks can be repaired without error by homologous recombination (gene conversion), commonly involving strand invasion of a sister chromatid, or by error-prone mechanisms such as NHEJ or single-strand annealing (28, 30).
In humans, chronic HBV infection causes an ongoing inflammatory response that results in oxidative damage to the DNA of liver cells (31, 32). Such damage can be converted to double-strand breaks during hepatocyte regeneration in response to cell death of liver tissue (27). As shown in this study, subsets of such putative double-strand breaks in virus-infected cells are expected to be genetically marked by integrated viral DNA, inserted by NHEJ. Previous reports have shown that oxidative damage leads to increased genomic levels of hepadnavirus integration (33, 34) in growing cell lines. Our study suggests that enhanced integration in these studies was caused by the generation of double-strand breaks that served as targets for integration. It seems likely, therefore, that double-strand breaks in cellular DNA, resulting from inflammation-induced DNA damage and regeneration, would be reflected in the level of integrated DNA in infected liver.
In previous studies, the frequency of integrated viral DNA in woodchuck livers chronically infected with woodchuck hepatitis virus was 1–2 orders of magnitude greater than that resulting from a transient infection. Moreover, the frequency of integrated DNAs in the liver did not change during clearance of virus by immune or antiviral therapy (6, 7). These results show that cellular genomic alterations acquired as a consequence of chronic or acute viral hepatitis accumulate during and persist after resolution of the infection. Judging by the frequencies of integrated DNA, we suppose that the amount of genetic injury incurred in the chronic infection would have been 1–2 orders of magnitude greater than that incurred in the transient infection if most integrations occurred at double-strand breaks. The fact that viral DNA integrations in chronic hepatitis B can be detected at frequencies as high as one or more copies per cell implies that hepatocytes and other cells in the liver have sustained even higher levels of mutation, placing DNA damage as a major component of the pathogenesis of hepatitis B.
Finally, we observed that, although the frequency of integrated DNA in cultured LMH cells was maintained through at least five transfers, the frequency of replicative intermediate DNA decreased rapidly, approximately in inverse proportion to the expansion of the original cell population. This decrease did not seem to be accounted for by selection against the virus-infected cells, because their progeny (identified by the presence of integrated DNA in some members) were maintained throughout the transfers with undiminished frequency. Although the DR1-13 mutation produces a partial defect in relaxed circular DNA synthesis, this defect is compensated by the 1165A mutation that enhances covalently closed circular DNA levels such that, in theory, a complete intracellular pathway of DNA replication should be sustained indefinitely. Nevertheless, intracellular replication was insufficient to maintain a stable frequency of infection in the dividing cells. This result suggests that individual dividing cells can be spontaneously “cured” of an infection when reinfection does not occur. This phenomenon is superficially similar to the clearance of transient infections in vivo, in which hepatocyte turnover and inhibition of reinfection by immune mechanisms are thought to play crucial roles (7, 35, 36). The exact mechanisms responsible for the loss of replicating virus, however, are not understood.
Supplementary Material
Acknowledgments
We thank William Mason and Simon Powell for a critical reading of the manuscript. This work was supported by Department of Health and Human Services Grant CA84017.
Abbreviations: HBV, hepatitis B virus; HCC, hepatocellular carcinoma; NHEJ, nonhomologous end joining; DHBV, duck HBV; EGFP, enhanced GFP; CMV, cytomegalovirus.
References
- 1.Beasley, R. P. (1988) Cancer 61, 1942–1956. [DOI] [PubMed] [Google Scholar]
- 2.Levy, L., Renard, C. A., Wei, Y. & Buendia, M. A. (2002) Ann. N.Y. Acad. Sci. 963, 21–36. [DOI] [PubMed] [Google Scholar]
- 3.Birrer, R. B., Birrer, D. & Klavins, J. V. (2003) Ann. Clin. Lab. Sci. 33, 39–54. [PubMed] [Google Scholar]
- 4.Summers, J. & Mason, W. S. (1982) Cell 29, 403–415. [DOI] [PubMed] [Google Scholar]
- 5.Brechot, C., Hadchouel, M., Scotto, J., Fonck, M., Potet, F., Vyas, G. N. & Tiollais, P. (1981) Proc. Natl. Acad. Sci. USA 78, 3906–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Summers, J., Jilbert, A. R., Yang, W., Aldrich, C. E., Saputelli, J., Litwin, S., Toll, E. & Mason, W. S. (2003) Proc. Natl. Acad. Sci. USA 100, 11652–11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Summers, J. & Mason, W. S. (2004) Proc. Natl. Acad. Sci. USA 101, 638–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasui, H., Hino, O., Ohtake, K., Machinami, R. & Kitagawa, T. (1992) Cancer Res. 52, 6810–6814. [PubMed] [Google Scholar]
- 9.Transy, C., Fourel, G., Robinson, W. S., Tiollais, P., Marion, P. L. & Buendia, M. A. (1992) Proc. Natl. Acad. Sci. USA 89, 3874–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen, L. J., Tennant, B. C., Seeger, C. & Ganem, D. (1993) Mol. Cell. Biol. 13, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fourel, G., Trepo, C., Bougueleret, L., Henglein, B., Ponzetto, A., Tiollais, P. & Buendia, M. A. (1990) Nature 347, 294–298. [DOI] [PubMed] [Google Scholar]
- 12.Gozuacik, D., Murakami, Y., Saigo, K., Chami, M., Mugnier, C., Lagorce, D., Okanoue, T., Urashima, T., Brechot, C. & Paterlini-Brechot, P. (2001) Oncogene 20, 6233–6240. [DOI] [PubMed] [Google Scholar]
- 13.Robinson, W. S. (1994) Annu. Rev. Med. 45, 297–323. [DOI] [PubMed] [Google Scholar]
- 14.Buendia, M. A. (1992) Adv. Cancer Res. 59, 167–226. [DOI] [PubMed] [Google Scholar]
- 15.Yang, W. & Summers, J. (1995) J. Virol. 69, 4029–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang, W. & Summers, J. (1999) J. Virol. 73, 9710–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Summers, J., Smith, P. M., Huang, M. J. & Yu, M. S. (1991) J. Virol. 65, 1310–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandart, E., Kay, A. & Galibert, F. (1984) J. Virol. 49, 782–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loeb, D. D., Hirsch, R. C. & Ganem, D. (1991) EMBO J. 10, 3533–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staprans, S., Loeb, D. D. & Ganem, D. (1991) J. Virol. 65, 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang, F., Han, M., Romanienko, P. J. & Jasin, M. (1998) Proc. Natl. Acad. Sci. USA 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenhoff, R. J. & Summers, J. (1994) J. Virol. 68, 5706–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong, S. S., Jensen, A. D., Chang, C. J. & Rogler, C. E. (1999) J. Virol. 73, 1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, Y., Lukacsovich, T. & Waldman, A. S. (1999) Mol. Cell. Biol. 19, 8353–8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, W., Mason, W. S. & Summers, J. (1996) J. Virol. 70, 4567–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, W. & Summers, J. (1998) J. Virol. 72, 8710–8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilenchik, M. M. & Knudson, A. G. (2003) Proc. Natl. Acad. Sci. USA 100, 12871–12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valerie, K. & Povirk, L. F. (2003) Oncogene 22, 5792–5812. [DOI] [PubMed] [Google Scholar]
- 29.Henderson, G. & Simons, J. P. (1997) Mol. Cell. Biol. 17, 3779–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothkamm, K., Kruger, I., Thompson, L. H. & Lobrich, M. (2003) Mol. Cell. Biol. 23, 5706–5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimoda, R., Nagashima, M., Sakamoto, M., Yamaguchi, N., Hirohashi, S., Yokota, J. & Kasai, H. (1994) Cancer Res. 54, 3171–3172. [PubMed] [Google Scholar]
- 32.Hagen, T. M., Huang, S., Curnutte, J., Fowler, P., Martinez, V., Wehr, C. M., Ames, B. N. & Chisari, F. V. (1994) Proc. Natl. Acad. Sci. USA 91, 12808–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen, J., Dandri, M., Burkle, A., Zhang, L. & Rogler, C. E. (1997) J. Virol. 71, 5455–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dandri, M., Burda, M. R., Burkle, A., Zuckerman, D. M., Will, H., Rogler, C. E., Greten, H. & Petersen, J. (2002) Hepatology 35, 217–223. [DOI] [PubMed] [Google Scholar]
- 35.Guo, J.-T., Zhou, H., Liu, C., Aldrich, C., Saputelli, J., Whittaker, T., Inmaculada Barrasa, M., Mason, W. & Seeger, C. (2000) J. Virol. 74, 1495–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieland, S., Spangenberg, H., Thimme, R., Purcell, R. & Chisari, F. (2004) Proc. Natl. Acad. Sci. USA 101, 2129–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.