Abstract
Mutations in SCN1A, the gene encoding the brain voltage-gated sodium channel α1 subunit (NaV1.1), are associated with at least two forms of epilepsy, generalized epilepsy with febrile seizures plus and severe myoclonic epilepsy of infancy (SMEI). We examined the functional properties of five SMEI mutations by using whole-cell patch-clamp analysis of heterologously expressed recombinant human SCN1A. Two mutations (F902C and G1674R) rendered SCN1A channels nonfunctional, and a third allele (G1749E) exhibited minimal functional alterations. However, two mutations within or near the S4 segment of the fourth repeat domain (R1648C and F1661S) conferred significant impairments in fast inactivation, including persistent, noninactivating channel activity resembling the pattern of channel dysfunction observed for alleles associated with generalized epilepsy with febrile seizures plus. Our data provide evidence for a range of SCN1A functional abnormalities in SMEI, including gain-of-function defects that were not anticipated in this disorder. Our results further indicate that a complex relationship exists between phenotype and aberrant sodium channel function in these inherited epilepsies.
Keywords: seizure, generalized epilepsy with febrile seizures plus, SCN1A, electrophysiology
Mutations in genes encoding neuronal voltage-gated sodium channels have been linked to inherited forms of epilepsy. Genetic defects in two pore-forming α subunits (encoded by SCN1A and SCN2A) and the accessory β1 subunit (encoded by SCN1B) have been discovered in four distinguishable clinical syndromes with overlapping features (1–6). Generalized epilepsy with febrile seizures plus (GEFS+) is an autosomal dominant disorder characterized by childhood febrile seizures that persist beyond age 6 years, as well as afebrile generalized or partial seizures of various types. In 1998, Wallace et al. (1) described a single missense mutation in SCN1B, the gene encoding the voltage-gated sodium channel β1 subunit, in a large GEFS+ pedigree. However, SCN1B mutations are rare causes of GEFS+ (7, 8). By contrast, mutations in SCN1A, the gene encoding the neuronal sodium channel α-subunit NaV1.1, have been identified in several GEFS+ families (2, 9–11).
SCN1A mutations occur also in severe myoclonic epilepsy of infancy (SMEI), a rare convulsive disorder characterized by febrile seizures with onset during the first year of life, followed by intractable epilepsy, impaired psychomotor development, and ataxia (12, 13). Seizures in this disorder typically do not respond to standard anticonvulsant pharmacotherapy. More than 80 heterozygous, predominantly de novo, SCN1A mutations have been reported in this disorder (3, 14–17). Because many of the SCN1A mutations discovered in SMEI probands are nonsense and frameshift alleles, loss of neuronal sodium channel function as the cause of this syndrome seems most plausible. This hypothesis is supported by the observation that certain missense mutations in this condition render SCN1A channels nonfunctional or severely impaired (18, 19), but whether all missense SCN1A mutations associated with SMEI cause loss of function is not known.
We report the biophysical characterization of five SCN1A missense mutations associated with SMEI. Consistent with the loss-of-function hypothesis, two of these mutations produced nonfunctional sodium channels. However, two other alleles exhibit persistent, noninactivating channel behavior closely resembling gain-of-function SCN1A mutations associated with GEFS+ (20). Our data suggest that a general correlation between channel defects and associated clinical phenotypes does not exist. These observations also suggest that other genetic, developmental, or environmental factors may interact with the biophysical defect to dictate the final clinical expression of neuronal sodium channelopathies.
Materials and Methods
Mutagenesis and Heterologous Expression of Human SCN1A. Site-directed mutagenesis of full-length (6,030 bp) human SCN1A cDNA was performed as described in ref. 20. All mutant cDNAs were fully sequenced before their use in transfection experiments. WT and mutant SCN1A channels were heterologously coexpressed with human accessory β1 and β2 subunits in human tsA201 cells as described in ref. 20. At least two different recombinant clones of each mutant cDNA were evaluated. Concentration of each plasmid DNA was determined spectrophotometrically (UV) and fluorometrically (PicoGreen, Molecular Probes).
Electrophysiology and Data Analysis. Whole-cell patch-clamp recording was used to characterize functional properties of WT and mutant sodium channels as described in refs. 18 and 20. Series resistance (2 ± 0.1 MΩ) was compensated 87–95% to ensure that the command potential was reached within microseconds and with a voltage error <3 mV. Specific voltage-clamp protocols assessing channel activation, inactivation, and recovery from inactivation are depicted in each figure. Persistent current was evaluated by 200-ms depolarizations to –10 mV in the presence and absence of 10 μM tetrodotoxin (TTX) (Sigma), a potent sodium-channel blocker. In experiments examining effects of internal acidification, pipette solutions were prepared with Mes buffer titrated to pH 6.2 instead of Hepes. The range of peak current amplitudes recorded at –10 mV in this study was 0.6–5.0 nA. Data analysis was performed by using clampfit 8.2 (Axon Instruments, Union City, CA), excel 2002 (Microsoft), and originpro 7.0 (OriginLab, Northampton, MA) software as described in ref. 18. All data were fit by using a nonlinear least-squares minimization method. Results are presented as means ± SEM, and statistical comparisons were made between data from mutant sodium channels and WT SCN1A by using the unpaired Student t test. Statistical significance was assumed for P < 0.05 and deemed not significant for P ≥ 0.05. In some figures, the standard error bars are smaller than the data symbols.
Results
We characterized the functional properties of five de novo SCN1A missense mutations associated with SMEI. The mutations were reported by Claes et al. (F1661S and G1749E) (16) or Ohmori et al. (F902C, R1648C, and G1674R; formerly reported as F891C, R1638C, and G1664R, respectively, based on alignments with a common splice variant lacking 33 nucleotides from exon 11) (15). All alleles reside within domain IV (D4) except F902C, which alters a residue in domain II. Mutation R1648C affects the same D4/S4 amino acid as an allele associated with GEFS+ (R1648H) (2).
Biophysical Properties of SCN1A Mutants. All mutant SCN1A channels were studied under identical conditions by using heterologous expression of recombinant human SCN1A in cultured human cells (tsA201) and whole-cell patch-clamp recording. Experiments were performed with coexpression of human β1 and β2 accessory subunits. Transient transfection of tsA201 cells with two mutants (F902C and G1674R) did not result in measurable sodium current (n = 10 for each mutant). By contrast, cells expressing the other three mutant channels exhibited ample sodium current to enable detailed biophysical analyses.
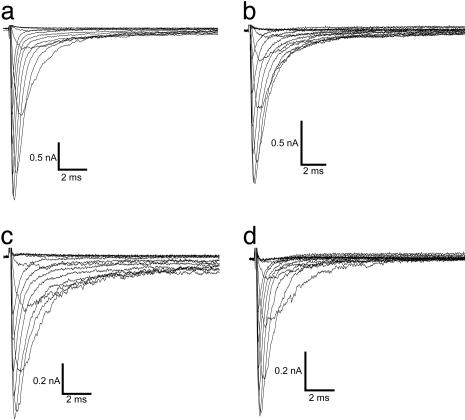
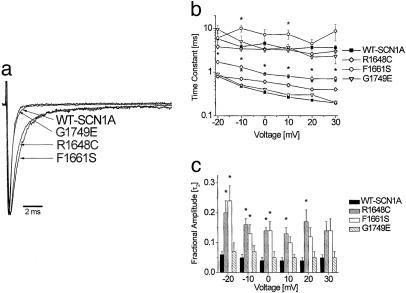
Fig. 1 illustrates representative whole-cell currents evoked by a series of depolarizing test potentials in cells expressing either WT-SCN1A or one of three functional mutant channels (R1648C, F1661S, and G1749E). In general, the three mutant channels examined in this experiment exhibited rapid activation similar to WT-SCN1A, but in some cases presented with subtle perturbations of fast inactivation properties. Mutants R1648C and F1661S inactivated more slowly and less completely than WT-SCN1A, whereas G1749E appeared no different from WT channels. These differences are more clearly illustrated in Fig. 2a, where current traces recorded at the same test potential (–10 mV) are normalized to peak current and superimposed. We quantified time constants for fast inactivation by fitting each time course with a two-exponential function and plotting the data against the test potential (Fig. 2b). Mutants R1648C and F1661S exhibited significant differences in time constants describing the faster component of inactivation at nearly all voltages, and G1749E was similar to WT-SCN1A. The slower component of fast inactivation was also significantly different for F1661S but only at a limited number of membrane potentials. Furthermore, there were significant increases in the proportion of inactivation proceeding with a slower time constant for R1648C and F1661S but not G1749E (Fig. 2c). These data indicate that significant impairments in fast inactivation are conferred by R1648C and F1661S, whereas minimal alteration of inactivation kinetics occurs in G1749E channels.
Fig. 1.
Functional properties of WT and mutant SCN1A sodium channels. Representative whole-cell currents recorded from tsA201 cells expressing WT (a) or mutant SCN1A channels R1648C (b), F1661S (c), and G1749E (d). Cells were stepped to various potentials between –80 and +40 mV in 10-mV increments from a holding potential of –120 mV (see Fig. 3b for pulse protocol). All experiments were performed in the whole-cell patch-clamp configuration at room temperature 24–72 h after transfection.
Fig. 2.
Voltage dependence of fast inactivation for WT and mutant channels. (a) Representative normalized whole-cell sodium currents from cells stepped to a potential of –10 mV from a holding potential of –120 mV. Peak current amplitudes were normalized. (b) Inactivation time constants of WT and mutant SCN1A currents. The decay phase of voltage-sensitive inward currents was fitted with a two-exponential function, It/Imax = A1·exp(–t/τ1) + A2·exp(–t/τ2) + C, where An and τn refer to fractional amplitude and time constant, respectively. (c) Fractional amplitudes of the slower component (designated as τ2) of fast inactivation plotted against voltage. Values significantly different from WT are indicated: *, P < 0.005; †, P < 0.05.
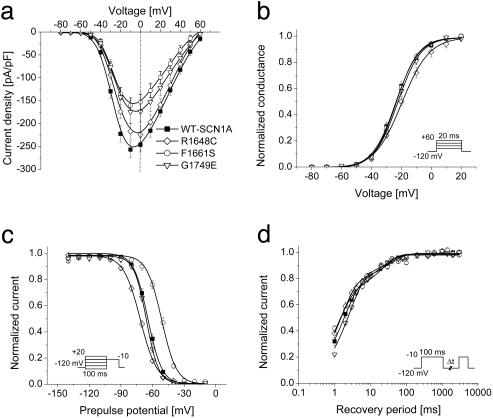
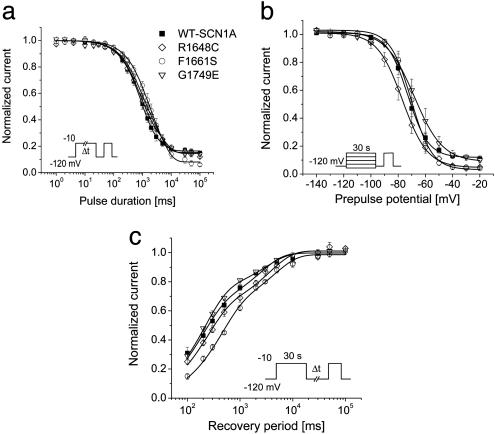
Current–voltage relationships, voltage dependence of activation, voltage dependence of steady-state channel availability, and recovery from fast inactivation were also examined (Fig. 3). The largest sodium current density was observed at approximately the same voltage (about –10 mV) for WT-SCN1A and the three functional mutants, but significantly lower current densities were observed for F1661S and G1749E. Activation was significantly shifted toward more depolarizing potentials and exhibited a less steep voltage dependence in cells expressing R1648C as compared with WT-SCN1A, whereas the other two functional mutants activated with the same voltage dependence as WT channels (Fig. 3b and Table 1). The voltage dependence of channel availability following a 100-ms depolarization was shifted in the hyperpolarized or depolarized direction for R1648C and F1661S, respectively (Fig. 3c and Table 1), but G1749E channel availability was not different from WT-SCN1A. Although inactivation is slower in R1648C and F1661S, recovery from inactivation triggered by a 100-ms depolarization (–10 mV) proceeded more quickly (illustrated by smaller time constants) as compared with WT-SCN1A (Fig. 3d and Table 1). These findings indicate significant alterations of fast inactivation for R1648C and F1661S but not G1749E.
Fig. 3.
Inactivation and activation properties of SMEI-associated SCN1A mutants. (a) Current–voltage relationships of whole-cell currents. Currents were elicited by test pulses to various potentials (see b Inset) and normalized to cell capacitance (WT, n = 17; R1648C, n = 7; F1661S, n = 8; G1749E, n = 8). F1661S current density is significantly smaller than WT between –30 and +50 mV (P < 0.05). G1749E current density is significantly smaller than WT between –20 and +30 mV (P < 0.05). (b) Voltage dependence of activation. The voltage dependence of channel activation was estimated by measuring peak sodium current during a variable test potential from a holding potential of –120 mV. The current at each membrane potential was divided by the electrochemical driving force for sodium ions and normalized to the maximum sodium conductance. (c) Voltage dependence of inactivation. The two-pulse protocol illustrated by Inset was used to examine channel availability after conditioning at various potentials. Currents were normalized to the peak current amplitude. (d) Recovery from fast inactivation. Channels were inactivated by a 100-ms pulse then stepped to –120 mV for various durations. Currents were normalized to the peak current amplitude measured during the inactivation pulse and fitted to a two-exponential function, It/Imax = A1·[1 – exp(–t/τ1) + A2·[1 – exp(–t/τ2), generating fast and slow recovery time constants. Fit parameters for all experiments are provided in Table 1.
Table 1. Biophysical parameters for activation and fast inactivation.
| Voltage dependence of activation
|
Voltage dependence of fast inactivation
|
Recovery from fast inactivation
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Channel | V1/2, mV | k | n | V1/2, mV | k | n | τ1, ms (amplitude) | τ2, ms (amplitude) | n |
| WT-SCN1A | -23.6 ± 1.2 | 7.4 ± 0.3 | 17 | -64.2 ± 1.1 | -5.8 ± 0.1 | 18 | 3.8 ± 0.5 (83 ± 2%) | 96 ± 16 (17 ± 2%) | 19 |
| R1648C | -19.7 ± 0.6* | 9.2 ± 0.6† | 7 | -71.3 ± 0.2§ | -7.7 ± 0.2§ | 7 | 1.5 ± 0.1* (77 ± 1.5%) | 33 ± 5* (21 ± 1.5%) | 7 |
| F1661S | -23.5 ± 0.4 | 7.5 ± 0.3 | 8 | -52.0 ± 0.4§ | -7.1 ± 0.4§ | 8 | 1.3 ± 0.1‡ (71 ± 2%†) | 31 ± 5* (28 ± 2%‡) | 8 |
| G1749E | -22.0 ± 1.6 | 7.6 ± 0.3 | 8 | -66.4 ± 1.1 | -6.0 ± 0.3* | 8 | 2.5 ± 0.2 (81 ± 3%) | 61 ± 27 (19 ± 2%) | 8 |
Values presented are mean ± SEM. Values statistically significantly different from WT-SCN1A are marked as follows: *, P < 0.05; †, P < 0.01; ‡, P < 0.005; §, P < 0.001.
R1648C and F1661S Channels Exhibit Noninactivating Sodium Current.
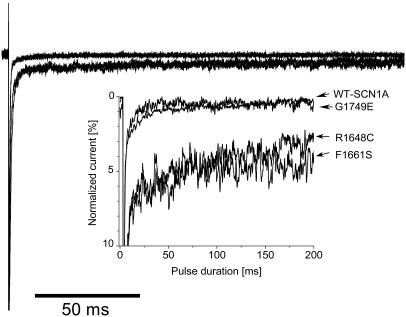
We previously reported that three SCN1A mutations associated with GEFS+, including R1648H, cause persistent, noninactivating whole-cell current, and we interpreted these findings as evidence of a gain-of-function sodium channel defect (20). By contrast, many mutations associated with SMEI are nonsense or frameshift alleles, suggesting a likely loss-of-function disease mechanism. We examined the three functional missense SMEI mutations for evidence of persistent TTX-sensitive current. Both R1648C and F1661S, but not G1749E, exhibited significant levels of noninactivating current measured at the end of a 200-ms depolarization. When expressed as the percentage of peak current amplitude (Fig. 4), the magnitudes of persistent current were WT, 0.3 ± 0.1% (n = 9); G1749E, 0.5 ± 0.1% (n = 4); R1648C, 3.6 ± 0.3% (n = 8, P < 0.05); and F1661S, 3.8 ± 0.3% (n = 5, P < 0.005). Thus, persistent, noninactivating sodium current may be observed with SCN1A mutations associated with either GEFS+ or SMEI. This unexpected result implies that no simple correlation exists between the clinical phenotype and biophysical behavior associated with missense SCN1A mutations.
Fig. 4.
Noninactivating sodium currents. Sodium current was elicited by a 200-ms depolarization from –120 to 10 mV. TTX-sensitive currents were obtained by digital subtraction of sodium currents recorded before and after TTX addition. Peak sodium currents were normalized. Inset shows an expanded y axis scaled to emphasize the relative proportion of noninactivating current.
Alterations in Slow Inactivation. We also tested cells expressing either WT or mutant channels for differences in slow inactivation properties (Fig. 5). In these experiments, the effects of fast inactivation were eliminated by a brief recovery pulse to –120 mV. Entry into the slow inactivated state proceeded with a similar biexponential time course for WT-SCN1A and all three functional mutants (Fig. 5a). Time constants for onset of slow inactivation were not significantly different from WT-SCN1A except for G1749E (τ1, P < 0.005) (Table 2). Steady-state channel availability following 30-s depolarizations to various potentials was similar in WT and mutants, but the voltage dependence was less steep for R1648C and G1749E (Fig. 5b). Recovery from slow inactivation following a 30-s depolarization proceeded in a similar biexponential time course for all channels, except that F1661S exhibited significantly larger time constants (Table 2), suggesting impaired recovery from slow inactivation.
Fig. 5.
Slow inactivation properties of WT and mutant SCN1A channels. (a) Onset of slow inactivation. Cells were depolarized to –10 mV for durations ranging from 1 ms to 100 s, allowed to recover from fast inactivation at –120 mV for 50 ms, and subjected to a –10-mV test pulse. (b) Steady-state slow inactivation after a 30-s depolarization to potentials between –140 and –10 mV. (c) Recovery from slow inactivation. Cells were conditioned at –10 mV for 30 s, allowed to recover from inactivation at –120 mV for 0.1–100 s, and immediately tested at –10 mV. Because the intermediate recovery period always exceeded 100 ms, effects of fast inactivation were negligible. All data were fitted to a two-exponential (see Fig. 3 legend) or a Boltzmann function (I/Imax = {1 + exp[(V – V1/2)/k]}–1), where V denotes the stepping potential, V1/2 denotes the stepping potential where half-maximal slow inactivation is achieved, and k is the slope factor of the fitting curve. Fit parameters for all experiments are provided in Table 2.
Table 2. Biophysical parameters for slow inactivation.
| Onset of slow inactivation
|
Voltage dependence of slow inactivation
|
Recovery from slow inactivation
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Channel | τ1, ms (amplitude) | τ2, ms (amplitude) | Residual current, % | n | V1/2, mV | k | Residual current, % | n | τ1, ms (amplitude) | τ2, ms (amplitude) | n |
| WT-SCN1A | 863 ± 113 (55 ± 5%) | 7,585 ± 2,352 (29 ± 5%) | 16 ± 2 | 11 | -72.5 ± 1.8 | 6.4 ± 0.3 | 12 ± 2 | 10 | 228 ± 26 (67 ± 2%) | 2,634 ± 246 (33 ± 2%) | 12 |
| R1648C | 833 ± 107 (55 ± 6%) | 4,492 ± 459 (30 ± 5%) | 15 ± 2 | 6 | -76.5 ± 2.7 | 8.6 ± 0.5† | 4 ± 1* | 5 | 232 ± 17 (61 ± 3%) | 4,098 ± 880 (40 ± 4%) | 6 |
| F1661S | 852 ± 257 (36 ± 12%) | 7,638 ± 3,380 (56 ± 12%*) | 7 ± 1* | 6 | -69.6 ± 2.7 | 6.0 ± 0.3 | 4 ± 1* | 5 | 422 ± 26† (62 ± 5%) | 5,057 ± 883† (36 ± 4%) | 5 |
| G1749E | 325 ± 57† (18 ± 4%†) | 2,368 ± 277† (67 ± 5%†) | 15 ± 2 | 7 | -67.6 ± 3.2 | 8.2 ± 0.5† | 10 ± 2 | 7 | 213 ± 13 (74 ± 2%) | 3,592 ± 620 (26 ± 2%) | 6 |
Values presented are mean ± SEM. Values statistically significantly different from WT are marked as follows: *, P < 0.05; †, P < 0.005.
Comparison of R1648H and R1648C. Two distinct amino acid substitutions of the same SCN1A residue (R1648) have been associated with either a mild (GEFS+: R1648H) or severe (SMEI: R1648C) epilepsy syndrome even though the two alleles share a similar biophysical disturbance (persistent sodium current). This observation prompted us to directly compare the degree of persistent sodium current exhibited by these two mutations. In a blinded, side-by-side comparison, the proportion of persistent TTX-sensitive current measured at the end of a 200-ms depolarization to –10 mV was 0.2 ± 0.05% for WT-SCN1A (n = 4), 1.8 ± 0.3% for R1648H (n = 6), and 3.6 ± 0.3% for R1648C (n = 5; P < 0.005 for the difference between R1648H and R1648C).
Finally, we tested whether lowering the internal pH might impart a positive charge to histidine (pKa ≈6.1) through titration of the imidazole side chain. Intracellular acidification may occur during prolonged seizures (21–23). Previous solvent accessibility studies performed with the human skeletal muscle sodium channel demonstrated that the fifth positive charge in the D4/S4 segment (designated as R5) corresponding to SCN1A-1648 is accessible to aqueous reagents applied to the cytoplasmic face of the membrane over a wide range of voltages (24). Lowering intracellular pH (pHi) to 6.2 exerted a significant effect on the proportion of persistent TTX-sensitive sodium current exhibited by R1648C (2.1 ± 0.3%, n = 6; P < 0.01 compared with pHi 7.3), but there were no significant effects on WT-SCN1A (0.57 ± 0.16, n = 4) or R1648H channels (1.1 ± 0.2%, n = 6; P = 0.1 compared with pHi 7.3). There were no significant effects on the degree of persistent current observed with external acidification.
Discussion
Mutations in genes encoding neuronal voltage-gated sodium channel subunits are responsible for a group of epilepsy syndromes with overlapping clinical characteristics but divergent clinical severity. Defining the molecular and physiological mechanisms underlying these disorders has advanced our understanding of epileptogenesis, improved our diagnostic framework, and may eventually facilitate development of new therapeutic strategies.
GEFS+ is usually a benign disorder associated with missense SCN1A or SCN1B mutations. Further evidence of genetic heterogeneity in GEFS+ has been demonstrated by discovery of mutations in genes encoding another sodium channel α subunit (SCN2A) (1, 4) and a γ-aminobutyric acid receptor subunit (GABRG2) (25, 26). SCN2A defects also occur in another mild epilepsy syndrome, benign familial neonatal–infantile seizures (BFNIS) (6). Neither GEFS+ nor BFNIS are associated with developmental delays or mental retardation. By contrast, SMEI and the related syndrome of intractable childhood epilepsy with generalized tonic–clonic (ICEGTC) seizures (5) represent the opposite extreme in clinical severity. ICEGTC seizures is a related severe neurological disorder differing from SMEI by the absence of myoclonic seizures and lower incidence of severe mental dysfunction. Some authors have referred to this syndrome as borderline SMEI (17, 27). The overlapping phenotypes and molecular genetic etiologies among the SCN1A-linked epilepsies lend support for the hypothesis that they represent a continuum of clinical disorders (28).
To correlate clinical phenotypes with the underlying sodium channel disorder, we and others have characterized the biophysical properties of various mutant voltage-gated sodium channels associated with epilepsy (18–20, 29–33). Early findings have suggested that in some cases of GEFS+, SCN1A mutations promote a gain of function, whereas mutations associated with SMEI disable channel function. However, a simple relationship between GEFS+ and SMEI mutation effects may not hold for all alleles as demonstrated by the recognition that some SCN1A GEFS+ mutations are nonfunctional or exhibit loss-of-function characteristics (18, 30). These findings emphasize the continued need for functional analyses of mutant sodium channels to explore the complete spectrum of genotype–phenotype correlations.
In this study, we have demonstrated that certain SCN1A mutations associated with SMEI exhibit biophysical characteristics classifiable as gain-of-function defects. This finding was unexpected given the predominance of predicted loss-of-function mutations in SMEI. Mutations R1648C and F1661S impair inactivation, leading to increased sodium conductance over time similar to GEFS+ mutations R1648H, T875M, and W1204R (20). We speculate that noninactivating sodium current in neurons will promote hyperexcitability in the nervous system either by lowering the activation threshold in excitatory neurons or by rendering inhibitory neurons inexcitable through sustained membrane depolarization. Which cellular mechanism predominates in the brain is unknown, but these biophysical observations motivate further experiments in animal models to determine the precise impact of sodium channel mutations.
Whereas gain-of-function is a prominent feature displayed by R1648C and F1661S, additional functional defects favor reduced channel availability. Specifically, reduced current density (G1749E and F1661S), depolarizing shifts in voltage dependence of activation (R1648C), and hyperpolarizing shifts in voltage dependence of steady-state inactivation (R1648C) were observed. Indeed, reduced current density is the only prominent defect exhibited by G1749E, which otherwise has near-normal biophysical properties. These findings suggest that some SCN1A mutants cause SMEI by a mixture of gain- and loss-of-function features. Biophysical defects that reduce channel availability may explain why some mutants exhibiting persistent current cause SMEI rather than GEFS+.
In this study we demonstrate that different amino acid substitutions of the same residue may give rise to similar biophysical defects yet be associated with clinical phenotypes of widely divergent severity. Mutation R1648H causes GEFS+, whereas R1648C is associated with SMEI. Why do mutations affecting the same SCN1A residue (Arg-1648) cause such disparity in the associated clinical phenotype? Direct comparisons of these two mutations revealed that R1648C exhibits larger persistent sodium current. Furthermore, differences in other biophysical properties also exist between R1648H and R1648C that may account for distinct clinical outcomes. Specifically, R1648C causes a more pronounced defect in fast inactivation (Fig. 2) than R1648H (20). The less severe inactivation defect observed for R1648H channels may suffice to explain its association with the more benign GEFS+. We did not observe a significant effect of lowering pHi on the behavior of R1648H, but the magnitude of persistent sodium current for R1648C was reduced with internal acidification. The sulfhydryl side chain of cysteine has a pKa of 8.3 and is not expected to be deprotonated to any significant extent in free solution, but it is conceivable that a small proportion of cysteine exists as a thiolate anion within the S4 helix. If this possibility were true, then lowering pHi could convert a greater proportion of Cys-1648 residues to neutral sulfhydryl residues that may affect channel inactivation less than the anionic form. It is also plausible that another titratable group in SCN1A is exposed in the R1648C because of allosteric changes in conformation of the protein.
The disparity in clinical severity between GEFS+ and SMEI probably requires explanations other than just differences in channel behavior. We would like to speculate that the severe neurological consequences of SMEI are caused by a combination of sodium channel dysfunction (either gain or loss of function) with predisposing genetic or developmental factors that lead to a great chance of neuronal injury. In this model, the sodium channel defect creates the initial seizure predisposition, but concomitant excitotoxicity is the direct cause for other neurological features of the disorder. Could certain individuals in the population be especially vulnerable to seizure-induced neuronal injury? The observation that mouse strains differ in susceptibility to hippocampal damage caused by excitotoxic agonists of the ionotropic glutamate receptor supports this idea (34). Perhaps carriers of SCN1A mutations with the greatest susceptibility to neuronal injury present with SMEI or intractable childhood epilepsy with generalized tonic-clonic seizures, whereas less susceptible carriers exhibit a more benign form of epilepsy (i.e., GEFS+) regardless of the specific biophysical properties of the mutant sodium channel. Additional experiments using tractable models of neuronal injury are warranted to investigate this hypothesis.
In conclusion, SCN1A sodium channel mutations associated with SMEI exhibit a broad range of abnormalities including loss- and gain-of-function phenotypes. A complex relationship exists between the biophysical properties of the mutant channel and seizure susceptibility, suggesting that other factors, including genetic and metabolic modifiers, impact on the ultimate clinical expression of the molecular defect. Further analysis of SCN1A mutations is warranted to more firmly establish genotype–phenotype relationships in these inherited epilepsies.
Acknowledgments
We thank Dr. Robert Dumaine for suggesting the pH experiments. This project was supported by National Institutes of Health Grants NS32387 (to A.L.G.) and DK061359 (to C.G.V.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GEFS+, generalized epilepsy with febrile seizures plus; pHi, intracellular pH; SMEI, severe myoclonic epilepsy of infancy; TTX, tetrodotoxin.
References
- 1.Wallace, R. H., Wang, D. W., Singh, R., Scheffer, I. E., George, A. L., Jr., Phillips, H. A., Saar, K., Reis, A., Johnson, E. W., Sutherland, G. R., et al. (1998) Nat. Genet. 19, 366–370. [DOI] [PubMed] [Google Scholar]
- 2.Escayg, A., MacDonald, B. T., Meisler, M. H., Baulac, S., Huberfeld, G., An-Gourfinkel, I., Brice, A., LeGuern, E., Moulard, B., Chaigne, D., et al. (2000) Nat. Genet. 24, 343–345. [DOI] [PubMed] [Google Scholar]
- 3.Claes, L., Del Favero, J., Ceulemans, B., Lagae, L., Van Broeckhoven, C. & De Jonghe, P. (2001) Am. J. Hum. Genet. 68, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugawara, T., Tsurubuchi, Y., Agarwala, K. L., Ito, M., Fukuma, G., Mazaki-Miyazaki, E., Nagafuji, H., Noda, M., Imoto, K., Wada, K., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 6384–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujiwara, T., Sugawara, T., Mazaki-Miyazaki, E., Takahashi, Y., Fukushima, K., Watanabe, M., Hara, K., Morikawa, T., Yagi, K., Yamakawa, K., et al. (2003) Brain 126, 531–546. [DOI] [PubMed] [Google Scholar]
- 6.Heron, S. E., Crossland, K. M., Andermann, E., Phillips, H. A., Hall, A. J., Bleasel, A., Shevell, M., Mercho, S., Seni, M. H., Guiot, M. C., et al. (2002) Lancet 360, 851–852. [DOI] [PubMed] [Google Scholar]
- 7.Wallace, R. H., Scheffer, I. E., Parasivam, G., Barnett, S., Wallace, G. B., Sutherland, G. R., Berkovic, S. F. & Mulley, J. C. (2002) Neurology 58, 1426–1429. [DOI] [PubMed] [Google Scholar]
- 8.Audenaert, D., Claes, L., Ceulemans, B., Lofgren, A., Van Broeckhoven, C. & De Jonghe, P. (2003) Neurology 61, 854–856. [DOI] [PubMed] [Google Scholar]
- 9.Escayg, A., Heils, A., MacDonald, B. T., Haug, K., Sander, T. & Meisler, M. H. (2001) Am. J. Hum. Genet. 68, 866–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abou-Khalil, B., Ge, Q., Desai, R., Ryther, R., Bazyk, A., Bailey, R., Haines, J. L., Sutcliffe, J. S. & George, A. L., Jr. (2001) Neurology 57, 2265–2272. [DOI] [PubMed] [Google Scholar]
- 11.Sugawara, T., Mazaki-Miyazaki, E., Ito, M., Nagafuji, H., Fukuma, G., Mitsudome, A., Wada, K., Kaneko, S., Hirose, S. & Yamakawa, K. (2001) Neurology 57, 703–705. [DOI] [PubMed] [Google Scholar]
- 12.Dravet, C., Bureau, M., Guerrini, R., Giraud, N. & Roger, J. (1992) in Epileptic Syndromes in Infancy, Childhood and Adolescence, eds. Rogers, J., Bureau, M., Dravet, C., Dreifuss, F. E. & Wolf, P. (Libbey, London), pp. 75–88.
- 13.Scheffer, I. E., Wallace, R., Mulley, J. C. & Berkovic, S. F. (2001) Brain Dev. 23, 732–735. [DOI] [PubMed] [Google Scholar]
- 14.Sugawara, T., Mazaki-Miyazaki, E., Fukushima, K., Shimomura, J., Fujiwara, T., Hamano, S., Inoue, Y. & Yamakawa, K. (2002) Neurology 58, 1122–1124. [DOI] [PubMed] [Google Scholar]
- 15.Ohmori, I., Ouchida, M., Ohtsuka, Y., Oka, E. & Shimizu, K. (2002) Biochem. Biophys. Res. Commun. 295, 17–23. [DOI] [PubMed] [Google Scholar]
- 16.Claes, L., Ceulemans, B., Audenaert, D., Smets, K., Lofgren, A., Del Favero, J., Ala-Mello, S., Basel-Vanagaite, L., Plecko, B., Raskin, S., et al. (2003) Hum. Mutat. 21, 615–621. [DOI] [PubMed] [Google Scholar]
- 17.Fukuma, G., Oguni, H., Shirasaka, Y., Watanabe, K., Miyajima, T., Yasumoto, S., Ohfu, M., Inoue, T., Watanachai, A., Kira, R., et al. (2004) Epilepsia 45, 140–148. [DOI] [PubMed] [Google Scholar]
- 18.Lossin, C., Rhodes, T. H., Desai, R. R., Vanoye, C. G., Wang, D., Carniciu, S., Devinsky, O. & George, A. L., Jr. (2003) J. Neurosci. 23, 11289–11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugawara, T., Tsurubuchi, Y., Fujiwara, T., Mazaki-Miyazaki, E., Nagata, K., Montal, M., Inoue, Y. & Yamakawa, K. (2003) Epilepsy Res. 54, 201–207. [DOI] [PubMed] [Google Scholar]
- 20.Lossin, C., Wang, D. W., Rhodes, T. H., Vanoye, C. G. & George, A. L., Jr. (2002) Neuron 34, 877–884. [DOI] [PubMed] [Google Scholar]
- 21.Tomlinson, F. H., Anderson, R. E. & Meyer, F. B. (1993) Epilepsy Res. 14, 123–137. [DOI] [PubMed] [Google Scholar]
- 22.Tomlinson, F. H., Anderson, R. E. & Meyer, F. B. (1992) Epilepsy Res. 13, 49–58. [DOI] [PubMed] [Google Scholar]
- 23.Young, R. S., Osbakken, M. D., Briggs, R. W., Yagel, S. K., Rice, D. W. & Goldberg, S. (1985) Ann. Neurol. 18, 14–20. [DOI] [PubMed] [Google Scholar]
- 24.Yang, N. B., George, A. L., Jr., & Horn, R. (1996) Neuron 16, 113–122. [DOI] [PubMed] [Google Scholar]
- 25.Baulac, S., Huberfeld, G., Gourfinkel-An, I., Mitropoulou, G., Beranger, A., Prud'homme, J. F., Baulac, M., Brice, A., Bruzzone, R. & LeGuern, E. (2001) Nat. Genet. 28, 46–48. [DOI] [PubMed] [Google Scholar]
- 26.Wallace, R. H., Marini, C., Petrou, S., Harkin, L. A., Bowser, D. N., Panchal, R. G., Williams, D. A., Sutherland, G. R., Mulley, J. C., Scheffer, I. E., et al. (2001) Nat. Genet. 28, 49–52. [DOI] [PubMed] [Google Scholar]
- 27.Ohmori, I., Ohtsuka, Y., Ouchida, M., Ogino, T., Maniwa, S., Shimizu, K. & Oka, E. (2003) Brain Dev. 25, 488–493. [DOI] [PubMed] [Google Scholar]
- 28.Singh, R., Andermann, E., Whitehouse, W. P., Harvey, A. S., Keene, D. L., Seni, M. H., Crossland, K. M., Andermann, F., Berkovic, S. F. & Scheffer, I. E. (2001) Epilepsia 42, 837–844. [DOI] [PubMed] [Google Scholar]
- 29.Spampanato, J., Escayg, A., Meisler, M. H. & Goldin, A. L. (2003) Neuroscience 116, 37–48. [DOI] [PubMed] [Google Scholar]
- 30.Spampanato, J., Escayg, A., Meisler, M. H. & Goldin, A. L. (2001) J. Neurosci. 21, 7481–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alekov, A., Rahman, M. M., Mitrovic, N., Lehmann-Horn, F. & Lerche, H. (2000) J. Physiol. (London) 529, 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cossette, P., Loukas, A., Lafreniere, R. G., Rochefort, D., Harvey-Girard, E., Ragsdale, D. S., Dunn, R. J. & Rouleau, G. A. (2003) Epilepsy Res. 53, 107–117. [DOI] [PubMed] [Google Scholar]
- 33.Alekov, A. K., Rahman, M. M., Mitrovic, N., Lehmann-Horn, F. & Lerche, H. (2001) Eur. J. Neurosci. 13, 2171–2176. [DOI] [PubMed] [Google Scholar]
- 34.Schauwecker, P. E. (2002) Exp. Neurol. 178, 219–235. [DOI] [PubMed] [Google Scholar]