Abstract
Sphingosine 1-phosphate (S1P) is a sphingolipid mediator that is involved in diverse biological functions. Local administration of S1P causes inflammation coupled to a large eosinophil (EO) recruitment in the rat-paw tissue. The inflammatory response is accompanied by an increase in S1P receptors, namely S1P1, S1P2, S1P3, and by an enhanced expression of CCR3, which is the main chemokine receptor known to be involved in EO function. Human EOs constitutively express S1P1 and, at a lower extent, S1P2, S1P3 receptors. S1P in vitro causes cultured human EO migration and an increase in S1P receptor mRNA copies and strongly up-regulates CCR3 and RANTES (regulated on activation, normal T cell-expressed and secreted) message levels; in particular CCR3 is up-regulated 18,000-fold by S1P. A blocking anti-CCR3 Ab inhibits S1P-induced chemotaxis, implying that S1P acts as specific recruiting signal for EOs not only through its own receptors but also through CCR3. These results show that S1P is involved in EO chemotaxis and contribute to shed light on the complex mechanisms underlying EO recruitment in several diseases such as asthma and some malignancies.
Keywords: CCR3, RANTES, eosinophil migration, inflammation
Sphingolipids are ubiquitously expressed in all eukaryotic cell membranes and much attention has been focused in this past decade on the role of the sphingolipid metabolites, ceramide, sphingosine, and in particular on its phosphorylated product sphingosine-1-phosphate (S1P). S1P is a lipid mediator that has been shown to act inside cells to regulate survival. The cloning and the characterization of different receptor isoforms, together with the finding that S1P is secreted by the platelets (1), has strengthened the notion that S1P acts also as an extracellular signaling molecule. It is now well established that S1P is the natural ligand for a family of G protein-coupled receptors originally known as the endothelial differentiation genes and recently renamed, S1P receptors (2) (S1PRs). To date, it is known that there are five specific receptors for S1P, that are S1P1, S1P2, S1P3, S1P4, and S1P5 (2). These receptors can couple to different G-proteins to elicit a wide array of cellular responses (3, 4) and modulate calcium release. S1P can activate monocytes (5, 6) endothelial cells (7, 8), mast cells (9, 10), and asthmatic smooth muscle cells (11) as well as mediate lymphocyte infiltration (12).
Sphingosine is converted to S1P by the enzyme sphingosine kinase (SphK). SphK is expressed both in humans and animals and two mammalian isozymes have been characterized as SphK1 and Sphk2. This enzyme is mainly located in the cytosol and, by using adenosine triphosphate as phosphate donor, catalyzes the phosphorylation of d-erythrosphingosine to S1P. Many external stimuli activate the enzyme SphK to generate S1P in particular cytokines such as tumor necrosis factor α (TNF-α) (13) and IL-1β (14) and cross-linking of the Ig receptor FcεRI (15). Thus, it has been proposed that S1P is not only involved in disorders such as cancer (16, 17) and atherosclerosis (18), but it is also in inflammation (6, 19). There are many studies that have indirectly suggested a role of S1P in inflammation, but at the present stage, studies mainly address the role of this mediator in vitro by using cells or isolated tissues. Here, we have determined whether exogenous S1P, in a pathophysiologically relevant concentration, can cause inflammation in vivo in rats. Administration of exogenous S1P causes an edema that is fast in onset and accompanied by a large eosinophil (EO) infiltrate. Similarly, i.p. administration of S1P rapidly recruits EOs into the peritoneal cavity. The edema formation is accompanied by a selective increase in the rat-paw tissue of CCR3. In vitro experiments performed by using highly purified human EOs demonstrated that EOs constitutively express higher mRNA levels of S1P1 and, to a lower extent, S1P2 and S1P3. Challenging of EOs in vitro with S1P causes chemotaxis coupled with a strong up-regulation of CCR3 and RANTES (regulated on activation, normal T cell-expressed and secreted) message. These data imply that, in inflammatory events in which S1P is released, EOs could be involved in an immune response that is most likely of the Th2 type.
Materials and Methods
EO Purification and Flow Cytometry Analysis. Blood samples were obtained from subjects who underwent clinical examination at the Allergy and Clinical Immunology Section at the Seconda Università di Napoli. All of the patients were informed fully of the study and gave written consensus to the procedure. EOs were isolated from the blood of patients as described (20), with minor modifications, by using immunomagnetic beads and the magnetic cell separation system (MACS). Diluted whole blood (1:5 dilution) was layered onto a Percoll gradient (d = 1.088 g/liter) and centrifuged at 400 × g for 30 min. The granulocyte pellet, enriched mainly in neutrophils and EOs, was harvested and depleted of erythrocytes by hypotonic saline lysis. At this point, the granulocyte pellet was incubated for 30 min at 4°C with anti-CD16 and anti-CD3 immunomagnetic beads, to remove neutrophils and contaminating lymphocytes, respectively. EOs were then eluted by passing the cells through the field of a permanent magnet. Freshly purified EOs were resuspended at 3 × 106 cells per ml in PBS containing 1% BSA. Flow cytometry analysis was performed on each EO preparation as follows: 50-μl aliquots of the single-cell preparations were incubated with FITC-conjugated anti-CD16, peridinin–chlorophyll–protein (perCP)-conjugated anti-CD3 and phycoerythrin (PE)-conjugated anti-CD19 mAb (to identify neutrophils or contaminating lymphocytes), or as negative control, FITC-conjugated isotype-matched Ab at a final concentration of 5 μg/ml for 1 h at 4°C in round-bottomed 96-well plates. After two washes in PBS, the cells were resuspended in PBS/0.5% BSA before analysis. Analysis was done by using FACSCalibur and cellquest software (Becton Dickinson). For the experiments, we used only preparations that were composed of >97% EOs (data not shown).
Real-Time Quantitative RT-PCR. Presence of S1PRs was screened by PCR, and we assessed that only S1P1, S1P2, and S1P3 were present (data not shown). Total mRNA from rat paws and from human purified EOs was extracted by using TRIzol reagent (GIBCO), according to the manufacturer's recommendations. Reverse transcription was performed, and 100 ng of the RNA samples described above were used for quantitative PCR. Samples were run in triplicate in 50-μl reactions by using an ABI PRISM 5700 sequence detector system (Applied Biosystems). Samples were incubated at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. SYBR-green oligonucleotides to detect both rat and human S1P1, S1P2, and S1P3 receptors and rat RANTES, CCR3, and eotaxin were specifically designed by using primer express software (Applied Biosystems) and validated for their specificity (Table 1). Relative quantification of target cDNA was determined by arbitrarily setting the control value at 100, and changes in cDNA content of a sample were expressed as a multiple thereof. Differences in cDNA input were corrected by normalizing signals obtained with primers specific for GAPDH. TaqMan-based predeveloped assay reagents were used for human Eotaxin, RANTES, and CCR3 by using a commercially available kit (Applied Biosystems). mRNA copy differences were corrected by using human GAPDH endogenous control predeveloped assay reagent (Applied Biosystems). To exclude nonspecific amplification and/or the formation of primer dimers, control reactions were performed in the absence of target cDNA. All of the experiments were run in triplicate.
Table 1. Oligonucleotides used in SYBR green experiments.
| Oligonucleotide | Sequence |
|---|---|
| Rat-S1P1-F | 5′-GGCCACCACCTACAAGCTCA-3′ |
| Rat-S1P1-R | 5′-CTTCCTTCCCGCAGAAACC-3′ |
| Rat-S1P2-F | 5′-CAGTGGCCAGGAACAGCAA-3′ |
| Rat-S1P2-R | 5′-TGCCGAGGAACAGGTACATG-3′ |
| Rat-S1P3-F | 5′-TGTCCGCTAGGAAGACGTTCA-3′ |
| Rat-S1P3-R | 5′-CCCTGAGGAACCACACTGTTG-3′ |
| Rat-GAPDH-F | 5′-ACCCATCACCATCTTCCAGGA-3′ |
| Rat-GAPDH-R | 5′-AGCCTTCTCCATGGTGGTGAA-3′ |
| Rat-CCR3-F | 5′-GGTCTTGCAGTATTGGCAGCA-3′ |
| Rat-CCR3-R | 5′-TGTCTTCTTCGCCCTCTGGAT-3′ |
| Rat-RANTES-F | 5′-GCCCACGTGAAGGAGTATTT-3′ |
| Rat-RANTES-R | 5′-TTGAACCCACTTCTTCTCTGG-3′ |
| Rat-scya 11-F | 5′-GCTTGGCAAAGAGATCTGTGC-3′ |
| Rat-scya 11-R | 5′-TTTGGTCCAGGTGCTTTGTG-3′ |
| Human-S1P1-F | 5′-GGGCCACCACCTACAAGCT-3′ |
| Human-S1P1-R | 5′-GCTGACAGGGCCACAAACAT-3′ |
| Human-S1P2-F | 5′-GCGCCATTGTGGTGGAAAA-3′ |
| Human-S1P2-R | 5′-CATTGCCGAGTGGAACTTGCT-3′ |
| Human-S1P3-F | 5′-GGTGATTGTGGTGAGCGTGTT-3′ |
| Human-S1P3-R | 5′-AGGCCACATCAATGAGGAAGA-3′ |
F, forward; R, reverse.
Human EO Chemotaxis. Chemotaxis assays were performed by using 24-well transwell plates with an 8-μm pore-size polycarbonate filter (Costar), as described (21). In brief, RPMI medium 1640 (GIBCO) containing either 8 ng/ml recombinant human eotaxin (PeproTech, London) or 100 nM S1P (Tocris Cookson, Bristol, U.K.) was placed in the lower chamber. We loaded 1 × 105 highly purified EOs in the upper chamber. Cells were incubated with a blocking anti-CCR3 Ab at 8 μg/ml (R & D Systems) for 30 min at 4°C or not incubated. All of the cells were resuspended at 1 × 106 cells per ml. Transwells were then incubated for 3 h at 37°C in a 5% CO2 humidified atmosphere. Cell migration was determined by counting cells that had migrated through the filter in five random high-power microscope fields (40 × 800). All experiments were performed in triplicate.
Rat Edema and Peritonitis. Male Wistar rats (Charles River Breeding Laboratories, Lecco, Italy) weighing 200–250g(n = 6) were lightly anaesthetized with enflurane 4%. Each rat received a subplantar administration of S1P (1–10 μg) or the vehicle (0.04% BSA) in a final volume of 100 μl. Paw volume was measured by using a hydropletismometer (Ugo Basile, Varese, Italy) immediately before subplantar injection and every 1 h for 6 h. The increase in paw volume was calculated by subtracting the basal value. To induce peritonitis S1P (3 μg per rat) or the vehicle (0.04% BSA) were injected in the rat peritoneal (n = 6) cavity in a final volume of 500 μl by using a 1-ml syringe. Rats were killed after 1 h, and the peritoneal exudates were recovered by washing the peritoneal cavity and a differential cell count performed after staining with eosin by an observer who was unaware of the treatment protocol.
Histology. Rat paws were fixed in neutral buffered formalin before being embedded in paraffin. Sections (4 μM) were stained with hematoxylin/eosin and analyzed under light microscopy by an observer unaware of the treatment protocol.
Statistical Analysis. Data are expressed as mean ± SEM. The level of statistical significance was determined by one-way ANOVA, followed by Bonferroni's t test for multiple comparisons by using prism software (GraphPad, San Diego).
Results
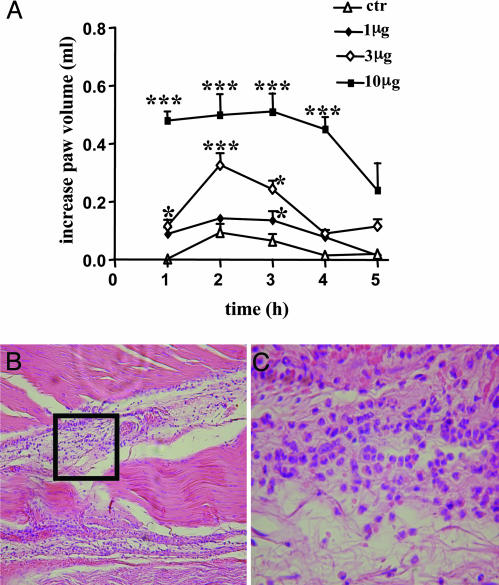
S1P Causes Inflammation and EO Recruitment in Rat in Vivo. The administration of S1P (1–10 μg) in rat paw (n = 6) causes a dose-dependent edema that peaks at the 2-h time point (Fig. 1A). Histological analysis showed the presence of a large EO infiltrate (Fig. 1 B and C). Examination of histological sections histological obtained by six different animals showed that, at the 2-h point, there were 27 ± 0.5 EOs for high-power field (HPF), whereas EOs were absent in the paw injected with the vehicle. Next, to assess whether the observed phenomenon is a feature only of the intraplantar injection, we i.p. administered S1P (3.10 μg) into rats (n = 6). The differentiated count performed on the exudates harvested 1 h after the i.p. administration showed that there was a consistent change in the cell distribution. Indeed, the vehicle (BSA) caused an increase in EOs/HPF of 3.8 ± 0.9%, whereas the doses of 3 and 10 μg of S1P caused an increase in EOs/HPF of 20 ± 3% and 30 ± 5%, respectively (P < 0.01 vs. vehicle).
Fig. 1.
S1P local administration causes edema formation and EO infiltration. (A) Subplantar administration of S1P (1, 3, and 10 μg per paw) causes a dose-dependent edema. Control animals received the vehicle used to dissolve S1P. *, P < 0.05; ***, P < 0.001 vs. control. (B) The histology shows that a large EO infiltrate (×10) is present in rat paw 1 h after the injection of S1P (10 μg). (C) Higher magnification (×40) of the area delimited by the square in B, where eosinophilia is clearly visible. Data are expressed as mean ± SEM.
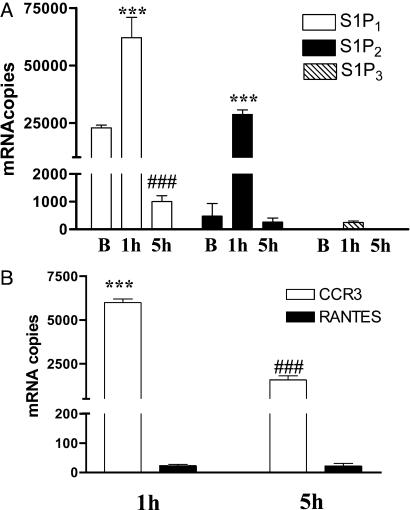
RT-PCR Quantitative Studies: Chemokines and Endothelial Differentiation Gene Receptors in Paw Tissue. Next, we addressed whether there is up-regulation of S1PRs in the paw tissues injected with S1P. RT-PCR quantitative analysis shows that, in basal condition, S1P1 is the most abundantly expressed, as compared with S1P2 and S1P3, whereas S1P4 and S1P5 are not detectable (Fig. 2A). After administration of S1P, there is an increase in the expression of S1P1 and S1P2 at a 2-h time point in rat-paw tissue. At the 5-h time point, whereas S1P2 level returns to basal value, S1P1 is significantly lower than its relative basal (Fig. 2 A). CCR3 message is increased, as opposed to RANTES (Fig. 2B), whereas eotaxin is undetectable in our experimental conditions (data not shown). The increased expression of CCR3 followed the edema profile and S1PRs expression, peaking at the 2-h time point and declining 5 h thereafter when the edema heals.
Fig. 2.
S1P increases S1PRs and CCR3 message in paw tissue. (A) S1P injection (10 μg) increases mRNA levels of S1P1, S1P2, and S1P3 receptors in the rat-paw tissue. Receptor expression reaches its maximum at the 1-h time point (***, P < 0.001 vs. its respective basal level). At the 5-h time point, when the edema heals, whereas S1P2 and S1P3 expression returns to basal level, S1P1 expression is significantly lower than its basal level (###, P < 0.01). (B) CCR3 and RANTES mRNA levels are undetectable in control tissue. After S1P injection (10 μg) CCR3 mRNA is strongly up-regulated (***, P < 0.001), as opposed to RANTES at the 1-h time point. CCR3 mRNA levels decrease significantly at the 5-h time point, when the edema heals (***, P < 0.001 vs. 1 h).
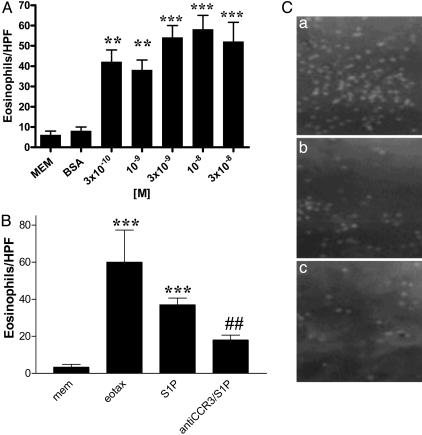
S1P Causes Chemotaxis of Human EOs. To further characterize the effects of S1P on EO recruitment, we prepared a set of in vitro experiments by using purified human EO. We find that S1P causes a dose-dependent chemotaxis in vitro of human EOs with a maximal increase in migration at 10–8 M (Fig. 3A). Because CCR3 is a key player in EO chemotaxis, we tested the effect of an anti-CCR3 Ab on S1P-induced EO chemotaxis, and we found that an anti-CCR3 mAb (8 μg/ml) significantly inhibited (≈50%) S1P-induced EO migration (Fig. 3 B and C).
Fig. 3.
S1P-induced EO migration. Dose-dependent migration of EOs after incubation with S1P (A); S1P-induced migration (10–8 M) is reduced significantly by anti-CCR3 neutralizing Ab (8 μg/ml) (B). Eotaxin has been used as a positive control. ***, P < 0.001 vs. MEM; ##, P < 0.01 vs. S1P. (C) Representative photos of EO chemotaxis in vitro. Eotaxin (a;8 μg/ml) or S1P (b;40ng/ml) causes EO chemotaxis, and incubation with anti-CCR3 antibodies reduces S1P-induced chemotaxis significantly (c).
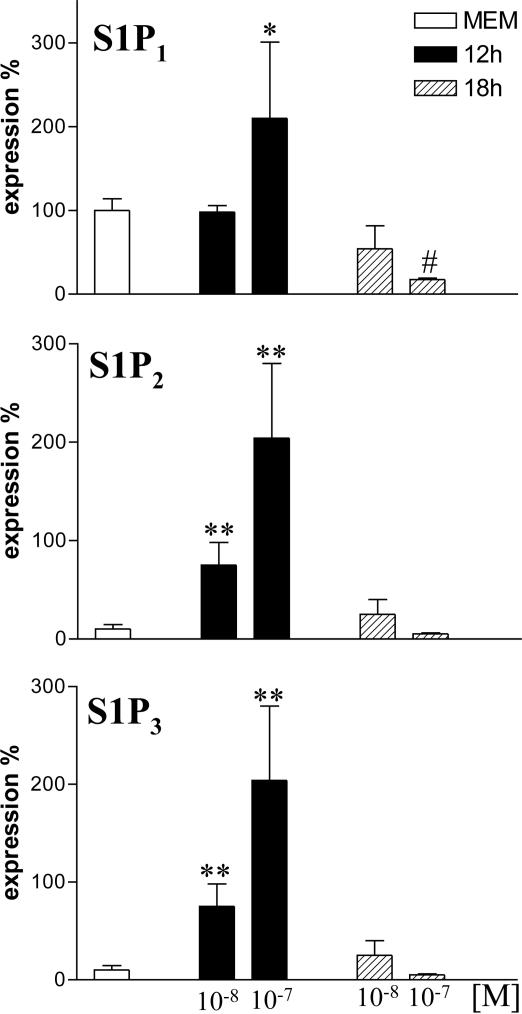
S1P Increases Message Expression of Its Receptors on Human EOs. Highly purified human EO (>97% purity, as assessed by flow cytometry) expresses S1P1, S1P2, and S1P3, whereas S1P4 and S1P5 are undetectable. Quantitative RT-PCR analysis shows that human EO in basal condition display higher levels of S1P1 than S1P2 and S1P3 (Fig. 4). Incubation of EO with S1P 10 or 100 nM for 12 h increases the expression of S1P1, S1P2, and S1P3 significantly (Fig. 4). Conversely, incubation for 18 h with 10 or 100 nM S1P reduces S1P1 expression vs. basal, whereas S1P2 and S1P3 are not different from their relative basal (Fig. 4).
Fig. 4.
S1P increases S1PRs expression on human EOs. Dose-dependent increase in S1P1, S1P2, and S1P3 expression on human purified EOs after incubation with MEM or S1P after 12 h. Incubation for 18 h causes a loss of expression. *, P < 0.05; **, P < 0.01 vs. MEM; #, P < 0.05 vs. S1P1 basal level.
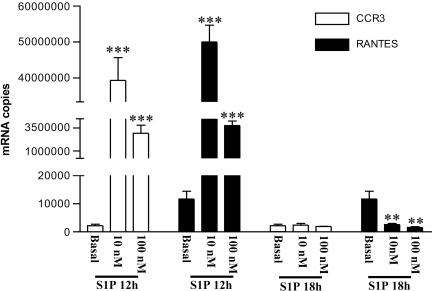
S1P Causes Increased Expression of CCR3 and RANTES in EOs in Vitro. S1P (10 and 100 nM) that is incubated for 12 h with human EO causes an increase in CCR3 and RANTES message, whereas eotaxin is undetectable in our experimental conditions (data not shown). S1P (10 nM) causes an increase of 18,000-fold for CCR3 and 4,000-fold for RANTES (Fig. 5). A longer time of incubation of S1P (10 and 100 nM) with EOs causes a loss of RNA message and a significant reduction of RANTES below the basal level (Fig. 5).
Fig. 5.
S1P increases the message for CCR3 and RANTES on human EOs. Dose-dependent increase in mRNA copies of CCR3 and RANTES after incubation for 12 h with S1P. Longer time of incubation (18 h) causes a loss in expression. *, P < 0.01; ***, P < 0.001 vs. their relative basal values.
Discussion
Local administration into the rat paw of S1P causes an edema characterized by a large EO infiltrate, which peaks at 2 h and it is resolved after 5 h. A consistent and rapid EO infiltrate is obtained also 1 h after i.p. administration of S1P. Thus, S1P displays a potent and selective recruiting activity for EOs in two different experimental models in vivo.
S1P explicates its activity through a receptorial mechanism that involves a class of G protein-coupled receptors that were recently renamed S1PRs. In control rat-paw tissues, we found high levels of mRNA message for S1P1, whereas the message for S1P2 receptor is ≈50 times less. S1P1, S1P2, and S1P3 mRNA copies increased at the 2-h point (when the edema peaks) and returned within basal levels at 5 h (when the edema resolves). S1P2 mRNA levels were enhanced ≈60 times over the basal value, indicating that this receptor is strongly up-regulated after S1P stimulation. Indeed, S1P1 values were enhanced only about three times because this receptor is highly expressed constitutively and most likely can account mainly for physiological, rather than pathological, signaling. Overall, this experimental evidence implies that up-regulation of these two receptors is crucial in the EO recruitment process that drives the observed inflammatory response.
It is widely accepted that EO recruitment depends on a complex cross-talk between several chemokines and cytokines, and among these, RANTES and eotaxin are the critical stimuli that modulate both EO trafficking and recruitment through CCR3 (22). Basal levels of CCR3, eotaxin, and RANTES are undetectable in our experimental conditions (i.e., in control paw tissue). After S1P administration in rat paw, CCR3 receptor tissue level shifts from undetectable to >5,000 mRNA copies, as assessed by quantitative RT-PCR. Both RANTES and eotaxin remained undetectable also in stimulated conditions. This consistent increase in CCR3 message, which is the receptor that mediates most of the EO functions (23, 24), correlates well with the large EO infiltrate driven by S1P paw injection.
S1P is stored in platelets, and these cells are the major source of S1P in plasma (1). It has been shown that the platelet–leukocyte interaction, as well as platelets alone, are involved in allergic inflammation and airway remodeling (25, 26). The exact mechanism by which platelet contributes to these processes in not known. However, it is known that platelet-derived growth factor receptor is tethered to S1P1 receptor, providing a platform for integrative signaling by these two types of receptors, resulting in activation and integration of downstream signal essentials for cell locomotion (27, 28). Our results suggest that S1P could represent the link among these cells that are involved in airway inflammation.
It is worth noting that the concentration of S1P used in this study is well within a pathophysiological range. Indeed, whereas S1P levels in plasma and serum range 200–900 nM (17), the amount injected in the paw to obtain edema coupled to EO migration ranges 8–25 nM. Thus, it is feasible that this concentration, being 30 times lower than the plasma levels, may be reached at a site of an injury.
Human EOs in resting condition constitutively express S1P1 and, to a lower extent, S1P2 and S1P3 message, which is similar to what happens in the rat-paw tissue. After stimulation in vitro with S1P for 12 h, there is a dose-related increase in S1P1, S1P2, and S1P3 receptor mRNA. Prolonged time of incubation leads to a reduction in expression of S1P1 below basal levels, whereas both S1P2 and S1P3 are not significantly different from their basal level. These data are not surprising because it is known that a prolonged exposition of cells to S1P leads to a reduction in biological activity (29). Next, we evaluated whether S1P, if it causes chemotaxis in vitro of purified human EOs already at the dose of 10–10 M, causes human EO chemotaxis in vitro in a dose-dependent manner, suggesting an important role for S1P in EO recruitment in human diseases. We then investigated the role of CCR3, eotaxin, and RANTES. The most striking result is that, in our experimental condition, we find an increase of ≈18,000 times in CCR3 message and 4,000 times in RANTES message, whereas the eotaxin level is undetectable. These data indicate that S1P, by specific up-regulation of CCR3 and RANTES in humans, drives EO recruitment. The striking up-regulation of CCR3 message observed in vivo and in vitro points toward a key role for S1P in regulating CCR3 expression in EOs. CCR3 is widely recognized as a key player in allergic inflammation, and it is known that lipids and chemically related substances can up-regulate gene expression. Thus, our data shed a light on a link between chemokine signaling and S1P, already well characterized for Th2-like responses in immune mediated events (30). However, it was not clear from our results whether S1P-induced chemotaxis through its receptors and in turn increased CCR3 message or whether it could somehow also bind CCR3. We found that an anti-CCR3 Ab, that selectively blocks the binding domain, reverses of ≈50% S1P-induced chemotaxis implying that S1P acts by means of the activation of its receptors and through CCR3. This hypothesis is not unlikely because both S1PRs and chemokine receptors are both G protein-coupled receptors. In addition, CCR3 is a promiscuous binder for chemokines and other factors that are not yet identified (22, 23, 30, 31) and S1P could be one of these unidentified factors. In our experiments, we did not detect eotaxin but we detected RANTES increase that is known to play a pivotal role, both in vivo and in vitro, in recruiting EOs at the site of inflammation through CCR3 binding (23, 24).
Despite the enormous effort of the research, the recruitment process of EOs in health and disease is still unclear. In particular, the eosinophilia that accompanies allergic diseases is still matter of debate (32). A recent study shows that S1P level increases in the airways of asthmatic patients after segmental allergen challenge (11). S1P levels reach an average concentration of >10 nM in bronchial alveolar lavage (BAL) fluid of asthmatics, compared with the baseline 1–2 nM concentration found in control subjects. The S1P level (10 nM) is the same concentration used in our in vitro study. Furthermore, in this study, CCR3 expression correlates with the EO number present in human BAL after segmental allergen challenge (33). Thus, these clinical results fit well with our in vitro and in vivo data, confirming that S1P also plays a role in the EO recruitment in vivo in humans. Recently, it has been shown (34) that S1P is involved in mast cell chemotaxis, further supporting a key role for this lipid in allergic inflammation.
In conclusion, we show that S1P acts as chemotactic agent for human EOs in vitro and causes EO recruitment in vivo in two different experimental models. This recruitment process depends on (i) an increase in S1P1, S1P2, and S1P3 mRNA levels; (ii) an increase in RANTES mRNA levels in human EOs only; and (iii) an increase in CCR3 mRNA levels. Together, these data may envisage a scenario in which, upon antigen stimulation, mast cells (8, 9), dendritic cells (35), and platelet aggregates (36) release S1P that recruits EOs, as well as determine an overall cytokines production that drives the Th2 response. On this basis, we suggest that an S1P antagonist, or interfering with S1P synthesis, could represent a valuable therapeutic tool in diseases such as allergies.
Acknowledgments
We thank Professor J. Gorski for helpful discussion; Drs. S. Carlomagno and L. Melillo for help in microchemotaxis analysis; Mrs. F. Papa for help in preparation of EOs; and all of the individuals who donated their blood for these experiments. This work was partially supported by Italian Ministry for the University and Scientific Research Grant PRIN 2003.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: S1P, sphingosine 1-phosphate; S1PR, S1P receptor; EO, eosinophil; SphK, sphingosine kinase.
References
- 1.Yatomi, Y., Ohmori, T., Rile, G., Kazama, F., Okamoto, H., Sano, T., Satoh, K., Kume, S., Tigyi, G., Igarashi, Y. & Ozaki, Y. (2000) Blood 96, 3431–3438. [PubMed] [Google Scholar]
- 2.Chun, J., Goetzl, E. J., Hla, T., Igarashi, Y., Lynch, K. R., Moolenaar, W., Pyne, S. & Tigyi, G. (2002) Pharmacol. Rev. 54, 265–269. [DOI] [PubMed] [Google Scholar]
- 3.Spiegel, S. & Milstien, S. (2003) Nat. Rev. Mol. Cell Biol. 4, 397–407. [DOI] [PubMed] [Google Scholar]
- 4.Spiegel, S., English, D. & Milstien, S. (2002) Trends Cell Biol. 12, 236–242. [DOI] [PubMed] [Google Scholar]
- 5.Van Brocklyn, J. R., Lee, M. J., Menzeleev, R., Olivera, A., Edsall, L., Cuvillier, O., Thomas, D. M., Coopman, P. J., Thangada, S., Liu, C. H., et al. (1998) J. Cell Biol. 142, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fueller, M., Wang, D., Tigyi, G. & Siess, W. (2003) Cell. Signalling 15, 367–375. [DOI] [PubMed] [Google Scholar]
- 7.Lee, M. J., Thangada, S., Claffey, K. P., Ancellin, N., Liu, C. H., Kluk, M., Volpi, M., Sha'afi, R. I. & Hla, T. (1999) Cell 99, 301–312. [DOI] [PubMed] [Google Scholar]
- 8.English, D., Kovala, A. T., Welch, Z., Harvey, K. A., Siddiqui, R. A., Brindley, D. N. & Garcia, J. G. (1999) J. Hematother. Stem Cell Res. 8, 627–634. [DOI] [PubMed] [Google Scholar]
- 9.Prieschl, E. E., Csonga, R., Novotny, V., Kikuchi, G. E. & Baumruker, T. (1999) J. Exp. Med. 190, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reischl, I. G., Coward, W. R. & Church, M. K. (1999) Biochem. Pharmacol. 58, 1841–1850. [DOI] [PubMed] [Google Scholar]
- 11.Ammit, A. J., Hastie, A. T., Edsall, L. C., Hoffman, R. K., Amrani, Y., Krymskaya, V. P., Kane, S. A., Peters, S. P., Penn, R. B., Spiegel, S. & Panettieri, R.A., Jr. (2001) FASEB J. 15, 1212–1214. [DOI] [PubMed] [Google Scholar]
- 12.Dorsam, G., Graeler, M. H., Seroogy, C., Kong, Y., Voice, J. K. & Goetzl, E. J. (2003) J. Immunol. 171, 3500–3507. [DOI] [PubMed] [Google Scholar]
- 13.Xia, P., Gamble, J. R., Rye, K. A., Wang, L., Hii, C. S., Cockerill, P., Khew-Goodall, Y., Bert, A. G., Barter, P. J. & Vadas, M. A. (1998) Proc. Natl. Acad. Sci. USA 95, 14196–14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikolova-Karakashian, M., Morgan, E. T., Alexander, C., Liotta, D. C. & Merrill, A. H., Jr. (1997) J. Biol. Chem. 272, 18718–18724. [DOI] [PubMed] [Google Scholar]
- 15.Choi, O. H., Kim, J. H. & Kinet J. P. (1996) Nature 380, 634–636. [DOI] [PubMed] [Google Scholar]
- 16.Cuvillier, O. & Levade, T. (2003) Pharmacol. Res. 47, 439–445. [DOI] [PubMed] [Google Scholar]
- 17.Perry, D. K & Kolesnick, R. N. (2003) Cancer Treat. Res. 115, 345–354. [DOI] [PubMed] [Google Scholar]
- 18.Okajima, F. (2002) Biochim. Biophys. Acta 1582, 132–137. [DOI] [PubMed] [Google Scholar]
- 19.Lee, H., Liao, J. J., Graeler, M., Huang, M. C. & Goetzl, E. J. (2002) Biochim. Biophys. Acta 1582, 175–177. [DOI] [PubMed] [Google Scholar]
- 20.Hansel, T. T., De Vries, I. J., Iff, T., Rihs, S., Wandzilak, M., Betz, S., Blaser, K. & Walker, C. (1991) J. Immunol. Methods 145, 105–110. [DOI] [PubMed] [Google Scholar]
- 21.Falk, W. R., Goodwin, R. H. J. & Leonard, E. J. (1980) J. Immunol. Methods 33, 239–247. [DOI] [PubMed] [Google Scholar]
- 22.Borish, L. C. & Steinke, J. W. (2003) J. Allergy Clin. Immunol. 111, S460–S475. [DOI] [PubMed] [Google Scholar]
- 23.Ono, S. J., Nakamura, T., Miyazaki, D., Ohbayashi, M., Dawson, M. & Toda, M. (2003) J. Allergy Clin. Immunol. 111, 1185–1199. [DOI] [PubMed] [Google Scholar]
- 24.Baggiolini, M. (1998) Nature 392, 565–568. [DOI] [PubMed] [Google Scholar]
- 25.Pitchford, S. C., Yano, S., Lever, R., Riffo-Vasquez, Y., Ciferri, S., Rose, M. J., Giannini, S., Momi, S., Spina, D., O'Connor, B., et al. (2003) J. Allergy Clin. Immunol. 112, 109–118. [DOI] [PubMed] [Google Scholar]
- 26.Pitchford, S. C., Riffo-Vasquez, Y., Sousa, A., Momi, S., Gresele, P., Spina, D. & Page, C. P. (2004) Blood 103, 639–647. [DOI] [PubMed] [Google Scholar]
- 27.Hobson, J. P., Rosenfeldt, H. M., Barak, L. S., Olivera, A., Poulton, S., Caron, M. G., Milstien, S. & Spiegel, S. (2001) Science 291, 1800–1803. [DOI] [PubMed] [Google Scholar]
- 28.Alderton, F., Rakhit, S., Kong, K. C., Palmer, T., Sambi, B., Pyne, S. & Pyne, N. J. (2001) J. Biol. Chem. 276, 28578–28585. [DOI] [PubMed] [Google Scholar]
- 29.Graeler, M., Shankar, G. & Goetzl, E. J. (2002) J. Immunol. 169, 4084–4087. [DOI] [PubMed] [Google Scholar]
- 30.Lilly, C. M. & Daugherty, B. L. (2001) Am. J. Respir. Cell Mol. Biol. 25, 673–675. [DOI] [PubMed] [Google Scholar]
- 31.Van der Kleij, D. & Yazdanbakhsh, M. (2003) Eur. J. Immunol. 33, 2953–2963. [DOI] [PubMed] [Google Scholar]
- 32.Adamko, D., Odemuyiwa, S. O. & Moqbel, R. (2003) Curr. Opin. Pharmacol. 3, 227–232. [DOI] [PubMed] [Google Scholar]
- 33.Liu, L.Y., Jarjour, N. N., Busse, W. W. & Kelly, E. A. (2003) J. Allergy Clin. Immunol. 112, 556–562. [DOI] [PubMed] [Google Scholar]
- 34.Jolly, P. S., Bekats, M., Olivera, A., Gonzalez-Espinosa, C., Proia, R. L., Rivera, J. & Spiegel, S. (2004) J. Exp. Med. 199, 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Idzko, M., Panther, E., Corinti, S., Morelli, A., Ferrari, D., Herouy, Y., Dichmann, S., Mockenhaupt, M., Gebicke-Haerter, P., Di Virgilio, F., et al. (2002) FASEB J. 16, 625–627. [DOI] [PubMed] [Google Scholar]
- 36.English, D., Welch, Z., Kovala, A. T., Harvey, K., Volpert, O. V., Brindley, D. N. & Garcia, J.G. (2000) FASEB J. 14, 2255–2265. [DOI] [PubMed] [Google Scholar]