Abstract
Receptor activator of NF-κB ligand (RANKL) plays an essential role in osteoclast formation and bone resorption. Although genetic and biochemical studies indicate that RANKL regulates osteoclast differentiation by activating receptor activator of NF-κB and associated signaling molecules, the molecular mechanisms of RANKL-regulated osteoclast differentiation have not yet been fully established. We investigated the role of the transcription factor c-Jun, which is activated by RANKL, in osteoclastogenesis using transgenic mice expressing dominant-negative c-Jun specifically in the osteoclast lineage. We found that the transgenic mice manifested severe osteopetrosis due to impaired osteoclastogenesis. Blockade of c-Jun signaling also markedly inhibited soluble RANKL-induced osteoclast differentiation in vitro. Overexpression of nuclear factor of activated T cells 1 (NFAT1) (NFATc2/NFATp) or NFAT2 (NFATc1/NFATc) promoted differentiation of osteoclast precursor cells into tartrate-resistant acid phosphatase–positive (TRAP–positive) multinucleated osteoclast-like cells even in the absence of RANKL. Overexpression of NFAT1 also markedly transactivated the TRAP gene promoter. These osteoclastogenic activities of NFAT were abrogated by overexpression of dominant-negative c-Jun. Importantly, osteoclast differentiation and induction of NFAT2 expression by NFAT1 overexpression or soluble RANKL treatment were profoundly diminished in spleen cells of the transgenic mice. Collectively, these results indicate that c-Jun signaling in cooperation with NFAT is crucial for RANKL-regulated osteoclast differentiation.
Introduction
The amount of bone remodeling is controlled by the balance between bone formation and bone resorption (1–3). Many osteopenic diseases, including osteoporosis, rheumatoid arthritis, Paget disease, and lytic bone metastases of malignancies are characterized by progressive and excessive bone resorption by osteoclasts, which are multinucleated giant cells that originate from hematopoietic cells (2). A TNF family member, receptor activator of NF-κB ligand (RANKL), which is expressed as a membrane-bound protein in osteoblasts and stromal cells, promotes the differentiation of osteoclast precursor cells into osteoclasts (4, 5). Gene-targeted mice deficient in RANKL expression show severe osteopetrosis with complete absence of osteoclast formation (5). These findings indicate that RANKL is an essential factor responsible for osteoclast differentiation.
RANKL mediates its biological effects through receptor activator of NF-κB (RANK), which is expressed in osteoclast precursors (6). RANK interacts with the intracellular molecule, TNF receptor–associated factor 6 (TRAF6), which plays an important role in the activation of downstream signaling pathways, including NF-κB, p38 kinase, and c-Jun N-terminal kinase (JNK) (2). Targeted disruption of RANK (6), TRAF6 (7, 8), or NF-κB (9) genes in mice induces osteopetrosis, indicating the indispensable roles of these molecules in regulating the differentiation and function of osteoclasts. Since treatment with p38 inhibitors suppresses osteoclastogenesis in vitro (10), p38 seems to be involved in controlling osteoclast differentiation.
RANKL also stimulates JNK, and subsequently elicits the activation of the transcription factor c-Jun (11). c-Jun forms activator protein-1 (AP-1) complexes with c-Fos, an essential transcription factor for osteoclast formation (12). These findings suggest a potential role of c-Jun signaling in osteoclasts. However, in contrast to NF-κB signaling, the physiological role of c-Jun signaling in RANKL-induced osteoclast differentiation and the subsequent transcriptional events that regulate osteoclastogenesis remain unclear.
In the present study, we investigated the role of c-Jun signaling in osteoclastogenesis and the molecular mechanisms in which c-Jun is involved. Our data demonstrate that activation of c-Jun signaling is essential for RANKL-regulated osteoclast formation in vivo and in vitro. Furthermore, we show that c-Jun regulates osteoclast differentiation in cooperation with nuclear factor of activated T cells (NFAT). Therefore, the coupling of c-Jun signaling with NFAT family is crucial for transcriptional events during osteoclastogenesis.
Results
Since c-Jun–deficient mice are embryonic lethal (13), investigating the role of c-Jun signaling in osteoclasts using c-Jun–deficient mice is complicated. To evaluate the physiological role of c-Jun signaling in osteoclast differentiation in vivo, we generated transgenic mice (TRAP-DN–c-Jun TG) in which dominant-negative c-Jun lacking the transcriptional activation domain (14, 15) was expressed under control of the mouse tartrate-resistant acid phosphatase (TRAP) gene promoter, which is specific for the osteoclast lineage (16, 17). RT-PCR analysis indicated specific expression of dominant-negative c-Jun in the osteoclast lineage (Figure 1A). As shown in Figure 1B, the body size of TRAP-DN–c-Jun TG mice was much smaller than that of wild-type littermates. Incisors and molars of TRAP-DN–c-Jun TG mice failed to erupt despite formation of tooth germs. TRAP-DN–c-Jun TG mice showed increased radiodensity of long bones, jaw bones, and vertebrae compared with control mice (Figure 1C). The bone marrow cavities of TRAP-DN–c-Jun TG mice were filled with unresorbed trabecular bones (Figure 1D). More importantly, TRAP staining demonstrated very few osteoclasts in the long bones of TRAP-DN–c-Jun TG mice (Figure 1D). Consistently, osteoclast formation of spleen cells derived from TRAP-DN–c-Jun TG mice was markedly lower than that of wild-type littermates (Figure 1E). In contrast, the spleen cells of TRAP-DN–c-Jun TG mice differentiated into macrophage-like cells just as well as did those of wild-type mice (Figure 1F). The differentiation of osteoblasts, which play a central role in bone formation, appears to be intact in TRAP-DN–c-Jun TG mice (Figure 1G). Thus, TRAP-DN–c-Jun TG mice manifested typical osteopetrosis due to an impairment of osteoclastogenesis. These results show that c-Jun is an essential transcription factor for osteoclast differentiation in vivo.
Figure 1.
Osteopetrotic phenotype of TRAP-DN–c-Jun TG mice. (A) Expression of DN–c-Jun transgene in TRAP-DN–c-Jun TG mice. Total RNA isolated from brain (lanes 1 and 7), kidney (lanes 2 and 8), liver (lanes 3 and 9), lung (lanes 4 and 10), spleen cells (lanes 5 and 11), or BMMΦ (lanes 6 and 12) of wild-type (WT; lanes 1–6) and TRAP-DN–c-Jun TG (TG; lanes 7–12) littermate mice were subjected to RT-PCR analysis using specific primers for DN–c-Jun (top panel) or β-actin (bottom panel). (B) Appearance of wild-type and TRAP-DN–c-Jun TG littermate mice. No tooth had erupted in TRAP-DN–c-Jun TG mice. (C) Radiological analyses in TRAP-DN–c-Jun TG mice. TRAP-DN–c-Jun TG mice showed increased radiodensity of long bones, vertebrae, and jaw compared with wild-type littermates. (D) H&E and TRAP staining of long bones of WT or TRAP-DN–c-Jun TG littermate mice. Left panels, lower magnification (H&E, ×50; TRAP, ×100); right panels, higher magnification (H&E, ×100; TRAP, ×400). (E) Impaired osteoclastogenesis of TRAP-DN–c-Jun TG mice in vitro. Spleen cells isolated from wild-type or TRAP-DN–c-Jun TG littermate mice were incubated with M-CSF and sRANKL for 6 days, and TRAP+ multinucleated osteoclast-like (ocl) cells were counted. (F) Macrophage-like cell differentiation determined by NSE staining in the culture of spleen cells in α-MEM–10% FCS with M-CSF. (G) Osteoblast differentiation determined by ALP staining (left panel) or evaluated for ALP activity (right panel) in the culture of primary osteoblasts isolated from calvariae in α-MEM–10% FCS with BMP2.
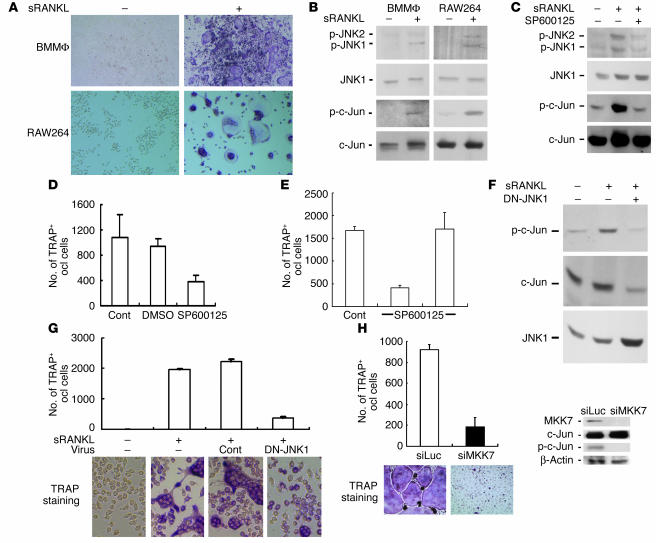
To understand the molecular mechanisms by which c-Jun regulates RANKL-induced osteoclastogenesis, we next examined the activation of JNK and c-Jun signaling in osteoclast precursors, bone marrow macrophage (BMMΦ) (18), and RAW264 cells (6) in the presence of soluble RANKL (sRANKL). Treatment with sRANKL induced differentiation of BMMΦ and RAW264 cells into TRAP+ multinucleated osteoclast-like cells (Figure 2A) and activated JNK1, JNK2, and c-Jun in these cells (Figure 2B). Treatment with a specific JNK inhibitor, SP600125 (19), profoundly abrogated the activation of JNK and c-Jun (Figure 2C), and markedly inhibited the formation of TRAP+ osteoclast-like cells induced by sRANKL in mouse bone marrow cultures (Figure 2D). The inhibitor was not acutely toxic to the cells, since its inhibitory effect was abrogated by washing it out of the cultures at 1 day (Figure 2E). These data suggest that sustained activation of JNK and c-Jun is necessary for RANKL-induced osteoclastogenesis.
Figure 2.
Inhibition of osteoclast differentiation by blockade of JNK activation. (A) Osteoclast differentiation of BMMΦ and RAW264 cells was determined by TRAP staining. (B) Lysates of BMMΦ or RAW264 cells were analyzed by immunoblotting using anti–phospho-JNK (anti–p-JNK), JNK1, phospho–c-Jun (p–c-Jun), or c-Jun antibody. (C) Lysates of BMMΦ cells treated with sRANKL and SP600125 (5 μM) as indicated, were analyzed by immunoblotting using anti–phospho-JNK, JNK1, phospho–c-Jun, or c-Jun antibody. (D) Suppression of osteoclastogenesis by JNK inhibitor. Mouse bone marrow cells were incubated with M-CSF and sRANKL in the presence of DMSO or SP600125 (5 μM), and TRAP+ multinucleated osteoclast-like cells were counted. Cont, control (E) The effect of SP600125 on osteoclastogenesis is reversible after 1 day. BMMΦ cells were incubated with M-CSF and sRANKL in the absence (left bar) or presence (middle and right bars) of SP600125 (5 μM). SP600125 was removed after 24 hours by washing carefully (right bar). After 6 days of incubation, TRAP+ multinucleated osteoclast-like cells were counted. (F) Lysates of RAW264 cells infected with control or dominant-negative–JNK1 (DN-JNK1) adenovirus were analyzed by immunoblotting using phospho–c-Jun, c-Jun, or anti-JNK1 antibody. (G) Suppression of osteoclastogenesis by DN-JNK1. RAW264 cells infected with control or DN-JNK1 adenovirus were incubated with sRANKL, and TRAP+ multinucleated osteoclast-like cells were counted. (H) Inhibition of osteoclast differentiation by suppression of c-Jun activation. Mouse spleen cells infected with retroviruses carrying either luciferase siRNA (siLuc) or MKK7 siRNA (siMKK7) were incubated with M-CSF and sRANKL, and TRAP+ multinucleated osteoclast-like cells were counted. The cell lysates were analyzed by immunoblotting as indicated.
To further investigate the importance of c-Jun signaling in RANKL-induced osteoclast differentiation, we interfered with c-Jun and upstream signaling to c-Jun using dominant-negative and small interfering RNA (siRNA) knockdown approaches. Overexpression of dominant-negative JNK1 abolished c-Jun phosphorylation (Figure 2F) and markedly inhibited osteoclast differentiation of RAW264 cells in response to sRANKL (Figure 2G). Likewise, siRNA-mediated knockdown of MAPK kinase 7 (MKK7), the upstream kinase for JNK, also indicated the necessity of c-Jun activation in osteoclast formation of spleen cells (Figure 2H). Furthermore, overexpression of dominant-negative c-Jun markedly suppressed sRANKL-induced transcriptional activation of AP-1 (Figure 3A) and osteoclast differentiation of RAW264 cells (Figure 3B). Finally, suppression of c-Jun expression by siRNA consistently and markedly inhibited osteoclast differentiation in a primary culture system (Figure 3C). In contrast, dominant-negative mutant of activating transcription factor-2 (ATF-2), which is also activated by sRANKL (Figure 3D), did not affect osteoclast differentiation of RAW264 cells (Figure 3E). Taken together, these data strongly suggest that c-Jun signaling specifically mediates the effect of RANKL and promotes osteoclastogenesis.
Figure 3.
Requirement of c-Jun and c-Fos for RANKL-induced osteoclastogenesis. (A) Inhibition of AP-1 transcriptional activity by DN–c-Jun or DN–c-Fos. RAW264 cells were transfected AP-1 reporter construct together with DN–c-Jun or DN–c-Fos, and incubated with sRANKL. Luciferase activity in cell lysates was measured. (B) Suppression of osteoclast differentiation by DN–c-Jun. RAW264 cells infected with control or DN–c-Jun adenovirus were incubated with sRANKL for 6 days, and TRAP+ multinucleated osteoclast-like cells were counted. (C) Inhibition of osteoclast differentiation by suppression of c-Jun expression. Mouse spleen cells infected with retroviruses carrying either luciferase siRNA or c-Jun siRNA (siJun) were incubated with M-CSF and sRANKL for 6 days, and TRAP+ multinucleated osteoclast-like cells were counted. The cell lysates were analyzed by immunoblotting using anti–c-Jun or anti–β-actin antibody. (D) Activation of ATF-2 by sRANKL. The lysates of RAW264 cells stimulated with sRANKL were analyzed by immunoblotting using anti–phospho–ATF-2 (anti–p-ATF-2) or anti–ATF-2 antibody. (E) No effects of DN–ATF-2 on osteoclast formation. RAW264 cells infected with control or DN–ATF-2 adenovirus were incubated with sRANKL for 6 days, and TRAP+ multinucleated osteoclast-like cells were counted. (F) Inhibition of osteoclastogenesis by DN–c-Fos. RAW264 cells infected with control or DN–c-Fos adenovirus were incubated with sRANKL for 6 days, and TRAP+ multinucleated osteoclast-like cells were counted. (G) No induction of osteoclastogenesis by overexpression of c-Jun and c-Fos. RAW264 cells infected with control, TRAF6, or c-Jun/c-Fos adenovirus were incubated with or without sRANKL for 6 days, and TRAP+ multinucleated osteoclast-like cells were counted.
Since c-Jun regulates the transcription of target genes by forming the AP-1 complexes with c-Fos, we determined the role of c-Fos in RANKL-induced osteoclastogenesis. As shown in Figure 3A, overexpression of dominant-negative c-Fos markedly blocked sRANKL-induced AP-1 transcriptional activity. The osteoclastogenic activity of sRANKL was also markedly inhibited by overexpression of dominant-negative c-Fos (Figure 3F). These data indicate that both c-Fos and c-Jun are required for RANKL to regulate osteoclast differentiation. However, we were unable to induce differentiation of RAW264 cells into osteoclast-like cells by overexpressing c-Jun together with c-Fos (Figure 3G), suggesting that complexes of c-Jun and c-Fos need a partner transcription factor or co-activator to promote osteoclastogenesis.
c-Jun/c-Fos complexes associate with NFAT family proteins, and subsequently regulate the transcription of specific genes in T cells and some other types of cells (20–22). Therefore, we investigated the involvement of NFAT in osteoclastogenesis and the relationship between RANKL signaling and NFAT. First, we evaluated the transcriptional activity of NFAT during osteoclast differentiation by performing a reporter assay using a luciferase reporter plasmid driven by tandem NFAT binding sites. In RAW264 cells differentiated into TRAP+ multinucleated cells by treatment with sRANKL, transcriptional activity of NFAT was markedly elevated compared with undifferentiated RAW264 cells (Figure 4A). Moreover, NFAT1 (NFATc2/NFATp) was expressed in BMMΦ, RAW264 cells, and differentiated osteoclasts (Figure 4, B–D), and stimulation with sRANKL induced the nuclear localization of NFAT1 in RAW264 cells (Figure 4C). Furthermore, overexpression of TRAF6, which mimics the effects of RANKL (Figure 3G), dramatically increased the transcriptional activity of NFAT1 (Figure 4E). These data suggest that RANKL signals activate NFAT1 function and consequently regulate osteoclastogenesis. To support this idea, treatment with cyclosporin A, a well-known inhibitor for the calmodulin-dependent phosphatase, calcineurin, which dephosphorylates and activates the NFAT family (22, 23), strikingly inhibited osteoclast differentiation in mouse bone marrow cultures (Figure 4F). A more selective NFAT inhibitor, VIVIT peptide (MAGPHPVIVITGPHEE) tagged with green fluorescent protein (GFP-VIVIT) (23), markedly suppressed osteoclast differentiation of RAW264 cells (Figure 4G). Collectively, these results suggest the importance of NFAT1 in RANKL-induced osteoclast differentiation.
Figure 4.
Activation of NFAT1 by RANKL signals and requirement of NFAT for osteoclastogenesis. (A) Activation of NFAT transcriptional activity during osteoclast differentiation. RAW264 cells incubated with or without sRANKL were transfected with a luciferase reporter construct containing three copies of NFAT-responsive elements. Luciferase activity in cell lysates was measured. TRAP (+), TRAP+ osteoclast-like cells treated with sRANKL. RLU, relative light units. (B) Expression of NFAT1 in BMMΦ cells. The lysates of BMMΦ cells were analyzed by immunoblotting using anti-NFAT1 antibody. (C) Nuclear translocation of NFAT1 by sRANKL. RAW264 cells were stimulated with or without sRANKL, and cytoplasmic and nuclear extracts were analyzed by immunoblotting with anti-NFAT1 antibody. (D) Expression of NFAT1 in osteoclasts. Mouse spleen cells were incubated with M-CSF and sRANKL for 6 days and analyzed by immunostaining (upper panel) or immunoblotting (lower panel) using NFAT1 antibody. (E) Transcriptional activation of NFAT1 by TRAF6. COS-7 cells were transfected with a luciferase reporter construct containing three copies of NFAT-responsive elements together with either NFAT1 or TRAF6, or both. Luciferase activity in cell lysates was measured. (F) Suppression of osteoclastogenesis by cyclosporin A. Mouse bone marrow cells were incubated with M-CSF and sRANKL for 6 days in the presence or absence of cyclosporin A (CsA) as indicated. TRAP+ multinucleated osteoclast-like cells were counted under a microscope. (G) Inhibition of osteoclastogenesis by NFAT inhibitor, VIVIT peptide. RAW264 cells infected with control or GFP-VIVIT adenovirus at 20 or 50 MOI were incubated with sRANKL for 6 days, and TRAP+ multinucleated osteoclast-like cells were counted.
To examine the importance of NFAT1 function in osteoclast differentiation, we overexpressed NFAT1 in RAW264 cells using adenoviral vectors. When overexpressed in RAW264 cells, NFAT1 was able to translocate into the nucleus even in the absence of sRANKL (Figure 5A). Importantly, overexpression of NFAT1 promoted differentiation of RAW264 cells or mouse spleen cells into TRAP+ multinucleated osteoclast-like cells (Figure 5, B and C). In parallel with this result, NFAT1 overexpression dramatically increased TRAP gene promoter activity (Figure 5D). A constitutively active mutant of NFAT1, which lacks a negative-regulatory domain present in its N-terminal region, further increased the TRAP gene promoter activity (Figure 5D). In contrast, another mutant NFAT1 (ØNC), which lacks the transactivation domains present in the N- and C-terminal regions, failed to increase TRAP gene promoter activity (Figure 5D). Analysis using serial-deletion mutants of the TRAP gene promoter and a DNA pull-down assay demonstrated that NFAT1 regulates TRAP gene promoter activity through the NFAT-binding element present at –523 to –507 in the TRAP gene promoter (Figure 5, E and F). Both basal and NFAT-induced reporter activity were markedly increased by deletion of the –881 to –737 region, suggesting the presence of a negative-regulatory element in this region, but decreased again by further removal of the –737 to –681 region (Figure 5E). Since the fold induction of luciferase activity by NFAT1 was similar between TRAP737-Luc and TRAP681-Luc, the –681 to –737 region may contain a positive-regulatory element that is independent of NFAT. Collectively, these data suggest that NFAT1 is an important transcription factor that regulates osteoclastogenesis.
Figure 5.
Induction of osteoclast differentiation by NFAT1 overexpression. (A) Cytoplasmic and nuclear extracts of RAW264 cells infected with NFAT1 adenovirus were analyzed by immunoblotting with anti-NFAT1 antibody. (B) RAW264 cells were infected with NFAT1 adenovirus at 10, 20, or 50 MOI, and TRAP+ multinucleated osteoclast-like cells were counted. (C) Mouse spleen cells infected with control or constitutively active NFAT1 retrovirus were cultured with M-CSF, and TRAP+ multinucleated osteoclast-like cells were counted. (D) RAW264 cells were transfected with TRAP gene promoter fused to a luciferase reporter construct together with wild-type, constitutively active (CA) or δNC mutant of NFAT1. Luciferase activity in cell lysates was measured. (E) Requirement of putative NFAT-binding element present in the TRAP gene promoter (–523 to –507) for transactivation by NFAT1. RAW264 cells were transfected with serial-deletion mutants of the TRAP gene promoter together with or without NFAT1, and luciferase activities were measured. The sequence of the putative NFAT-binding element in the TRAP gene promoter is indicated. (F) Binding of NFAT1 to NFAT-binding element present in TRAP gene promoter (–523 to –507). Lysates of COS-7 cells transfected with or without NFAT1 were precipitated (Ppt) with the oligonucleotide containing NFAT-binding element (NFAT-BE) from the TRAP gene promoter, and analyzed by immunoblotting using anti-NFAT1 antibody. (G) The lysates of RAW264 cells infected with NFAT1 adenovirus or incubated with sRANKL were determined by immunoblotting with anti-NFAT2 antibody. (H) Induction of osteoclast differentiation by NFAT2 overexpression. RAW264 cells were infected with control or NFAT1 or NFAT2 adenovirus and determined by TRAP staining.
Recently, NFAT2 (NFATc1/NFATc) has been proposed as a target of RANKL and a crucial transcription factor for osteoclast differentiation (24, 25). To understand the relationship between NFAT1 and NFAT2 during osteoclastogenesis, we determined whether NFAT1 is involved in the regulation of NFAT2 expression. We found that overexpression of NFAT1 induced NFAT2 expression in the absence of sRANKL (Figure 5G). In addition, overexpression of NFAT2 also induced osteoclast differentiation of RAW264 cells (Figure 5H). These data suggest that NFAT1 upregulates NFAT2 expression during osteoclast differentiation.
To investigate the molecular mechanisms by which c-Jun regulates osteoclast differentiation, we next examined the functional relationship between c-Jun and NFAT during osteoclast differentiation. As shown in Figure 6A, osteoclast-like cell formation induced by NFAT1 or NFAT2 was clearly enhanced by overexpression of c-Jun and c-Fos. In contrast, overexpression of either dominant-negative c-Jun or dominant-negative c-Fos markedly blocked osteoclast differentiation of RAW264 cells induced by NFAT1 or NFAT2 (Figure 6, B and C). These data suggest that the collaboration of c-Jun/c-Fos complexes and NFAT is important for the regulation of osteoclastogenesis. To verify the importance of the partnership between c-Jun and NFAT at the transcriptional level, we tested the effect of a fusion protein of JNK1 and MKK7 (MKK7-JNK1), which constitutively activates AP-1 (26), on TRAP gene promoter activity. Overexpression of MKK7-JNK1 alone was unable to transactivate the TRAP gene promoter (Figure 6D). On the other hand, overexpression of MKK7-JNK1 enhanced the transactivation of the TRAP gene promoter by NFAT1 (Figure 6D). Moreover, overexpression of either dominant-negative c-Jun or dominant-negative c-Fos blocked transactivation of the TRAP gene promoter by NFAT1 (Figure 6E). These results strongly suggest that functional cooperation between c-Jun and NFAT is necessary for the stimulation of osteoclast differentiation.
Figure 6.
Partnership between c-Jun/c-Fos complexes and NFAT in osteoclast differentiation. (A) Enhancement of NFAT1- or NFAT2-induced osteoclastogenesis by overexpression of c-Jun and c-Fos. RAW264 cells were infected with adenoviruses as indicated and incubated for 6 days. TRAP+ multinucleated osteoclast-like cells were counted. (B and C) Suppression of NFAT1- or NFAT2-induced osteoclastogenesis by DN–c-Jun or DN–c-Fos. RAW264 cells were infected with NFAT1 or NFAT2 adenovirus (50 MOI) together with control, DN–c-Jun, or DN–c-Fos adenovirus (50 MOI), and TRAP+ multinucleated osteoclast-like cells were counted. (D) Cooperative role of AP-1 and NFAT1 in transactivation of TRAP gene promoter. RAW264 cells were transfected with TRAP gene promoter fused to luciferase reporter construct together with NFAT1, MKK7-JNK1, or both. Luciferase activity in cell lysates was measured 48 hours after transfection. (E) Suppression of NFAT1-induced TRAP gene promoter activation by DN–c-Jun or DN–c-Fos. RAW264 cells were transfected with the TRAP gene promoter fused to a luciferase reporter construct together with DN–c-Jun or DN–c-Fos. Luciferase activity in cell lysates was measured 48 hours after transfection.
To further understand the importance of functional cooperation between c-Jun and NFAT during osteoclast differentiation, we examined the ability of NFAT1 to induce osteoclastogenesis and NFAT2 expression in spleen cells isolated from TRAP-DN–c-Jun TG mice. Overexpression of NFAT1 induced osteoclast-like cell formation and NFAT2 expression in spleen cells isolated from wild-type mice (Figure 7, A and B), but failed to induce osteoclast-like cell formation and NFAT2 expression in spleen cells of TRAP-DN–c-Jun TG mice (Figure 7, A and B). In addition, induction of NFAT2 by sRANKL was also abolished in spleen cells of TRAP-DN–c-Jun TG mice (Figure 7C). These results indicate that NFAT1–c-Jun cooperation is required for NFAT1 function and NFAT2 expression during osteoclast differentiation induced either by RANKL or by NFAT1 itself.
Figure 7.
Impairment of osteoclastogenesis and NFAT2 induction by NFAT1 overexpression in the spleen cells of TRAP-DN–c-Jun TG mice. (A) Spleen cells isolated from wild-type littermates or TRAP-DN-TG (TG) mice were infected with control or constitutively active NFAT1 retrovirus, and cultured for 7 days in the presence of M-CSF. TRAP+ multinucleated osteoclast-like cells were counted. (B) Spleen cells isolated from WT littermates or TG mice were infected with control or constitutively active NFAT1 retrovirus and cultured for 5 days in the presence of M-CSF. Cell lysates were analyzed by immunoblotting using anti-NFAT2 or anti–β-actin antibody. (C) Spleen cells isolated from WT littermates or TG mice were cultured for 5 days in the presence of M-CSF alone or both M-CSF and sRANKL. Cell lysates were analyzed by immunoblotting using anti-NFAT2 or anti–β-actin antibody.
Discussion
Although genetic studies using gene targeting technology have clearly demonstrated that RANK, TRAF6, and NF-κB, which transduce the signals of RANKL, are essential molecules for osteoclast differentiation (2), the mechanisms by which RANKL regulates osteoclastogenesis have not been fully clarified and thus our overall understanding of osteoclast differentiation is incomplete. In particular, the role of MAPKs, including JNK and p38 kinase, both of which are elicited by RANKL (11), in osteoclastogenesis is elusive. In the present study, we showed that transgenic mice expressing dominant-negative c-Jun exhibited osteopetrosis, most likely as the result of impaired osteoclastogenesis, because very few osteoclasts were present in the long bones of the transgenic mice and osteoclast progenitor cells isolated from the spleens of transgenic mice formed few osteoclasts in the presence of sRANKL. Consistent with these data, we also found that the blockade of the JNK and c-Jun pathway by treatment with a JNK inhibitor, SP600125, suppressed osteoclast differentiation induced by sRANKL. In addition, sRANKL-induced osteoclast differentiation was markedly inhibited by overexpression of dominant-negative JNK1 or dominant-negative c-Jun. Furthermore, suppression of c-Jun or MKK7 expression using siRNA technology dramatically inhibited osteoclastogenesis. Collectively, our genetic and biochemical results indicate that the activation of c-Jun is indispensable for RANKL-dependent osteoclast differentiation of the progenitor cells.
Recent studies demonstrated that RANKL induced NFAT2 expression in BMMΦ or RAW264 cells, and showed the necessity of NFAT2 in osteoclastogenesis in vitro (24, 25). Most notably, introduction of NFAT2 promoted the differentiation of BMMΦ cells into TRAP+ osteoclast-like cells even in the absence of sRANKL (25). These findings provided evidence that NFAT2 is an important transcription factor for osteoclast formation. To advance these previous studies, we investigated the molecular basis by which NFAT regulates osteoclastogenesis. We show here that overexpression of NFAT1, which is expressed in both osteoclast progenitors and mature osteoclasts, induces expression of NFAT2 and osteoclast differentiation of RAW264 cells. Consistent with our results, Zhou et al. reported regulation of the NFAT2 gene promoter by NFAT1 in the T-cell lineage (27). We have also demonstrated that induction of NFAT2 by NFAT1 overexpression or sRANKL treatment was markedly diminished in spleen cells isolated from TRAP-DN–c-Jun TG mice. It is therefore likely that NFAT1 functions as the upstream regulator for NFAT2 during osteoclastogenesis and that c-Jun is necessary for NFAT1 to induce NFAT2 expression. More interestingly, we also clearly showed that osteoclastogenic activity of NFAT1 and NFAT2 was enhanced by overexpression of c-Jun and c-Fos, but inhibited by overexpression of either dominant-negative c-Jun or c-Fos, indicating that the partnership between c-Jun/c-Fos and the NFAT family is crucial for osteoclast differentiation. In support of this notion, we demonstrated that overexpression of NFAT1 was unable to promote osteoclastogenesis in the spleen cells of TRAP-DN–c-Jun TG mice. Collectively, our results suggest that sequential activation of the NFAT family and cooperation between c-Jun and the NFAT family are important for RANKL-induced osteoclast differentiation.
The NFAT family is well known to be controlled by calcineurin, which dephosphorylates the NFAT family and subsequently induces its nuclear localization (20, 28). We have provided evidence that the RANKL/TRAF6 signal activates NFAT function during osteoclast differentiation. First, treatment with sRANKL induced nuclear localization of NFAT1 in RAW264 cells. Second, treatment with sRANKL or overexpression of TRAF6 upregulated the transcriptional activity of NFAT1. Recently, two groups reported that, in osteoclasts, RANKL elicits intracellular Ca2+ elevation (25, 29), which is necessary for activation of calcineurin (22, 28). Taken together, it seems likely that RANKL activates the NFAT family through TRAF6 and calcineurin. To support this notion, Koga et al. reported the cooperative role of immunoreceptor tyrosine-based activation motif-harboring adapters with RANKL signaling (30). The discovery of a protein kinase that links TRAF6 to calcineurin might establish the mechanism by which RANKL controls the NFAT family.
Identification of the NFAT family as important transcription factors for osteoclastogenesis provides a new therapeutic target for bone diseases such as osteoporosis or rheumatoid arthritis. As VIVIT peptide has been developed as an optimized peptide that inhibits the interaction between the NFAT family and calcineurin but does not affect the phosphatase activity of calcineurin (23, 28) the small peptide composed of only 16 amino acids is a more specific inhibitor for the NFAT family than FK506 or cyclosporin A. Indeed, we have found that overexpression of GFP-VIVIT successfully blocks osteoclastogenesis induced by sRANKL. Although an appropriate drug delivery system into osteoclast progenitor cells must be developed, the peptide could be a useful and powerful agent for bone diseases associated with bone destruction by osteoclasts.
In conclusion, we have established an essential role for c-Jun signaling in RANKL-induced osteoclast differentiation. We have also shown the crucial role of c-Jun in the regulation of expression and function of the NFAT family, which is important for osteoclastogenesis. Thus, we believe that these findings provide new insights into the molecular basis of osteoclastogenesis and a rationale for development of novel therapeutic agents for bone diseases associated with excessive and progressive osteoclastic bone resorption.
Methods
Cell and reagents.
A monocytic cell line, RAW264, and COS-7 cell line were purchased from the RIKEN cell bank (Tsukuba, Ibaraki, Japan), and cultured in α-MEM containing 10% FCS (HyClone Laboratories, Logan, Utah, USA). sRANKL and M-CSF were purchased from PeproTech EC Ltd. (Rocky Hill, New Jersey, USA) and Green Cross Corp. (Osaka, Japan), respectively. Anti-JNK1, anti–phospho-JNK, anti–phospho–c-Jun, and anti–phospho–ATF-2 antibodies were purchased from Cell Signaling Technology, Inc. (Beverly, Massachusetts, USA). Anti–c-Jun, anti–c-Fos, anti-NFAT1, anti-NFAT2, anti-MKK7, anti–ATF-2, and anti–β-actin antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, California, USA).
Constructs and transfection.
Dominant-negative c-Fos cDNA (31) and an NFATx3 luciferase construct (32) were gifts from Charles Vinson (National Cancer Institute, Bethesda, Maryland, USA) and David J. McKean (Mayo Clinic, Rochester, Minnesota, USA), respectively. c-Jun cDNA was obtained from the RIKEN gene bank (Tsukuba, Ibaraki, Japan). NFAT1 cDNA, NFAT2 cDNA, GFP-VIVIT construct, JNK1 cDNA, MKK7-JNK1 cDNA, AP-1 luciferase construct, and the TRAP gene promoter have been described previously (16, 33, 34). To generate a dominant-negative mutant form of c-Jun (14), a PCR product that encoded the C-terminal domain of c-Jun (amino acids 123–334) lacking the transactivation domain was synthesized and then subcloned into pcDNA3 (Invitrogen Corp., Carlsbad, California, USA) tagged with a Flag epitope in the N terminus. To generate dominant-negative JNK1, Lys55 of JNK1 was point-mutated into methionine by performing in vitro mutagenesis. To generate a dominant-negative mutant of ATF-2 (35), a PCR product that encoded the C-terminal domain of ATF-2 (amino acids 108–505) lacking the transactivation domain was synthesized and then subcloned into pcDNA3 (Invitrogen Corp.) tagged with a Myc epitope in the N terminus. The sequences of these mutants were confirmed by DNA sequence analysis. Transfection of RAW264 cells was carried out using Superfect (QIAGEN Inc., Valencia, California, USA) according to the manufacturer’s protocol.
Generation and analysis of transgenic mice.
Dominant-negative c-Jun cDNA tagged with a Flag epitope was fused to 1.8 kb of the mouse TRAP gene promoter (16, 17). The transgenic mice were generated by pronuclear injection methods into mouse strain BDF1 (17). Genomic DNA isolated from the tail was analyzed by PCR and Southern blotting analysis using specific primers or the probes for the transgene, respectively. The expression of transgene was also confirmed by RT-PCR analysis. Bones were fixed in 10% buffered formalin, decalcified, and subjected to staining with H&E or for TRAP activity (17). Spleen cells isolated from wild-type or TRAP-DN–c-Jun TG mice were analyzed by the in vitro osteoclast differentiation assay (11). All the experiments were performed with sex- and age (4- to 6-week-old)-matched mice under protocols approved by the Osaka University Graduate School of Dentistry animal care committee.
RT-PCR analysis.
Total RNA was isolated from brain, kidney, liver, lung, or spleen cells, which were cultured in the absence or presence of M-CSF and sRANKL, and treated with RNase-free DNase (Wako Pure Chemicals Industries, Osaka, Japan). After denaturation of the total RNA at 70°C for 10 minutes, cDNAs were synthesized with oligo-dT primer and reverse transcriptase (QIAGEN Inc.). PCR amplification was performed by using specific primers for dominant-negative c-Jun tagged with a Flag epitope (forward primer: 5′-AAGGACGATGATGACAAGGG-3′; reverse primer: 5′-AGTTGCTGAGGTTGGCGTAG-3′). PCR products were loaded onto agarose gels and stained with ethidium bromide. After PCR products were subcloned into TA-cloning vector (Invitrogen Corp.), DNA sequences of PCR products were determined.
Osteoclast differentiation in vitro.
Bone marrow cells or spleen cells were isolated from C57BL/6 mice (Nihon SLC Co., Hamamatsu, Shizuoka, Japan) and incubated with M-CSF (30 ng/ml) and sRANKL (100 ng/ml) for 6 days (11). After 6 days, the cells were analyzed by TRAP staining (11), and TRAP+ multinucleated cells were counted as osteoclasts (11). BMMΦ cells were generated as described (18) and incubated in the presence of sRANKL (100 ng/ml) and M-CSF (30 ng/ml) for 6 days; then, the TRAP+ multinucleated osteoclast-like cells were counted. RAW264 cells were incubated in the presence of sRANKL (20 ng/ml) for 6 days, and the TRAP+ multinucleated osteoclast-like cells were counted.
Osteoblast differentiation in vitro.
The calvariae were isolated from 2- or 3-day neonatal mice and digested with 0.1% collagenase (Wako Pure Chemicals Industries) and 0.2% dispase (Invitrogen Corp.) for 7 minutes at 37°C. The digested calvariae were sequentially digested four times with 0.1% collagenase and 0.2% dispase for 7 minutes at 37°C. The last three groups of fractionated cells were collected and used as the primary osteoblasts. The cells were incubated with 100 ng/ml bone morphogenetic protein-2 (BMP2) for 7 days, and determined by alkaline phosphatase (ALP) analysis as described previously (36).
Nonspecific esterase staining.
Spleen cells isolated from mice were cultured with M-CSF (10 ng/ml) for 6 days and stained by nonspecific esterase (NSE) staining kit (Sigma-Aldrich, St. Louis, Missouri, USA) according to the manufacturer’s protocol (8).
Immunoblotting analysis.
The cell lysates or nuclear extracts were isolated from BMMΦ or RAW264 cells, and protein concentration was determined as previously described (37). Equivalent amounts of protein were loaded for SDS-PAGE, and immunoblotting was performed using specific antibodies for JNK1, c-Jun, c-Fos, phospho-JNK, phospho–c-Jun, NFAT1, NFAT2, β-actin, and Myc epitope. In all immunoblotting analyses, we also confirmed that equal amounts of proteins were loaded by staining the transferred membranes with Ponceau S.
Generation of adenovirus.
The recombinant adenovirus carrying NFAT1, NFAT2, GFP-VIVIT, c-Jun, c-Fos, or the dominant-negative form of JNK1, c-Jun, c-Fos, or ATF-2 was constructed by homologous recombination between the expression cosmid cassette and the parental virus genome in 293 cells as previously described (38). The viruses were confirmed to retain no proliferative activity in the cells other than 293 cells, because they lack crucial regions for proliferation of adenovirus, E1A, and E1B (39). Titers of the viruses were determined using the modified point assay (39).
Generation of retrovirus.
A retrovirus packaging cell line, platinum-E (Plat-E) (40), was transfected with a retrovirus vector, pMXs (40), carrying constitutively active NFAT1 cDNA. The cultured media were collected 48 hours after transfection and stored as retrovirus stocks. Spleen cells were cultured in the presence of 30 ng/ml M-CSF for 4 days and subsequently infected with retrovirus in the presence of polybrene (4 μg/ml) (Sigma-Aldrich) for 4 hours.
Reporter assay.
The luciferase reporter constructs driven by the TRAP gene promoter or AP-1 or NFAT-responsive elements were described previously (16, 32, 34). Transfection and luciferase assay were performed as described (34).
Oligonucleotide pull-down assay.
Nuclear extracts were incubated with biotinylated oligonucleotide containing the NFAT-binding element present in the TRAP gene promoter (5′-AATCCTCGGAGAAAATGCATCATCTTTCC-3′) for 2 hours. Interaction of the oligonucleotides with NFAT1 protein was determined by immunoblotting using anti-NFAT1 antibody as described previously (37).
Immunocytochemistry.
Cells were washed three times with ice-cold PBS and fixed with 3.8% paraformaldehyde-PBS, permeabilized by incubation for 15 minutes with 0.1% Triton-PBS. The cells were blocked with 1% BSA-PBS, incubated with anti-NFAT1 antibody for 2 hours, and washed six times with 0.1% Triton-PBS, and finally incubated with Rhodamine-conjugated anti-mouse IgG antibody (Jackson ImmunoResearch Laboratories Inc., West Grove, Pennsylvania, USA). The cells were extensively washed with PBS and visualized using a fluorescence microscope (Carl Zeiss Vision GmbH, Hallbergmoos, Germany).
Generation of siRNA.
siRNA constructs were generated in a retroviral vector pSINsi-H1 (Takara Bio Inc., Otsu, Shiga, Japan). The target sequences are as follow: mouse MKK7, 5′-GGAAGAGAATAAGCGCATT-3′ (41); mouse c-Jun, 5′-GTGCCTACGGCTACAGTAA-3′; luciferase, 5′-GTTGGCACCAGCAGCGCAC-3′. These siRNA constructs were transfected into Plat-E cells, and the retroviruses containing the siRNA were infected into mouse spleen cells as described earlier.
Statistical analysis.
Data were analyzed by ANOVA and represent mean ± SD (n = 4). All experiments were performed at least three times with identical results.
Acknowledgments
We thank G. David Roodman for providing the TRAP gene promoter constructs and critical reading of the manuscript, Charles Vinson for providing dominant-negative c-Fos, David J. McKean for providing the NFATx3 luciferase construct, Masahiro Fujii for providing the c-Fos cDNA, and Toshio Kitamura for providing the Plat-E cell line and pMXs vector. This work was supported in part by grants-in-aid for scientific research A 11307041 (to T. Yoneda), B 15390560 (to R. Nishimura), and C 10671739 (to R. Nishimura), a grant-in-aid for scientific research on priority areas B 12137205 (to T. Yoneda), and a grant from the 21st Century COE Program (to T. Yoneda and R. Nishimura), all from the Ministry of Education, Science, Sports, and Culture; and by the Senri Life Science Foundation (to R. Nishimura).
Footnotes
See the related Commentary beginning on page 463.
Nonstandard abbreviations used: activating transcription factor-2 (ATF-2); activator protein-1 (AP-1); alkaline phosphatase (ALP); bone marrow macrophage (BMMΦ); bone morphogenetic protein-2 (BMP2); c-Jun N-terminal kinase (JNK); green fluorescent protein fused to the sequence encoding MAGPHPVIVITGPHEE (GFP-VIVIT); MAPK kinase 7 (MKK7); nonspecific esterase (NSE); nuclear factor of activated T cells (NFAT); platinum-E (Plat-E); receptor activator of NF-κB (RANK); receptor activator of NF-κB ligand (RANKL); small interfering RNA (siRNA); soluble RANKL (sRANKL); tartrate-resistant acid phosphatase (TRAP); TNF receptor–associated factor 6 (TRAF6); TRAP dominant-negative c-Jun transgenic (TRAP-DN–c-Jun TG).
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Suda T, et al. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr. Rev. 1999;20:345–357. doi: 10.1210/edrv.20.3.0367. [DOI] [PubMed] [Google Scholar]
- 2.Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289:1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 3.Wagner EF, Karsenty G. Genetic control of skeletal development. Curr. Opin. Genet. Dev. 2001;11:527–532. doi: 10.1016/s0959-437x(00)00228-8. [DOI] [PubMed] [Google Scholar]
- 4.Yasuda H, et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong YY, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 6.Li J, et al. RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. . Proc. Natl. Acad. Sci. U. S. A. 2000;97:1566–1571. doi: 10.1073/pnas.97.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lomaga MA, et al. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naito A, et al. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells. 1999;4:353–362. doi: 10.1046/j.1365-2443.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 9.Franzoso G, et al. Requirement for NF-kappaB in osteoclast and B-cell development. Genes Dev. 1997;11:3482–3496. doi: 10.1101/gad.11.24.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumoto M, Sudo T, Saito T, Osada H, Tsujimoto M. Involvement of p38 mitogen-activated protein kinase signaling pathway in osteoclastogenesis mediated by receptor activator of NF-kappa B ligand (RANKL) J. Biol. Chem. 2000;275:31155–31161. doi: 10.1074/jbc.M001229200. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi N, et al. Segregation of TRAF6-mediated signaling pathways clarifies its role in osteoclastogenesis. EMBO J. 2001;20:1271–1280. doi: 10.1093/emboj/20.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grigoriadis AE, et al. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994;266:443–448. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- 13.Johnson RS, van Lingen B, Papaioannou VE, Spiegelman BM. A null mutation at the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev. 1993;7:1309–1317. doi: 10.1101/gad.7.7b.1309. [DOI] [PubMed] [Google Scholar]
- 14.Dong Z, et al. AP-1/jun is required for early Xenopus development and mediates mesoderm induction by fibroblast growth factor but not by activin. J. Biol. Chem. 1996;271:9942–9946. doi: 10.1074/jbc.271.17.9942. [DOI] [PubMed] [Google Scholar]
- 15.Lenczowski JM, et al. Lack of a role for Jun kinase and AP-1 in Fas-induced apoptosis. Mol. Cell. Biol. 1997;17:170–181. doi: 10.1128/mcb.17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy SV, et al. Characterization of the mouse tartrate-resistant acid phosphatase (TRAP) gene promoter. J. Bone Miner. Res. 1995;10:601–606. doi: 10.1002/jbmr.5650100413. [DOI] [PubMed] [Google Scholar]
- 17.Schwartzberg PL, et al. Rescue of osteoclast function by transgenic expression of kinase-deficient Src in src–/– mutant mice. Genes Dev. 1997;11:2835–2844. doi: 10.1101/gad.11.21.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi K, et al. Tumor necrosis factor α stimulates osteoclast differentiation by a mechanism independent of the ODF/RANKL-RANK interaction. J. Exp. Med. 2000;191:275–286. doi: 10.1084/jem.191.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett BL, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 21.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells [review] Cell. 2002;109(Suppl.):S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 22.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 23.Aramburu J, et al. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- 24.Ishida N, et al. Large scale gene expression analysis of osteoclastogenesis in vitro and elucidation of NFAT2 as a key regulator. J. Biol. Chem. 2002;277:41147–41156. doi: 10.1074/jbc.M205063200. [DOI] [PubMed] [Google Scholar]
- 25.Takayanagi H, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 26.Zheng C, Xiang J, Hunter T, Lin A. The JNKK2-JNK1 fusion protein acts as a constitutively active c-Jun kinase that stimulates c-Jun transcription activity. J. Biol. Chem. 1999;274:28966–28971. doi: 10.1074/jbc.274.41.28966. [DOI] [PubMed] [Google Scholar]
- 27.Zhou B, et al. Regulation of the murine Nfatc1 gene by NFATc2. J. Biol. Chem. 2002;277:10704–10711. doi: 10.1074/jbc.M107068200. [DOI] [PubMed] [Google Scholar]
- 28.Kiani A, Rao A, Aramburu J. Manipulating immune responses with immunosuppressive agents that target NFAT. Immunity. 2000;12:359–372. doi: 10.1016/s1074-7613(00)80188-0. [DOI] [PubMed] [Google Scholar]
- 29.Komarova SV, Pilkington MF, Weidema AF, Dixon SJ, Sims SM. RANK ligand-induced elevation of cytosolic Ca2+ accelerates nuclear translocation of nuclear factor kappa B in osteoclasts. J. Biol. Chem. 2003;278:8286–8293. doi: 10.1074/jbc.M206421200. [DOI] [PubMed] [Google Scholar]
- 30.Koga T, et al. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature. 2004;428:758–763. doi: 10.1038/nature02444. [DOI] [PubMed] [Google Scholar]
- 31.Moitra J, et al. Life without white fat: a transgenic mouse. Genes Dev. 1998;12:3168–3181. doi: 10.1101/gad.12.20.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hedin KE, et al. δ-opioid receptors expressed by Jurkat T cells enhance IL-2 secretion by increasing AP-1 complexes and activity of the NF-AT/AP-1-binding promoter element. J. Immunol. 1997;159:5431–5440. [PubMed] [Google Scholar]
- 33.Macian F, et al. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 2002;109:719–731. doi: 10.1016/s0092-8674(02)00767-5. [DOI] [PubMed] [Google Scholar]
- 34.Ito M, et al. JSAP1, a novel jun N-terminal protein kinase (JNK)-binding protein that functions as a scaffold factor in the JNK signaling pathway. Mol. Cell. Biol. 1999;19:7539–7548. doi: 10.1128/mcb.19.11.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monzen K, et al. Smads, TAK1, and their common target ATF-2 play a critical role in cardiomyocyte differentiation. J. Cell Biol. 2001;153:687–698. doi: 10.1083/jcb.153.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimura R, et al. Smad5 and DPC4 are key molecules in mediating BMP-2-induced osteoblastic differentiation of the pluripotent mesenchymal precursor cell line C2C12. J. Biol. Chem. 1998;273:1872–1879. doi: 10.1074/jbc.273.4.1872. [DOI] [PubMed] [Google Scholar]
- 37.Nishimura R, Hata K, Harris SE, Ikeda F, Yoneda T. Core-binding factor α1 (Cbfa1) induces osteoblastic differentiation of C2C12 cells without interactions with Smad1 and Smad5. Bone. 2002;31:303–312. doi: 10.1016/s8756-3282(02)00826-8. [DOI] [PubMed] [Google Scholar]
- 38.Hata K, et al. Differential roles of Smad1 and p38 kinase in regulation of peroxisome proliferator–activating receptor γ during bone morphogenetic protein 2-induced adipogenesis. Mol. Biol. Cell. 2003;14:545–555. doi: 10.1091/mbc.E02-06-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyake S, et al. Efficient generation of recombinant adenoviruses using adenovirus DNA-terminal protein complex and a cosmid bearing the full-length virus genome. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1320–1324. doi: 10.1073/pnas.93.3.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 41.Deng Y, Ren X, Yang L, Lin Y, Wu X. A JNK-dependent pathway is required for TNFα-induced apoptosis. Cell. 2003;115:61–70. doi: 10.1016/s0092-8674(03)00757-8. [DOI] [PubMed] [Google Scholar]