Abstract
Heme is a common factor linking several metabolic perturbations in Alzheimer's disease (AD), including iron metabolism, mitochondrial complex IV, heme oxygenase, and bilirubin. Therefore, we determined whether heme metabolism was altered in temporal lobes obtained at autopsy from AD patients and age-matched nondemented subjects. AD brain demonstrated 2.5-fold more heme-b (P < 0.01) and 26% less heme-a (P = 0.16) compared with controls, resulting in a highly significant 2.9-fold decrease in heme-a/heme-b ratio (P < 0.001). Moreover, the strong Pearson correlation between heme-a and heme-b measured in control individuals (r2 = 0.66, P < 0.002, n = 11) was abolished in AD subjects (r2 = 0.076, P = 0.39, n = 12). The level of ferrochelatase (which makes heme-b in the mitochondrial matrix) in AD subjects was 4.2 times (P < 0.04) that in nondemented controls, suggesting up-regulated heme synthesis. To look for a possible connection between these observations and established mechanisms in AD pathology, we examined possible interactions between amyloid β (Aβ) and heme. Aβ(1–40) and Aβ(1–42) induced a redshift of 15–20 nm in the spectrum of heme-b and heme-a, suggesting that heme binds Aβ, likely to one or more of the histidine residues. Lastly, in a tissue culture model, we found that clioquinol, a metal chelator in clinical trials for AD therapy, decreased intracellular heme. In light of these observations, we have proposed a model of AD pathobiology in which intracellular Aβ complexes with free heme, thereby decreasing its bioavailability (e.g., heme-a) and resulting in functional heme deficiency. The model integrates disparate observations, including Aβ, mitochondrial dysfunction, cholesterol, and the proposed efficacy of clioquinol.
Keywords: mitochondria, heme-a, iron, clioquinol, ferrochelatase
Heme (ferriprotoporphyrin IX) metabolism appears altered in the brains of Alzheimer's disease (AD) patients. Heme oxygenase (HO) increases in AD (1, 2), and the level of bilirubin (one of the products of heme degradation by HO) is increased in AD patients (3). Mitochondrial complex IV, the only enzyme in cells that contains heme-a (see below) (4), declines in AD (5–8). Heme-a is rate limiting for the assembly of complex IV (9). Furthermore, an inhibitor of muscarinic acetylcholine receptor binding, which increased in AD brain, was suggested to be heme (10, 11). Therefore, we studied heme metabolism in AD brain and nondemented age-matched normal subjects.
Heme-b, the product of ferrochelatase (FC) (also known as protoheme), is produced in the mitochondrial matrix (12, 13) and is the precursor for heme-c and heme-a. The structure of heme-c is similar to that of heme-b, but it is covalently attached to a few specific proteins (reviewed in ref. 14). Heme-b and heme-a exist in two major pools in the cell, free and protein-associated. Conversion of heme-b to heme-a requires farnasylation and oxidation (15). Farnesyl-pyrophsphate (FPP) is the precursor for the farnesyl moiety in heme-a, cholesterol, dolichol, farnesylation of specific proteins, and coenzyme Q (16). Thus, the heme-a maturation pathway appears to compete with several metabolic pathways for the FPP.
Physiologically relevant metals, such as iron, zinc, or copper, can accelerate amyloid β (Aβ) aggregation in vitro (17). Clioquinol, which chelates zinc, copper, and iron (all of these metals are essential for heme synthesis; see Discussion), decreases the incidence of senile plaques in a mouse model transgenic for amyloid precursor protein (APP) (18) and slows cognitive decline in severe cases of AD (19). It has been suggested that clioquinol inhibits Aβ aggregation in vitro and in vivo (17, 18).
The three histidine residues in Aβ can potentially bind heme. Heme was shown to inhibit in vitro aggregation of Aβ(1–40) and Aβ(1–42) and protected neuronal cells against Aβ's toxicity (20). Additionally, APP inhibits HO (21). Heme deficiency induced by inhibition of FC causes dimerization of APP (22), selective decrease in heme-a, and therefore, loss of complex IV (23). These observations suggest that heme metabolism can influence both APP and Aβ. Furthermore, a potential heme regulatory motif (24) appears in the C-terminal sequence of APP.
In this study we measured the levels of heme-b and heme-a, and their ratio, in addition to FC in frozen temporal lobes of nondemented age-matched normal and AD subjects. We found an AD-dependent change in heme metabolism. Furthermore, clioquinol inhibited heme synthesis in cultivated cells. We found that Aβ binds heme-b and heme-a, which may be relevant for Aβ aggregation. Aβ binding heme may limit the bioavailability of heme and induce a condition of heme deficiency. The possible relevance of these results to AD pathology, cholesterol, and the therapeutic use of clioquinol is discussed.
Materials and Methods
Materials. Materials include 5-chloro-7-iodo-8-hydroxy-quinolone (clioquinol), Aβ(1–40) (amide form), Aβ(1–40), Aβ(1–42), protease inhibitors, and EDTA (Sigma). Rabbit anti-human FC (catalogue no. AB-FC-1) was from EnVirtue Biotechnologies (Winchester, VA). Rabbit polyclonal anti-human Hb (ab2708–500) was from Abcam (Cambridgeshire, U.K.). Heme-a was a gift from E. Hegg (University of Utah, Salt Lake City), and heme-b was purchased from Frontier Scientific (Logan, UT). Triflouroacetic acid, acetonitrile, acetone, and hydrochloric acid were HPLC grade from Aldrich. Gel reagents and other chemicals were the highest grade of purity available.
Source of Tissue. Temporal lobes from AD patients and age-matched normal controls were from the Alzheimer's Research Center of the HealthPartners Research Foundation (provided by W.H.F.). Children's Hospital Oakland Research Institute approved the use of human tissue.
HPLC Analysis of Heme. Frozen brain sections from the temporal lobe (≈5 g) were homogenized in 20 ml of ice-cold PBS/1 mM EDTA/0.5% Tween 20/protease inhibitors and stored in 1-ml aliquots at –80°C. Heme was extracted from 40 μl of homogenate (or volume equivalent to 500 μg of protein) with 300 μlof HCl/acetone (mixing 250 μl of HCl into 10 ml of acetone) (25). Samples were vortexed and incubated at ice for 2 min, 300 μl of acetonitrile was added with additional vortexing, and the pH was adjusted to ≈5.5 by the addition of 30 μl of 1 M Hepes (pH 7). This method extracts heme-b and heme-a but not heme-c, which is covalently bound to proteins. The extract was then centrifuged at 14,000 × g for 2 min to remove proteins, and 50 μl of the top organic phase was taken for analysis by reversed-phase HPLC on a column of Bond-Clone-C18, 300 × 3.9 mm (Phenomenex, Belmont, CA) with two mobile phases containing 30% (A) and 70% (B) acetonitrile in 0.08% trifluoroacetic acid. Each cycle of separation was as follows: wash with 100% buffer A for 2 min, within 5 min reach 100% buffer B, wash with buffer B until 20 min, and then return to buffer A within 20 min. Elution of heme-b (12–13 min) and heme-a (16–17 min) was monitored at 400 nm.
Determining Hb and FC by Western Blotting. Autopsies are contaminated with Hb, which contains heme-b, leading to overestimation of total heme in the tissue. Therefore, Hb in the homogenate was quantified with an antibody specific for human Hb (Abcam) and standard human Hb. Samples (60 μg of protein per well) and Hb standard (500 and 1,000 ng of Hb per well equivalent to 20 and 40 ng of heme-b per well) were simultaneously separated on 12% SDS/PAGE, immunobloted, and quantified by densitometry. Heme contributed by Hb in each sample was subtracted from the total heme content measured by HPLC to give the intrinsic heme content of the brain tissue. FC was determined by Western blotting (23).
Experiments on Clioquinol. Clioquinol was freshly prepared in 0.1 M NaOH, sterilized, and used immediately to treat the cells. Human lung normal fibroblasts (IMR90) or astrocytoma (U373) were treated with 10 μM clioquinol for a week. The cells were then washed with 5 ml of ice-cold PBS, scraped off the plate into 2 ml of PBS, and palleted by centrifugation. Heme content of the cells was analyzed with HPLC and normalized for protein content. The excitation coefficient of heme-a is lower than that of heme-b, which limits the detection of heme-a in the cultured cells.
Binding of Aβ to Heme-b and Heme-a. Interaction of heme with Aβ was tested in 50 mM Hepes (pH 7.5). Aβ was prepared in double-distilled water at 0.6 mM (or at 1 mM for the amide form) final concentrations and stored at –20°C as recommended by the manufacturer. Aβ(1–42) was prepared in 0.1% trifluoroacetic acid at a final concentration of 0.22 mM. Heme-b was freshly prepared in 0.1 M NaOH and stored in the dark on ice. Heme-a was dissolved in acetonitrile/0.1% trifluoroacetic acid. The final volume of the reaction was 200 μl, and it was prepared in a 96-well plate. The final concentrations of heme-b and Aβ were 50 μM. It was difficult to achieve high concentrations of heme-a; therefore, it was tested at a concentration of 30 μM. The spectrum of heme, Aβ, and Aβ+heme was immediately measured between 350–750 nm at room temperature by using SpectraMAX 340 plate reader (Molecular Devices). The possible effect of zinc or copper on the binding of heme by Aβ was tested as described above by using a 50 and 100 μM final concentration of each metal. The metals were preincubated with Aβ for 5 min when added before heme. Heme was preincubated for 5 min with Aβ when added before the metals. The source for Zn2+ was ZnCl2, and the source for Cu2+ was CuSO4.
Statistical Analysis. Data are presented as the mean ± SEM unless otherwise stated. Graphing, regression, and statistical analyses were performed with prism 4.0 (GraphPad, San Diego). Differences are considered significant at P < 0.05.
Results
The mean age of AD patients was 77 ± 10 years (n = 12), whereas the mean age of the control group was 71 ± 11 years (n = 11). The difference in mean age between the groups was not statistically significant (P = 0.2). The mean postmortem delay interval was 12 ± 8 h (n = 12) in the AD group, whereas the mean postmortem delay interval was 16 ± 8 h (n = 11) in the control group. The difference in postmortem delay interval between the groups was not statistically significant (P = 0.2). Maurer et al. (5) demonstrated postmortem delay interval of up to 38 h was without influence on the activity of cytochrome c oxidase (complex IV) or complex I of the temporal cortex. Both genders were equally represented in each group.
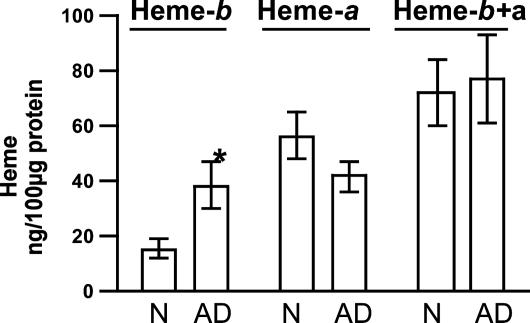
Content of Heme-b and Heme-a, and the Ratio of Heme-a/Heme-b in the Temporal Lobe from AD Patients Compared with Age-Matched Nondemented Controls. Acid/acetone extracts all forms of heme, and thus the data presented here cannot discriminate between free and protein-bound heme-b (or heme-a). Heme-b in the temporal lobe of nondemented control subjects was 15.8 ± 3.8 ng/100 μg of total protein (or 0.24 ± 0.06 nmol/mg of total protein), whereas heme-b content in AD patients was 38.93 ± 8.5 ng/100 μg of total protein (or 0.6 ± 0.13 nmol/mg of total protein) (Fig. 1). Thus, heme-b content increased in the temporal lobe of AD patients by 2.5-fold when compared with the age-matched controls (P < 0.01).
Fig. 1.
Levels of heme-b and heme-a in the temporal lobe of nondemented control and AD subjects. In the temporal lobe from AD patients, heme-b increased 2.5-fold, whereas heme-a shows a moderate decline of 26% (did not reach significance). Heme-b and heme-a were determined as described in Materials and Methods. Data for heme-b are mean ± SEM (nonparametric Mann–Whitney test, P < 0.01, n = 11 for control and n = 12 for AD subjects). Data are for heme-a are mean ± SEM (P = 0.16 Student's t test, n = 11 and 12 for control and AD subjects, respectively).
Heme-a content in the brain of nondemented control subjects was 57 ± 9 ng/100 μg of total protein (or 0.64 ± 0.1 nmol/mg of total protein). Heme-a content in the brain of AD patients was 42 ± 6 ng/100 μg total of protein (or 0.47 ± 0.07 nmol/mg of protein), which is 26% less than age-matched nondemented controls (Fig. 1). The decline in heme-a was not statistically significant (P = 0.16).
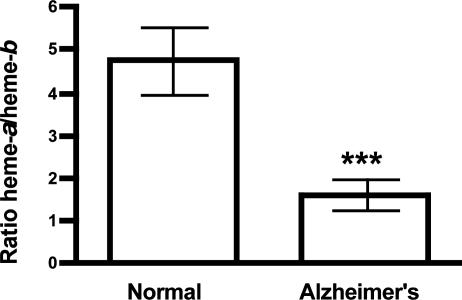
Total heme (heme-b and heme-a) did not change between both groups (Fig. 1). The ratio of heme-a/heme-b showed a 2.9-fold decrease in AD patients compared with nondemented controls (P < 0.001) (Fig. 2). The heme-a/heme-b ratio in normal subjects is ≈5 and ≈1.5 in AD patients.
Fig. 2.
The ratio heme-a/heme-b ratio in the temporal lobe of nondemented control and AD subjects. In the temporal lobe of AD patients the ratio heme-a/heme-b ratio decreased 2.9-fold compared with controls. The ratio heme-a/heme-b ratio was calculated from the data in Fig. 1. Data are mean ± SEM (nonparametric Mann–Whitney test, P < 0.001, n = 11 and 12 for control and AD subjects, rspectively).
Blood is a poor source of heme-a, as it exists only in the white blood cells and platelets, which are a negligible fraction of the total blood cells that usually contaminate brain autopsy tissue. Therefore, heme-a measured here is intrinsic to the brain tissue.
The Level of FC Increases in the Temporal Lobe in AD Patients. The level of the enzyme FC increased 4.2-fold (P < 0.04) in the temporal lobe of AD brain compared with nondemented controls (Fig. 3). FC in brain tissue of normal subjects was very low, except in 1 of 11 cases tested. FC in AD brains, however, was elevated in most of the cases. Protein density of FC in the gel in AD brains and age-matched controls, on average, was 70,080 ± 23,310 and 16,820 ± 8,017 (n = 11, P < 0.04), respectively. As noted previously for heme-a, blood contaminates autopsies and is a poor source of FC. Therefore, FC measured here is intrinsic to the brain tissue.
Fig. 3.
Levels of FC in the temporal lobe of nondemented control and AD subjects. Levels of FC increased in the temporal lobe in most of the AD subjects. A representative Western blot analysis for FC is shown. Numbers on the gel refer to coding of subjects. Protein density in FC band in the gel from AD and age-matched control subjects on average were 70,080 ± 23,310 and 16,820 ± 8,017 (n = 10, P < 0.04), respectively (mean ± SEM, nonparametric Mann–Whitney test, P < 0.04, n = 11 and 12 for control and AD subjects, respectively). FC determination was as described in Materials and Methods.
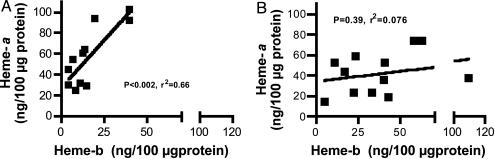
Positive Correlation Between Heme-a and Heme-b in Normal but Not in AD Subjects. A strong positive correlation between the content of heme-b and heme-a is seen in the temporal lobe from normal subjects (Fig. 4A)(P < 0.002, r2 = 0.66). The correlation between the content of heme-b and heme-a was weaker in AD subjects and did not reach statistical significance (Fig. 4B) (P = 0.39, r2 = –0.076).
Fig. 4.
Correlative analysis of heme-a vs. heme-b in the temporal lobe of nondemented control and AD subjects. A strong Pearson correlation between heme-a and heme-b in control (A) was abolished in AD (B) subjects. (A) A significant positive correlation between heme-a and heme-b is seen in the temporal lobe of normal subjects (P < 0.002, r2 = 0.66, n = 11). (B) The correlation between heme-a and heme-b in the temporal lobe of AD subjects was not significant (P < 0.39, r2 = 0.076, n = 12).
No correlation was found between the level of heme-b in the blood and the level of intrinsic heme-b in the tissue (P = 0.83, r2 = 0.002) (data not shown). The lack of correlation suggests that blood in the brain autopsy did not account for the increase in heme content in the temporal lobe in the AD subjects.
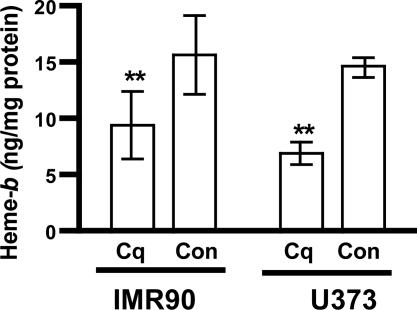
Clioquinol Inhibits Heme Synthesis in Human Normal Fibroblasts and Human Astrocytoma. Clioquinol chelation of zinc suggests that it may inhibit heme synthesis by inhibiting δ-aminolevulinic acid synthase, which is essential for heme synthesis. Clioquinol (10 μM) decreased heme content in human astrocytoma (U373) cells by ≈50% after 1 week of incubation (Fig. 5). Clioquinol also reduces heme in normal human fibroblasts (Fig. 5).
Fig. 5.
Clioquinol decreases heme in normal human fibroblasts (IMR90) and in human astrocytoma (U373) cells. Clioquinol (Cq) lowers heme in IMR90 and U373 cells. The cells were incubated with 10 μM Cq for a week. Heme was analyzed as described in Materials and Methods. One representative experiment of each cell type is shown (U373, Student's t test, P < 0.005; IMR90, Student's paired t test, P < 0.006). Con, control.
Clioquinol was toxic at low concentrations (0.1–1 μM) after longtime exposure (2 months), whereas doses higher than 5 μM applied for 1–2 weeks were mildly toxic (data not shown). No direct interaction between clioquinol and heme was observed based on lack of spectral change of heme in the presence of equimolar concentration of clioquinol at physiologic pH (data not shown). Decrease in heme is likely to contribute to clioquinol toxicity.
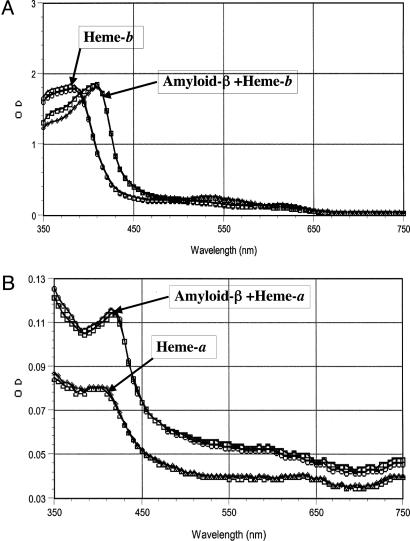
Heme-b and Heme-a Bind to Aβ and Prevent Zinc Binding. Mixing heme-b (or heme-a) with Aβ at molar ratios of 1:1 (Aβ(1–40) amide) or 1:3 (Aβ(1–40), Aβ(1–42)) lead to redshift in heme (≈20 nm to heme-b or ≈15 nm to heme-a, Fig. 6 A and B). The difference in the heme/Aβ molar ratios may be caused by fewer monomers in the preparation of the nonamidic Aβ forms, suggesting that heme binds to the monomer of Aβ. The redshift in the heme spectrum indicates that the iron moiety of heme is involved in the binding to Aβ. Histidine is the most likely amino acid to mediate this interaction. The interaction between Aβ and heme was instantaneous and occurred at room temperature. Zinc and copper ions, which are known to bind histidines in Aβ and induce its aggregation, were able to inhibit the formation of the complex heme–Aβ only if they were added to Aβ before the addition of heme. Neither metal ion had any affect on a preformed heme–Aβ complex (Table 1). Heme-a may bind to Aβ with even higher affinity than does heme-b because of its hydrophobic farnesyl moiety, which may bind to the hydrophobic regions of Aβ, causing an increase in absorbance at the visible range of the spectrum of heme-a–Aβ complex. A possible difference between the two types of heme complex with Aβ (i.e., heme-b–Aβ vs. heme-a–Aβ) has not been ruled out.
Fig. 6.
Spectral redshift to heme induced by Aβ.Aβ(1–40) or Aβ(1–42) induces a redshift of 18 and 15 nm in heme-b and heme-a, respectively. The spectral change of heme-b (A) or heme-a (B) induced by Aβ were determined as described in Materials and Methods.Aβ was used at a final concentration of 50 μM. The concentration of heme-b was 50 μM and 30 μM for heme-a. All chemicals were mixed in 50 mM Hepes (pH 7), and spectra were collected immediately at room temperature by using a SpectraMAX 340 plate reader.
Table 1. Summary of the redshift to heme caused by Aβ and the effect of zinc and copper.
| Heme-b, 50 μM
|
Aβ(1-40) (50 μM) added relative to metals
|
Zn2+, 100 μM
|
Cu2+, 100 μM
|
Redshift in heme spectrum
|
|
|---|---|---|---|---|---|
| Before | After | ||||
| + | + | - | - | - | 20 nm |
| + | + | - | + | - | 20 nm |
| + | + | - | - | + | 20 nm |
| + | + | - | + | + | 20 nm |
| + | - | + | + | - | Weak |
| + | - | + | - | + | Weak |
| + | - | + | + | + | Weak |
+, reagent included in the mixture. -, reagent was not included in the mixture. Zn2+, ZnCl2; Cu2+, CuSO4.
Discussion
We hypothesized that altered heme metabolism is central to many of the disparate observations in AD brain (22, 23); therefore, we studied heme in autopsies of the temporal lobe of AD patients. The increase in heme-b demonstrated in the temporal lobe of AD patients is a result of an intrinsic synthesis of heme in brain cells and not contamination from the heme in the blood. This conclusion is supported by the increase in FC in AD compared with normal subjects, the differential change in heme metabolism between AD and normal subjects, and the lack of correlation between heme-b in Hb and heme-b in the brain tissue (data not shown).
Because of technical difficulties in assaying heme-c, this study focused specifically on heme-b and heme-a. Heme-a decreased by 26% (not significant), whereas heme-b increased 250% in the temporal lobe of AD subjects. Lack of correlation between heme-a and heme-b, in conjunction with decline in the heme-a/heme-b ratio in AD subjects, suggests low efficiency of the heme-a maturation pathway. Interestingly, Parker et al. (7) have also demonstrated a 25% AD-dependent decline in cytochrome-a (which is in complex IV and the functional form of heme-a) in isolated mitochondria from whole brain. Therefore, the increase in heme-b synthesis could be a compensatory response to the decline in availability of heme-a. This result may explain, in part, the moderate but consistent decrease in heme-a.
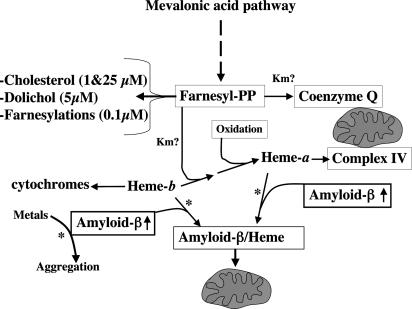
Production of heme-a is influenced by the availability of not only heme-b but also FPP moieties. FPP moieties produced by the mevalonate pathway is the precursor for cholesterol and various isoprenoids (16), including heme-a (Scheme 1). Therefore, a tight regulation of cholesterolgenesis is critical for maintaining levels of FPP high enough to allow synthesis of essential nonsterol isoprenoids (26, 27). Diversion of FPP into the noncholesterol isoprenoids products is also facilitated by the high affinity for FPP of the initial enzymes in these pathways relative to that of squalene synthetase (26). Statins are drugs used to regulate cholesterolgenesis in individuals with hypercholesterolemia, a disorder that is likely to overdraw much FPP for cholesterol production. Vulnerable neurons in AD brain were shown to contain levels of free cholesterol that were higher than levels in healthy neurons (28), and cholesterol accumulates in the senile plaques (29), suggesting oversynthesis of cholesterol. Statins were associated with lower risk for AD if given at middle age, although the exact mechanism of their action is not yet clear (30, 31). Another possible explanation for the effect of statins is regulating the synthesis of cholesterol while preserving the FPP group for nonsterol isoprenoids such as heme-a. However, it is important to emphasize that statins do not reach a high enough concentration in the brain to completely inhibit hydroxymethylglutaryl-CoA reductase; otherwise, cell death could occur (31). In the light of the complexity of the metabolism of cholesterol and its influence on the pool of FPP, more research on statins and nonisoprenoids in AD is required to understand the influence of statins on metabolism beyond cholesterol.
Scheme 1.
Possible metabolic consequences of heme-binding Aβ. Intracellular Aβ aggregation may be prevented by binding to free heme-b and heme-a, although a decline in their free pools could be a possible side effect. Increased cholesterolgenesis may compete with heme-a maturation on FPP (27), causing even more decrease in heme-a. Low heme-a limits assembly of complex IV and causes up-regulation of heme-b synthesis. If the rate of production of Aβ exceeds the capacity of heme synthesis or there is an increase in free metals, then excess Aβ reacts with free metals and forms aggregates (see Discussion). The Km values for FPP in FPP-dependent metabolic pathways are in parentheses.?, Km not determined; ↑, increased rate of production of Aβ; *, pathways not demonstrated in vivo.
Low intracellular levels of heme-a decrease the rate of assembly of new complex IV, keeping a low steady-state level of the complex (23). Decrease of only complex IV among the mitochondrial electron transport chain increases the release of free radicals by the mitochondria (23, 32). Deficiency for complex IV, which is the case in AD (5, 7), is likely to increase the production of free radicals in the mitochondrial matrix (33). Up-regulation of heme-b biosynthesis in the mitochondrial matrix could lead to elevated heme-b in this compartment (13). High levels of heme-b, whatever the possible metabolic advantage, are associated with a risk of oxidative damage, as heme is a prooxidant (34). Thus, excess heme in the mitochondrial matrix may exacerbate oxidative stress and accelerate the decay of the mitochondria that is seen in AD. The possible role of heme in oxidative damage in AD is suggested in refs. 10 and 11. HO, which in the endoplasmic reticulum degrades heme to bilirubin, is also up-regulated in AD patients (1–3), suggesting that heme should remain under control. This result may explain in part the lack of difference in total heme.
Previously, Howlett et al. (20) demonstrated that heme prevented aggregation of Aβ(1–40) and Aβ(1–42), suggesting that heme may interact with Aβ. Our data support that heme binds to the monomer form of Aβ. The heme–Aβ complex could be a new form of Aβ that may exist in AD brain in vivo. One (or more) of the hisitdines in the amino acid sequence of Aβ is likely to be the site for binding heme. The histidines in Aβ also bind to zinc, copper, or iron ions, causing its aggregation; therefore, binding heme to Aβ may prevent the aggregation (20). We found that zinc or copper ions did not affect the Aβ–heme complex at concentrations double that of heme (Table 1). Although the intracellular concentration of free heme is not known, it is conceivable that it reaches concentrations high enough to compete with the free forms of these metals for binding Aβ (Scheme 1). Nucleated cells contentiously produce heme, and heme synthesis can be up-regulated. The concentration of the metal ions in glial cells is estimated between 20 and 200 μM (35), whereas in neurons the intracellular zinc is largely localized to synaptic vesicles (36). The concentration of the free form of these metals is estimated in the low nM range for zinc (37) even lower for copper (38), and ≈200 nM for free iron (39). We hypothesize that heme binding to the monomer form of Aβ peptides may change the conformation of the peptide, its hydrophobicity, or mask the site that binds free metals (i.e., histidine), thus, inhibiting Aβ oligomerization (20) and keeping intracellular Aβ in monomer form (Scheme 1). Intracellular accumulation of Aβ has been hypothesized to form prefibrillar species (e.g., oligomers) (40), which are resistant to proteolysis. Recent research on AD implicates intracellular prefibrillar species of Aβ, rather than the senile plaques or monomers, in neurotoxicity and neuronal dysfunction of AD (41–43). Heme–Aβ complex could be safely turned over by cellular proteases (44) and by HO, which degrades the heme moiety.
We believe that largely the monomers of Aβ binds heme in the cytoslic pool of free heme because Aβ is mainly produced in the cytosol. The mitochondrial pool of free heme is unlikely to play a major role in binding Aβ, as there is no evidence that Aβ accumulates in the mitochondrial matrix. Binding of heme to Aβ could increase because of elevated production of Aβ or to an increase in the steady-state concentration of metals, which otherwise would bind to Aβ (e.g., zinc and copper). Binding of Aβ to heme may trap free heme-b and heme-a, creating a condition of functional deficiency of heme, consequently contributing to the decrease in complex IV (Scheme 1). We have demonstrated that whenever heme deficiency occurs, cells selectively lose heme-a, and as a result, mitochondrial complex IV (22, 23). Thus, increased heme synthesis might be a compensation for a decrease in free heme.
Increased production of heme-b may prevent metal-dependent aggregation of Aβ in the short term. As heme production and accumulation persist in the mitochondrial matrix, heme may exacerbate oxidative stress as described above. Therefore, modulation of heme synthesis may benefit AD patients by decreasing oxidative stress in the mitochondria. Clioquinol improved the cognitive function only in severe cases of AD (19). Clioquinol modulates heme synthesis in vitro, probably by chelating zinc (45), an essential ion for heme synthesis (12). If clioquinol modulates heme synthesis in severe AD patients, then it may decrease heme in the mitochondrial matrix, thus ameliorating the oxidative stress. Modulation of heme synthesis by clioquinol should also increase the availability of succinyl-CoA, a tricarboxylic acid cycle intermediate, which is the precursor for heme. Low oxidative stress in mitochondria and increase availability of succinyl-CoA are likely to increase the efficiency of ATP production, which appears to decline in AD (46, 47). ATP is essential for neuronal function and cognitive activity. Clioquinol may also directly dismantle the senile plaques by metal chelation as is suggested in ref. 18, an activity that would increase heme binding to Aβ over metal ions.
Altered heme metabolism may contribute to the metabolic changes in AD brain (48). The consequences of heme binding to Aβ have yet to be identified. As additional data are collected on heme metabolism and isolation of the heme–Aβ complex from human brain, a more complete picture of the role of heme in AD pathology will emerge. If altered heme metabolism precedes AD neurodegeneration, then the level of heme in white blood cells, the ratio heme-a/heme-b, and/or FC may serve as markers for identifying individuals predisposed to AD if these markers reflect changes in heme metabolism in the brain.
Acknowledgments
We thank B. N. Ames, F. Beal, E. C. Theil, A. Butterfield, and M. A. Smith for commenting on the manuscript and Elizabeth Bordayo, Kathleen Boyle, and Guyve Shalileh for technical assistance. This work was supported by the Bruce and Giovanna Ames Foundation and by a National Center on Minority Health and Health Disparities Center pilot grant (to H.A.).
Abbreviations: Aβ, amyloid β; AD, Alzheimer's disease; APP, amyloid precursor protein; FC, ferrochelatase; FPP, farnesyl-pyrophosphate; HO, heme oxygenase.
References
- 1.Schipper, H. M., Cisse, S. & Stopa, E. G. (1995) Ann. Neurol. 37, 758–768. [DOI] [PubMed] [Google Scholar]
- 2.Smith, M. A., Kutty, R. K., Richey, P. L., Yan, S. D., Stern, D., Chader, G. J., Wiggert, B., Petersen, R. B. & Perry, G. (1994) Am. J. Pathol. 145, 42–47. [PMC free article] [PubMed] [Google Scholar]
- 3.Kimpara, T., Takeda, A., Yamaguchi, T., Arai, H., Okita, N., Takase, S., Sasaki, H. & Itoyama, Y. (2000) Neurobiol. Aging 21, 551–554. [DOI] [PubMed] [Google Scholar]
- 4.Steffens, G. C., Biewald, R. & Buse, G. (1987) Eur. J. Biochem. 164, 295–300. [DOI] [PubMed] [Google Scholar]
- 5.Maurer, I., Zierz, S. & Moller, H. J. (2000) Neurobiol. Aging 21, 455–462. [DOI] [PubMed] [Google Scholar]
- 6.Cottrell, D. A., Blakely, E. L., Johnson, M. A., Ince, P. G. & Turnbull, D. M. (2001) Neurology 57, 260–264. [DOI] [PubMed] [Google Scholar]
- 7.Parker, W. D., Jr., Parks, J., Filley, C. M. & Kleinschmidt-DeMasters, B. K. (1994) Neurology 44, 1090–1096. [DOI] [PubMed] [Google Scholar]
- 8.Mutisya, E. M., Bowling, A. C. & Beal, M. F. (1994) J. Neurochem. 63, 2179–2184. [DOI] [PubMed] [Google Scholar]
- 9.Wielburski, A. & Nelson, B. D. (1984) FEBS Lett. 177, 291–294. [DOI] [PubMed] [Google Scholar]
- 10.Fawcett, J. R., Bordayo, E. Z., Jackson, K., Liu, H., Peterson, J., Svitak, A. & Frey, W. H., II (2002) Brain Res. 950, 10–20. [DOI] [PubMed] [Google Scholar]
- 11.Venters, H. D., Jr., Bonilla, L. E., Jensen, T., Garner, H. P., Bordayo, E. Z., Najarian, M. M., Ala, T. A., Mason, R. P. & Frey, W. H., II (1997) Brain Res. 764, 93–100. [DOI] [PubMed] [Google Scholar]
- 12.Ponka, P. (1999) Am. J. Med. Sci. 318, 241–256. [DOI] [PubMed] [Google Scholar]
- 13.Ponka, P., Borova, J. & Neuwirt, J. (1973) Biochim. Biophys. Acta 304, 715–718. [DOI] [PubMed] [Google Scholar]
- 14.Atamna, H., Walter, P. B. & Ames, B. N. (2002) Arch. Biochem. Biophys. 397, 345–353. [DOI] [PubMed] [Google Scholar]
- 15.Mogi, T., Saiki, K. & Anraku, Y. (1994) Mol. Microbiol. 14, 391–398. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein, J. L. & Brown, M. S. (1990) Nature 343, 425–430. [DOI] [PubMed] [Google Scholar]
- 17.Bush, A. I. (2003) Alzheimer Dis. Assoc. Disord. 17, 147–150. [DOI] [PubMed] [Google Scholar]
- 18.Cherny, R. A., Atwood, C. S., Xilinas, M. E., Gray, D. N., Jones, W. D., McLean, C. A., Barnham, K. J., Volitakis, I., Fraser, F. W., Kim, Y., et al. (2001) Neuron 30, 665–676. [DOI] [PubMed] [Google Scholar]
- 19.Ritchie, C. W., Bush, A. I., Mackinnon, A., Macfarlane, S., Mastwyk, M., MacGregor, L., Kiers, L., Cherny, R., Li, Q. X., Tammer, A., et al. (2003) Arch. Neurol. (Chicago) 60, 1685–1691. [DOI] [PubMed] [Google Scholar]
- 20.Howlett, D., Cutler, P., Heales, S. & Camilleri, P. (1997) FEBS Lett. 417, 249–251. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi, M., Dore, S., Ferris, C. D., Tomita, T., Sawa, A., Wolosker, H., Borchelt, D. R., Iwatsubo, T., Kim, S. H., Thinakaran, G., et al. (2000) Neuron 28, 461–473. [DOI] [PubMed] [Google Scholar]
- 22.Atamna, H., Killilea, D. W., Killilea, A. N. & Ames, B. N. (2002) Proc. Natl. Acad. Sci. USA 99, 14807–14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atamna, H., Liu, J. & Ames, B. N. (2001) J. Biol. Chem. 276, 48410–48416. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, L. & Guarente, L. (1995) EMBO J. 14, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown, K. R., Allan, B. M., Do, P. & Hegg, E. L. (2002) Biochemistry 41, 10906–10913. [DOI] [PubMed] [Google Scholar]
- 26.Brown, M. S. & Goldstein, J. L. (1980) J. Lipid Res. 21, 505–517. [PubMed] [Google Scholar]
- 27.Tansey, T. R. & Shechter, I. (2000) Biochim. Biophys. Acta 1529, 49–62. [DOI] [PubMed] [Google Scholar]
- 28.Distl, R., Meske, V. & Ohm, T. G. (2001) Acta Neuropathol. 101, 547–554. [DOI] [PubMed] [Google Scholar]
- 29.Mori, T., Paris, D., Town, T., Rojiani, A. M., Sparks, D. L., Delledonne, A., Crawford, F., Abdullah, L. I., Humphrey, J. A., Dickson, D. W. & Mullan, M. J. (2001) J. Neuropathol. Exp. Neurol. 60, 778–785. [DOI] [PubMed] [Google Scholar]
- 30.Hoglund, K., Wiklund, O., Vanderstichele, H., Eikenberg, O., Vanmechelen, E. & Blennow, K. (2004) Arch. Neurol. (Chicago) 61, 333–337. [DOI] [PubMed] [Google Scholar]
- 31.Meske, V., Albert, F., Richter, D., Schwarze, J. & Ohm, T. G. (2003) Eur. J. Neurosci. 17, 93–102. [DOI] [PubMed] [Google Scholar]
- 32.Sohal, R. S. (1993) Free Radical Biol. Med. 14, 583–588. [DOI] [PubMed] [Google Scholar]
- 33.Han, D., Williams, E. & Cadenas, E. (2001) Biochem. J. 353, 411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner, K. R., Sharp, F. R., Ardizzone, T. D., Lu, A. & Clark, J. F. (2003) J. Cereb. Blood Flow Metab. 23, 629–652. [DOI] [PubMed] [Google Scholar]
- 35.Tholey, G., Ledig, M., Mandel, P., Sargentini, L., Frivold, A. H., Leroy, M., Grippo, A. A. & Wedler, F. C. (1988) Neurochem. Res. 13, 45–50. [DOI] [PubMed] [Google Scholar]
- 36.Perez-Clausell, J. & Danscher, G. (1985) Brain Res. 337, 91–98. [DOI] [PubMed] [Google Scholar]
- 37.Canzoniero, L. M., Turetsky, D. M. & Choi, D. W. (1999) J. Neurosci. 19, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rae, T. D., Schmidt, P. J., Pufahl, R. A., Culotta, V. C. & O'Halloran, T. V. (1999) Science 284, 805–808. [DOI] [PubMed] [Google Scholar]
- 39.Konijn, A. M., Glickstein, H., Vaisman, B., Meyron-Holtz, E. G., Slotki, I. N. & Cabantchik, Z. I. (1999) Blood 94, 2128–2134. [PubMed] [Google Scholar]
- 40.Walsh, D. M., Tseng, B. P., Rydel, R. E., Podlisny, M. B. & Selkoe, D. J. (2000) Biochemistry 39, 10831–10839. [DOI] [PubMed] [Google Scholar]
- 41.Gong, Y., Chang, L., Viola, K. L., Lacor, P. N., Lambert, M. P., Finch, C. E., Krafft, G. A. & Klein, W. L. (2003) Proc. Natl. Acad. Sci. USA 100, 10417–10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selkoe, D. J. (2002) Science 298, 789–791. [DOI] [PubMed] [Google Scholar]
- 43.Mucke, L., Masliah, E., Yu, G. Q., Mallory, M., Rockenstein, E. M., Tatsuno, G., Hu, K., Kholodenko, D., Johnson-Wood, K. & McConlogue, L. (2000) J. Neurosci. 20, 4050–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leissring, M. A., Farris, W., Chang, A. Y., Walsh, D. M., Wu, X., Sun, X., Frosch, M. P. & Selkoe, D. J. (2003) Neuron 40, 1087–1093. [DOI] [PubMed] [Google Scholar]
- 45.Bush, A. I. (2002) Neurobiol. Aging 23, 1031–1038. [DOI] [PubMed] [Google Scholar]
- 46.Blass, J. P. (2001) J. Neurosci. Res. 66, 851–856. [DOI] [PubMed] [Google Scholar]
- 47.Valla, J., Berndt, J. D. & Gonzalez-Lima, F. (2001) J. Neurosci. 21, 4923–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perry, G., Nunomura, A., Raina, A. K., Aliev, G., Siedlak, S. L., Harris, P. L., Casadesus, G., Petersen, R. B., Bligh-Glover, W., Balraj, E., et al. (2003) Neurochem. Res. 28, 1549–1552. [DOI] [PubMed] [Google Scholar]