Abstract
Guizhi decoction (GZD), a well-known traditional Chinese medicine (TCM) prescription consisting of Ramulus Cinnamomi, Radix Paeoniae Alba, Radix Glycyrrhizae, Fructus Jujubae and Rhizoma Zingiberis Recens, is usually used for the treatment of common colds, influenza, and other pyretic conditions in the clinic. However, the absorbed ingredients and metabolic compounds of GZD have not been reported. In this paper, a method incorporating rapid resolution liquid chromatography (RRLC) with quadrupole-time-of-flight mass spectrometry (Q-TOF-MS) was used to identify ingredients after oral administration of GZD. Identification of the primary components in GZD, drug-containing serum and urine samples was carried out in order to investigate the assimilation and metabolites of the decoction in vivo. By comparing the total ion chromatograms (TICs) of GZD, a total of 71 constituents were detected or characterized. By comparing TICs of blank and dosed rat plasma, a total of 15 constituents were detected and identified as prototypes according to their retention time (tR) and MS, MS/MS data. Based on this, neutral loss scans of 80 and 176 Da in samples of rat plasma and urine helped us to identify most of the metabolites. Results showed that the predominant metabolic pathways of (epi) catechin and gallic acid were sulfation, methylation, glucuronidation and dehydroxylation; the major metabolic pathways of flavone were hydrolysis, sulfation and glucuronidation. Furthermore, degradation, oxidation and ring fission were found to often occur in the metabolism process of GZD in vivo.
Keywords: Guizhi decoction, rapid resolution liquid chromatography with quadrupole-time-of-flight mass spectrometry (RRLC-Q-TOF-MS), identification, metabolites
1. Introduction
Traditional Chinese medicine (TCM) prescriptions, usually made up of several medicinal herbals according to certain mass ratios guided by traditional Chinese medicine theory, have been used for centuries for the treatment of disorders or diseases in ancient and modern China [1,2,3]. It is well-accepted that a great deal of complex ingredients exist in compound Chinese traditional medicine (CCTM) and are effective through their preventive, therapeutic and synergistic effects [4,5]. Furthermore, the efficacy of a CCTM does not simply equal the sum of the efficacies of all active components [6,7]. Therefore, for the sake of investigating the material basis of the preventive and therapeutic effects of CCTM, it is crucial and necessary for us to make the absorbed constituents and metabolites clear in CCTM.
Guizhi decoction (GZD) is a common TCM formula composed of Ramulus Cinnamomi, Radix Paeoniae Alba, Radix Glycyrrhizae, Fructus Jujubae and Rhizoma Zingiberis Recens, which was recorded in a classic clinical TCM book titled Shanghan Lun (Treatise on Febrile Diseases) [8]. In the clinic, GZD is widely used in different systems of the body, including circulation, immunity, procreation, endocrine, digestion, nerves, etc. [9]. Modern pharmacological researches and the clinic experience have proved that GZD usually serve as antibacterial, analgesic, anti-anaphylaxis and antipyretic therapy as well as for the adjustment of enterokinesia, immune function, blood pressure and cardiac autonomic nervous balance [10,11,12]. However, up to now, there has been little reference to the absorbed components and metabolic compounds of GZD in the literature. Research on the Rhizoma Zingiberis Recens, an important drug of GZD, is particularly scarce.
Based on previous study [13,14,15], the constituents of GZD have been tentatively identified. Furthermore, we have focused on detecting and identifying the compounds absorbed in vivo and metabolites in GZD. Here, SD rats were chosen as the experimental animal. Drug-containing serum and urine samples were obtained. Preparation of GZD was conducted according to a certain ratio. Then, RRLC-Q-TOF-MS was used to detect the prototype compounds and metabolites after oral administration of GZD. The related metabolic investigation of GZD could provide useful information for further study.
2. Results
2.1. Analysis the Constituents of GZD
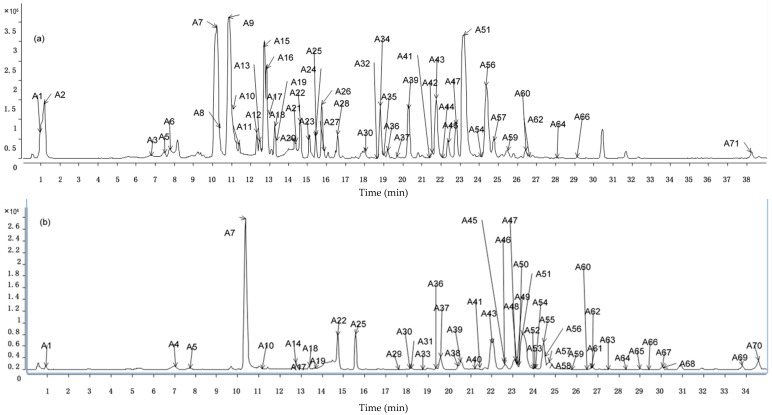
In order to identify the constituents of Guizhi decoction (GZD), the rapid resolution liquid chromatography with quadrupole-time-of-flight mass spectrometry (RRLC-Q-TOF-MS) was used in positive and negative ion modes. Furthermore, the MS data including tR, values of m/z and MS/MS data was got from RRLC-Q-TOF-MS analysis. The total ion chromatograms (TICs) of GZD in electrospray ionization (ESI) negative and positive modes are listed in Figure 1.
Figure 1.
Rapid resolution liquid chromatography with quadrupole-time-of-flight mass spectrometry (RRLC-Q-TOF-MS) chromatograms of prepared Guizhi decoction (GZD). (a) Total ion chromatograms in negative mode; (b) Total ion chromatograms in positive mode.
The structures of most compounds in GZD were tentatively characterized by comparing their TOF-MS data, referring to related literature and comparing reference standards. Finally, a total of 71 compounds including 16 compounds originating from Radix Paeoniae Alba, one from Ramulus Cinnamomi, 33 from Radix Glycyrrhizae, four from Fructus Jujubae and 17 from Rhizoma Zingiberis Recens in GZD were detected by RRLC-Q-TOF-MS in negative or positive ion mode. In addition, 60 compounds of GZD had a known structure, and the others were isomers of part compounds from the explicit structures. The related information about identified components is listed in Table 1.
Table 1.
Identification and detection of chemical compounds of Guizhi decoction (GZD) by rapid resolution liquid chromatography with quadrupole-time-of-flight mass spectrometry (RRLC-Q-TOF-MS).
| No. | tR (min) | Identification | Formula | Negative Ion (m/z) | Positive Ion (m/z) | Source | ||
|---|---|---|---|---|---|---|---|---|
| Quasi-Molecular (ppm) | MS/MS (m/z) | Quasi-Molecular (ppm) | MS/MS (m/z) | |||||
| A1 | 0.945 | cAMP | C10H12N5O6P | 328.0459 | 134.0463 | 330.0590 | 136.0618 | ZJ |
| A2 | 1.194 | Gallic acid | C7H6O5 | 169.0133 | 125.0239 | - | - | P |
| A3 | 6.804 | Epicatechin | C15H14O6 | 289.0728 | 245.0824, 203.0730, 125.0824,109.0306 | - | - | P |
| A4 | 7.035 | Oxypaeoniflorin isomer I | C23H28O12 | - | - | 497.1649 | 197.0811, 179.0701, 151.0751, 121.0285 | P |
| A5 | 7.685 | Catechin | C15H14O6 | 289.0718 | 245.0827, 203.0720, 151.0398 | 291.0858 | 207.0669, 179.0705, 147.0432, 139.0388 | P |
| A6 | 7.762 | Oxypaeoniflorin | C23H28O12 | 495.1508 | 465.1381, 333.0985, 255.0670, 177.0554 | - | - | P |
| A7 | 10.199 | Albiflorin | C23H28O11 | 525.1630 [M+HCOO−H]− | 121.0294 | 481.1706 | 197.0811 | P |
| A8 | 10.391 | Liquiritigenin-7-O-glucoside-4’-O-apiosyl-O-glucoside | C32H40O18 | 711.2131 | 549.1615 | - | - | G |
| A9 | 10.856 | Paeoniflorin | C23H28O11 | 525.1619 [M+HCOO−H]− | 449.1453, 327.1082, 165.0541, 121.0291 | - | - | P |
| A10 | 11.140 | Mudanpioside I | C23H28O11 | 479.1664 | 121.0290 | 481.1703 | 179.0709 | P |
| A11 | 11.405 | Oxypaeoniflorin isomer II | C23H28O12 | 495.1513 | 137.0243 | - | - | P |
| A12 | 12.305 | Naringenin-7-O-glucoside | C21H22O10 | 433.1154 | 271.0612, 151.0034, 119.0503 | - | - | G |
| A13 | 12.423 | Liquiritigenin-7-O-glucoside-4’-O-apiosyl-O-glucoside isomer I | C32H40O18 | 711.2131 | 549.1615 | - | - | G |
| A14 | 12.704 | 4-Shogoal | C15H20O3 | - | - | 249.1500 | 177.0920, 137.0604 | ZR |
| A15 | 12.710 | Liquiritin | C21H22O9 | 417.1189 | 255.0654 | - | - | G |
| A16 | 12.810 | Liquiritin-apioside | C26H30O13 | 549.1589 | 255.0662 | - | - | G |
| A17 | 12.825 | Rutin | C27H30O16 | 609.1451 | 301.1452 | 611.1645 | 303.0517 | ZJ |
| A18 | 13.245 | Galloylpaeoniflorin | C30H32O15 | 631.1660 | 465.1385, 313.0556, 271.0454 | 633.1814 | 315.0705, 179.0700, 153.0179, 127.0390 | P |
| A19 | 13.404 | Kaempferol-3-O-rutinoside | C27H30O15 | 593.1537 | 285.0394 | 595.1698 | 287.0563 | ZJ |
| A20 | 14.277 | Galloylpaeoniflorin isomer I | C30H32O15 | 631.1667 | 477.0935, 271.0604, 121.0290 | - | - | P |
| A21 | 14.351 | Naringenin-5-O-glucoside | C21H22O10 | 433.1154 | 271.0609, 151.0032 | - | - | G |
| A22 | 14.706 | Paeoniflorin isomer I | C23H28O11 | 525.1630 [M+HCOO−H]− | 121.0291 | 481.1709 | 197.0809, 105.0339 | P |
| A23 | 15.056 | Lactiflorin | C23H26O10 | 507.1519 [M+HCOO−H]− | 461.1430, 283.0631, 177.0555, 121.0292 | - | - | P |
| A24 | 15.400 | Lsoliquiritin-apioside | C26H30O13 | 549.1620 | 255.0662 | - | - | G |
| A25 | 15.538 | Ononin | C22H22O9 | 475.1256 [M+HCOO−H]− | 267.0651, 252.0416 | 431.1336 | 269.0809 | G |
| A26 | 15.714 | Lsoliquiritin | C21H22O9 | 417.1150 | 225.0659, 135.0086, 119.0500 | - | - | G |
| A27 | 15.813 | 5-hydroxylliquiritin | C21H22O10 | 433.1146 | 271.0607 | - | - | G |
| A28 | 16.544 | Liquiritigenin | C15H12O4 | 255.0671 | 199.0508, 135.0094 | - | - | G |
| A29 | 17.605 | 10-Gingerdiol | C21H36O4 | - | - | 376.1810 | 259.1711, 137.0609 | ZR |
| A30 | 18.112 | Licorice saponin G2 | C42H62O17 | 837.3917 | 351.0576 | 839.4062 | 663.3724, 469.3306 | G |
| A31 | 18.125 | 8-Gingerdione | C19H28O4 | - | - | 321.2217 | 177.0923, 137.0604 | ZR |
| A32 | 18.638 | Pentagalloylglucose | C41H32O26 | 939.1109 | - | - | - | P |
| A33 | 18.679 | 10-Gingerol | C21H34O4 | - | - | 373.1668 [M+Na]+ | 351.2658, 207.1017, 177.0557, 137.0605 | ZR |
| A34 | 18.781 | Benzoylpaeoniflorin | C41H32O26 | 629.1876, [M+HCOO−H]− | 553.1716, 431.1349, 165.0558, 121.0291 | - | - | P |
| A35 | 18.977 | Apioglycyrrhizin | C42H62O16 | 821.3949 | - | - | - | G |
| A36 | 19.334 | Benzoylpaeoniflorin isomer | C30H32O12 | 629.1881 [M+HCOO−H]− | 121.0293 | 607.1770 [M+Na]+ | 319.1183, 267.0860, 197.0808, 151.0340 | P |
| A37 | 19.542 | Licorice saponin A3 | C48H72O21 | 983.4484 | 821.3958, 351.0559 | 985.4642 | 809.4310, 615.3888, 453.3359 | G |
| A38 | 20.278 | Acetoxyglycyrrhizin acid | C44H64O18 | - | - | 881.4165 | 705.3835, 511.3421 | G |
| A39 | 20.503 | Licorice saponin G2 isomer I | C42H62O17 | 837.3904 | - | 839.4058 | 663.3730, 469.3310 | G |
| A40 | 21.153 | 8-Gingerdiol | C19H32O4 | - | - | 326.1860 | 137.0605 | ZR |
| A41 | 21.354 | Formononetin | C16H12O4 | 267.0661 | 252.0415, 223.5401 | 269.0806 | 254.0850, 225.0557 | G |
| A42 | 21.510 | Isoliquiritigenin | C15H12O4 | 255.0671 | 135.0094, 119.0498 | - | - | G |
| A43 | 21.767 | Licorice saponin E2 | C42H60O16 | 819.3816 | 351.0548 | - | - | G |
| A44 | 21.960 | Licorice saponin G2 isomer II | C42H62O17 | 837.3907 | - | 839.4058 | 645.3617, 469.3312, 451.3203 | G |
| A45 | 22.636 | Licorice saponin G2 isomer III | C42H62O17 | - | - | 839.4060 | 487.3406, 469.3307, 451.3193 | G |
| A46 | 22.636 | 22β-Acetoxyllicoricesaponin C2 | C44H64O17 | 863.4066 | - | 865.4217 | 495.3466 | G |
| A47 | 23.051 | Licorice saponin G2 isomer IV | C44H62O17 | 837.3918 | - | 839.4069 | 487.3415, 469.3307, 451.3215 | G |
| A48 | 23.182 | 6-Paradol | C17H26O3 | - | - | 279.1969 | 177.0940, 163.0755, 145.0661, 137.0601 | ZR |
| A49 | 23.182 | 8-Dehydrogingerdione | C19H26O4 | - | - | 319.1886 | 177.0916, 163.0751, 145.0659, 137.0604 | ZR |
| A50 | 23.208 | 6-Gingerdiol | C17H28O4 | - | - | 297.2057 | 177.0923, 163.0752, 137.0595 | ZR |
| A51 | 23.416 | Glycyrrhizic acid | C42H62O16 | 821.3952 | - | 823.4109 | 647.3780, 453.3368 | G |
| A52 | 23.935 | Licorice saponin G2 isomer IV | C42H62O17 | - | - | 839.4046 | 487.3423, 469.3307, 451.3203 | G |
| A53 | 23.961 | 6-Gingerol | C17H26O4 | - | - | 295.1916 | 163.0757, 137.0605 | ZR |
| A54 | 24.117 | Uralenol | C20H18O7 | 369.1353 | 229.0864, 139.0395 | 371.1493 | 315.0874, 175.0398 | G |
| A55 | 24.403 | 6-Shogaol | C17H24O3 | - | - | 277.1790 | 177.0675, 145.0647, 137.0598 | ZR |
| A56 | 24.482 | LS-K2 | C42H62O16 | 821.3942 | - | 823.4411 | 647.3785, 453.3361 | G |
| A57 | 24.664 | Apioglycyrrhizin | C42H62O16 | 821.3949 | - | 823.4111 | 647.3780, 453.3362 | G |
| A58 | 24.755 | 6-Gingerdione | C17H24O4 | - | - | 293.1773 | 177.0557, 145.0295, 137.0604 | ZR |
| A59 | 25.756 | LS-J2 | C42H64O16 | 823.4130 | 351.0554 | 825.4303 | 453.3377 | G |
| A60 | 26.458 | LS-C2 | C42H62O15 | 805.4300 | 351.0565 | 825.4300 [M+H2O+H]+ | 437.3435, 353.0731 | G |
| A61 | 26.640 | 10-Gingerdione | C21H32O4 | - | - | 349.1784 | 177.0917, 137.0591 | ZR |
| A62 | 26.718 | Glycycoumarin | C21H20O6 | 367.1184 | 309.0402, 297.0405 | 369.1335 | 313.0725, 285.0769 | G |
| A63 | 27.472 | 7-Shogaol | C18H26O3 | - | - | 291.1974 | 177.0918, 137.0606 | ZR |
| A64 | 28.278 | Licoricone | C22H22O6 | 381.1340 | 351.0869, 323.0593 | 383.1508 | 327.0872, 299.0934 | G |
| A65 | 28.954 | 10-Dehydrogingerdione | C21H30O4 | - | - | 347.2188 | 177.0835, 137.0611 | ZR |
| A66 | 29.344 | Isoglycyrol | C21H18O6 | 365.1039 | 307.0247, 295.0247 | 367.1265 | 339.1254, 311.0562 | G |
| A67 | 30.020 | 8-Shogaol | C19H28O3 | - | - | 305.2118 | 177.0921, 137.0604 | ZR |
| A68 | 30.020 | 8-Gingerol | C19H30O4 | - | - | 345.2042 | 177.0908, 137.0596 | ZR |
| A69 | 33.791 | 10-Shogaol | C21H32O3 | - | - | 333.2431 | 177.0910, 137.0595 | ZR |
| A70 | 34.519 | Cinnamic acid | C9H8O2 | - | - | 149.0235 | 121.0283 | C |
| A71 | 38.121 | Oleanolic acid | C30H38O3 | 455.3533 | - | - | - | ZJ |
Note: tR (min): Retention time; P: Radix Paeoniae Alba; G: Radix Glycyrrhizae; C: Ramulus Cinnamomi; ZJ: Fructus Jujubae and Rhizoma; ZR: Zingiberis Recens; -: Not existed.
2.2. Analysis of Prototype Compounds in Plasma Sample
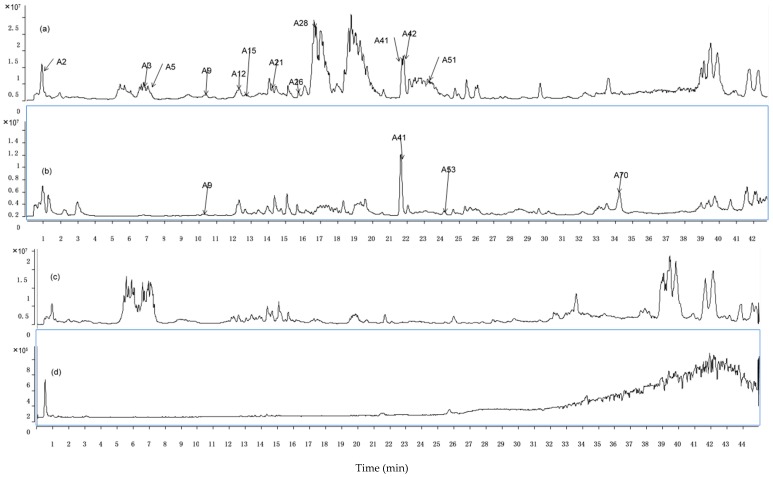
For the sake of analyzing the prototype components in plasma, we compared the TICs of dosed and blank rat blood. Compared with the normal control group, a total of 15 constituents were identified from rat plasma at dosed group according to their TOF-MS data (m/z, MS/MS, tR et al.). Among these 15 compounds, there are five compounds originating from Radix Paeoniae Alba (paeoniflorin, albiflorin, epicatechin, catechin and gallic acid), eight from Radix Glycyrrhizae (liquiritin, isoliquiritin, liquritigenin, isoliqurigenin, glycyrrhizic acid, naringenin-5-O-glucoside, naringenin-7-O-glucoside and formononetin), one from Ramulus Cinnamomi (cinnamic acid) and one from Rhizoma Zingiberis Recens (6-gingerol). The TICs of those 15 compounds and blank rat plasma are shown in Figure 2. In addition, prototype compounds of GZD including its related TOF-MS data can be found in Table 1.
Figure 2.
RRLC-Q-TOF-MS chromatograms of 15 prototype components in dosed and blank rat plasma. (a) Total ion chromatogram (TIC) of dosed rat plasma in negative mode; (b) TIC of dosed rat plasma in positive mode; (c) TIC of blank rat plasma in negative mode; and (d) TIC of blank rat plasma in positive mode.
2.3. Analysis of Metabolites in Blood and Urine Samples
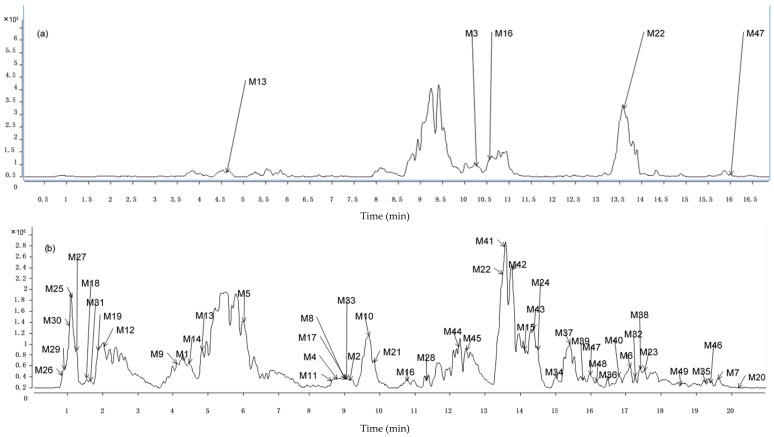
As we know, after intragastric administration of the drug, the compounds originating from the drug were metabolized by intestinal bacteria in the intestine [16]. Then, they were absorbed into plasma so they can be metabolized further by all kinds of drug metabolism enzymes in liver. In principle, there are two metabolic reactions which are called phase I and phase II reactions. Through the phase I reactions including oxidation, reduction, and hydrolysis [17], the prototype components could be converted into aglycone, oxidized aglycone or reduced aglycone. After that, phase II reactions can convert the products of phase I into metabolites. In addition, the phase II reactions were focused on conjugating with glucuronide and sulfate [18,19,20,21]. In order to screen metabolites which were mainly conjugated with glucuronidation and sulfation, we use base peak chromatograms (BPCs) with neutral loss scans of 176 and 80 Da to find compounds existing in the rat plasma (Figure 3a) and urine samples (Figure 3b) in negative mode. A total of 47 peaks were identified which highly promoted the metabolite profiling of GZD. One metabolite was identified in positive mode. Comparing the TICs and, referring to references [22,23], another four metabolites were detected. The results showed that 52 components were tentatively detected as metabolites of GZD. All of the available information about the metabolites is shown in Table 2 and Table 3.
Figure 3.
Base peak chromatograms (BPCs) with neutral loss scanning of 176 and 80 Da in negative mode to find constituents existing in (a) Rat plasma samples; (b) Rat urine samples.
Table 2.
Identification and detection of metabolites of GZD in rat plasma and urine samples by RRLC-Q-TOF-MS.
| No. | tR (min) | Identification | Formula | Urine | Plasma | Negative Ion (m/z) | Possible Original Compound | Source | |
|---|---|---|---|---|---|---|---|---|---|
| Quasi-Molecular (ppm) | MS/MS (m/z) | ||||||||
| M1 | 4.494 | Catechin-O-glucuronide | C21H22O12 | + | - | 465.0785 | 289.0622, 245.0724 | (Epi) catechin-related | P |
| M2 | 9.026 | 3’-O-Methyl (epi)catechin 5-O-glucuronide | C21H24O12 | + | - | 479.0965 | 303.0976, 175.0176 | (Epi) catechin-related | P |
| M3 | 10.220 | 3’-O-Methyl (epi)catechin 7- or 4’-O-glucuronide | C21H24O12 | - | + | 479.0948 | 303.0876, 175.0242 | (Epi) catechin-related | P |
| M4 | 8.861 | 5-(3,4-Dihydroxyphenyl)-γ-valerolactone glucuronide | C17H20O10 | + | - | 383.0757 | 207.0587, 163.0691 | (Epi) catechin-related | P |
| M5 | 5.949 | 5-(3,4-Dihydroxyphenyl)-γ-valerolactone sulfate | C11H12O7S | + | - | 287.0027 | 207.0580, 163.0685 | (Epi) catechin-related | P |
| M6 | 17.003 | 5-(3-Methoxyl-4-hydroxyphenyl)-valerolactone glucuronide | C18H22O10 | + | - | 397.0907 | 221.0719 | (Epi) catechin-related | P |
| M7 | 19.489 | 5-(3-Methoxyl-4-hydroxyphenyl)-valerolactone sulfate | C12H14O7S | + | - | 301.0187 | 221.0739, 206.0503 | (Epi) catechin-related | P |
| M8 | 9.305 | 5-(3-Hydroxyphenyl)-γ-valerolactone sulfate | C11H12O6S | + | - | 271.0092 | 191.0624 | (Epi) catechin-related | P |
| M9 | 4.276 | 4-Hydroxy phenylpropionic acid sulfate | C9H10O6S | + | - | 244.9955 | 165.0596 | (Epi) catechin-related | P |
| M10 | 9.549 | 3-Hydroxy phenylpropionic acid sulfate | C9H10O6S | + | - | 245.0320 | 165.0846 | (Epi) catechin-related | P |
| M11 | 8.547 | 3,4-Dihydroxy phenylpropionic acid sulfate | C9H10O7S | + | - | 260.9888 | 181.0409, 166.0193 | (Epi) catechin-related | P |
| M12 | 2.022 | 3-Hydroxy-4-methoxy-phenylpropionic acid sulfate | C10H12O7S | + | - | 275.0040 | 195.0717 | (Epi) catechin-related | P |
| M13 | 5.027 | 3-Methoxy-4-hydroxy-phenylpropionic acid sulfate | C10H12O7S | + | + | 275.0042 | 195.0504 | (Epi) catechin-related | P |
| M14 | 4.511 | m-Coumaric acid sulfate | C9H8O6S | + | - | 242.9793 | 163.0324 | (Epi) catechin-related | P |
| M15 | 14.077 | p-Coumaric acid sulfate | C9H8O6S | + | - | 243.0173 | 163.0694 | (Epi) catechin-related | P |
| M16 | 10.666 | Ferulic acid sulfate | C10H10O7S | + | + | 273.0259 | 193.0845 | (Epi) catechin-related | P |
| M17 | 9.057 | 3-Hydroxy-4-methoxyphenylacetic acid sulfate | C9H10O7S | + | - | 261.0254 | 181.0718 | (Epi) catechin-related | P |
| M18 | 1.661 | 3,4-Dihydroxy phenylacetic acid sulfate | C8H8O7S | + | - | 247.0111 | 167.0575 | (Epi) catechin-related | P |
| M19 | 1.713 | Protocatechuic acid-3- or -4-O-sulfate | C7H6O7S | + | - | 232.9967 | 153.0423 | (Epi) catechin-related | P |
| M20 | 20.086 | Benzoyl glucuronide | C13H14O8 | + | - | 297.0939 | 121.0658 | (Epi) catechin-related | P |
| M21 | 9.92 | 3-Hydroxyhipuric acid | C9H9NO4 | + | - | 194.0298 | 150.0462 | (Epi) catechin-related | P |
| M22 | 13.393 | Gallic acid glucuronide | C13H14O11 | + | + | 345.1354 | 169.1232 | Gallic acid-related | P |
| M23 | 17.521 | 4-O-Methylgallic acid | C8H8O5 | + | - | 183.0887 | 168.0197, 139.0268 | Gallic acid-related | P |
| M24 | 14.378 | 4-O-Methylgallic acid glucuronide | C14H16O11 | + | - | 359.1115 | 191.0628, 183.0944 | Gallic acid-related | P |
| M25 | 1.202 | 2-Deoxy-pyrogallol-1-O-sulfate | C6H6O5S | + | - | 188.9718 | 125.0010, 109.0087 | Gallic acid-related | P |
| M26 | 0.845 | Pyrogallol-2-O-sulfate | C6H6O6S | + | - | 204.9665 | 125.0126 | Gallic acid-related | P |
| M27 | 1.254 | Pyrogallol-1-O-sulfate | C6H6O6S | + | - | 204.9665 | 124.9982 | Gallic acid-related | P |
| M28 | 30.462 | 2-O-Methylpyrogallol glucuronide | C13H16O9 | + | - | 315.2336 | 171.1027, 139.1119 | Gallic acid-related | P |
| M29 | 0.888 | 2-O-Methylpyrogallol sulfate | C7H8O6S | + | - | 218.9790 | 139.0419, 124.0168 | Gallic acid-related | P |
| M30 | 1.228 | 1-O-Methylpyrogallol-3-O-sulfate | C7H8O6S | + | - | 218.9809 | 139.0277 | Gallic acid-related | P |
| M31 | 2.021 | 1-O-Methylpyrogallol-2-O-sulfate | C7H8O6S | + | - | 218.9809 | 139.0273 | Gallic acid-related | P |
| M32 | 17.146 | Paeonimetabolin I glucuronide isomer I or II | C16H22O10 | + | - | 373.1311 | 197.1181 | Paeoniflorin-related | P |
| M33 | 8.939 | C10H14O3 sulfate | C10H14O6S | + | - | 260.9913 | 181.0443 | Paeoniflorin-related | P |
| M34 | 14.952 | C10H18O2 glucuronide | C16H26O8 | + | - | 345.1354 | 169.1223 | Paeoniflorin-related | P |
| M35 | 19.135 | Naringenin | C15H12O5 | + | - | 271.0419 | 151.0032 | Naringenin-O-glucoside -related | G |
| M36 | 16.194 | Naringenin-O-glucuronide-O-sulfate | C21H20O14S | + | - | 527.0264 | 447.0951, 351.0192, 271.0628 | Naringenin-O-glucoside-related | G |
| M37 | 15.292 | Naringenin-4’-O-glucuronide | C21H20O11 | + | - | 447.1025 | 271.0887, 175.0160 | Naringenin-O-glucoside-related | G |
| M38 | 17.312 | Naringenin-7-O-glucuronide | C21H20O11 | + | - | 447.1025 | 271.0887, 175.0160, 150.9851 | Naringenin-O-glucoside-related | G |
| M39 | 13.133 | Naringenin-O-sulfate | C15H12O8S | + | - | 350.9982 | 271.0390, 150.9829 | Naringenin-O-glucoside-related | G |
| M40 | 16.690 | Liquiritigenin-O-glucuronide-O-sulfate | C21H20O13S | + | - | 511.0306 | 431.0900, 335.0135 | Liquiritigenin-related | G |
| M41 | 13.386 | 7,4’-Dihydroxyflavone-O-sulfate | C15H10O7S | + | - | 332.9885 | 253.0431 | Liquiritigenin-related | G |
| M42 | 13.619 | Liquiritigenin-4’-O-sulfate | C15H12O7S | + | - | 335.0018 | 255.0569, 134.9995 | Liquiritigenin-related | G |
| M43 | 14.230 | Liquiritigenin-7-O-sulfate | C15H12O7S | + | - | 335.0018 | 255.0658, 135.0088 | Liquiritigenin-related | G |
| M44 | 12.532 | Liquiritigenin-4’-O-glucuronide | C21H20O10 | + | - | 431.0754 | 255.0498, 113.0137 | Liquiritigenin-related | G |
| M45 | 12.924 | Liquiritigenin-7-O-glucuronide | C21H20O10 | + | - | 431.0754 | 255.0499, 113.0132 | Liquiritigenin-related | G |
| M46 | 29.631 | Isoliquiritigenin-O-sulfate | C15H12O7S | + | - | 335.2224 | 255.2111 | Isoliquiritigenin-related | G |
| M47 | 16.023 | Isoliquiritigenin-4’-O-glucuronide | C21H20O10 | + | + | 431.0754 | 255.0672, 113.0247 | Isoliquiritigenin-related | G |
| M48 | 16.441 | Isoliquiritigenin-7-O-glucuronide | C21H20O10 | + | - | 431.0754 | 255.0559, 113.0163 | Isoliquiritigenin-related | G |
| M42 | 13.619 | Liquiritigenin-4’-O-sulfate | C15H12O7S | + | - | 335.0018 | 255.0569, 134.9995 | Liquiritigenin-related | G |
| M43 | 14.230 | Liquiritigenin-7-O-sulfate | C15H12O7S | + | - | 335.0018 | 255.0658, 135.0088 | Liquiritigenin-related | G |
| M44 | 12.532 | Liquiritigenin-4’-O-glucuronide | C21H20O10 | + | - | 431.0754 | 255.0498, 113.0137 | Liquiritigenin-related | G |
| M45 | 12.924 | Liquiritigenin-7-O-glucuronide | C21H20O10 | + | - | 431.0754 | 255.0499, 113.0132 | Liquiritigenin-related | G |
| M46 | 29.631 | Isoliquiritigenin-O-sulfate | C15H12O7S | + | - | 335.2224 | 255.2111 | Isoliquiritigenin-related | G |
| M47 | 16.023 | Isoliquiritigenin-4’-O-glucuronide | C21H20O10 | + | + | 431.0754 | 255.0672, 113.0247 | Isoliquiritigenin-related | G |
| M48 | 16.441 | Isoliquiritigenin-7-O-glucuronide | C21H20O10 | + | - | 431.0754 | 255.0559, 113.0163 | Isoliquiritigenin-related | G |
| M49 | 18.463 | Fomononetin-O-sulfate | C16H12O7S | + | - | 347.0012 | 267.0637 | Fomononetin-related | G |
Note: tR (min): Retention time; P: Radix Paeoniae Alba; G: Radix Glycyrrhizae; C: Ramulus Cinnamomi; ZR: Zingiberis Recens; +: Detected; -: Not detected.
Table 3.
Table 2 supplementary.
| No. | tR (min) | Identification | Formula | Urine | Plasma | Positive Ion (m/z) | Possible Original Compound | Source | |
|---|---|---|---|---|---|---|---|---|---|
| Quasi-molecular (ppm) | MS/MS (m/z) | ||||||||
| M50 | 34.133 | Glycyrrhetinic acid | C30H46O4 | + | + | 471.3544 | 317.2164 | Glycyrrhizin-related | G |
| M51 | 23.150 | 6-Gingerol-O-glucuronide | C23H34O10 | + | - | 471.1015 | 295.0564 | 6-Gingerol-related | ZR |
| M52 | 0.524 | Hippuric acid | C9H8NO3 | + | + | 178.1015 | - | Cinnammic acid-related | C |
Note: tR (min): retention time; G: Radix Glycyrrhizae; C: Ramulus Cinnamomi; ZR: Zingiberis Recens; +: Detected; -: Not detected.
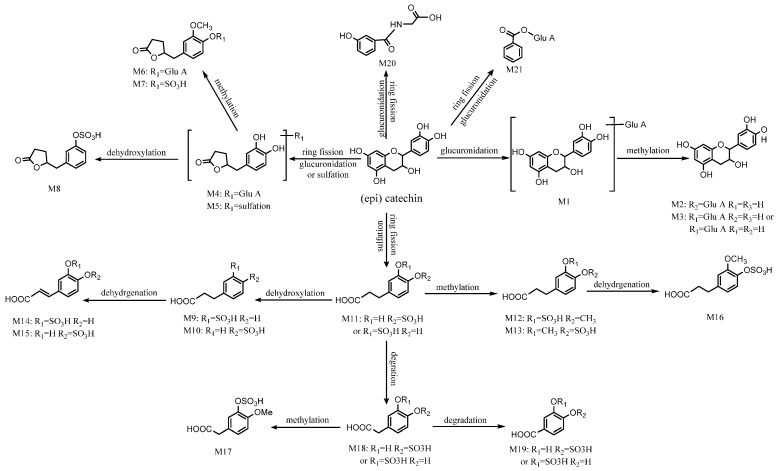
2.3.1. Characterization of (epi) Catechin-Related Metabolites
(Epi) catechin-related metabolites are the main metabolic constituents of Radix Paeoniae Alba. Referring to Liang’s et al. researches [24] and comparing the mass spectrometry data, 21 constituents altogether were tentatively assigned in rat plasma and urine samples as metabolites, which were derived from (epi) catechin. The details are summarized in Table 2. The potential metabolism profile of (epi) catechin-related metabolites is presented in Figure 4.
Figure 4.
The potential metabolic profile of (epi) catechin-related metabolites.
2.3.2. Characterization of Gallic Acid-Related Metabolites
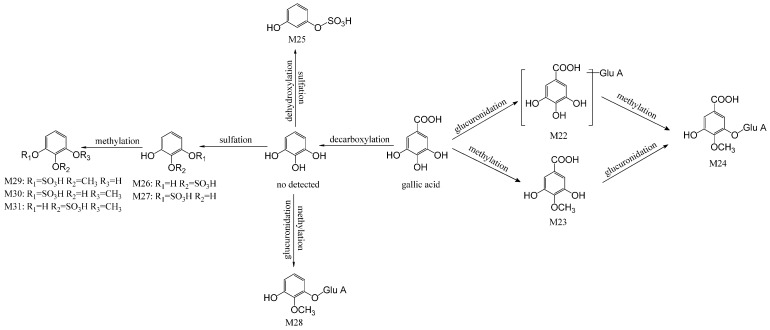
The metabolites of gallic acid-related are also the main metabolites of Radix Paeoniae Alba. By referring to Liang’s et al. and Yan’s et al. studies [24,25], glucuronidation and sulfation after deglycosylation was the principal metabolic pathway of gallic acid. Ten compounds altogether were identified in animal’s plasma and urine samples as metabolites, which were derived from gallic acid. Among them, nine metabolites were characterized as glucuronide conjugates or sulfate conjugates. The available information about gallic acid-related metabolites is presented in Table 2, and a proposed metabolic pathway of gallic acid in rat is displayed in Figure 5.
Figure 5.
The potential metabolic profile of gallic acid-related metabolites.
2.3.3. Characterization of Flavone-Related Metabolites
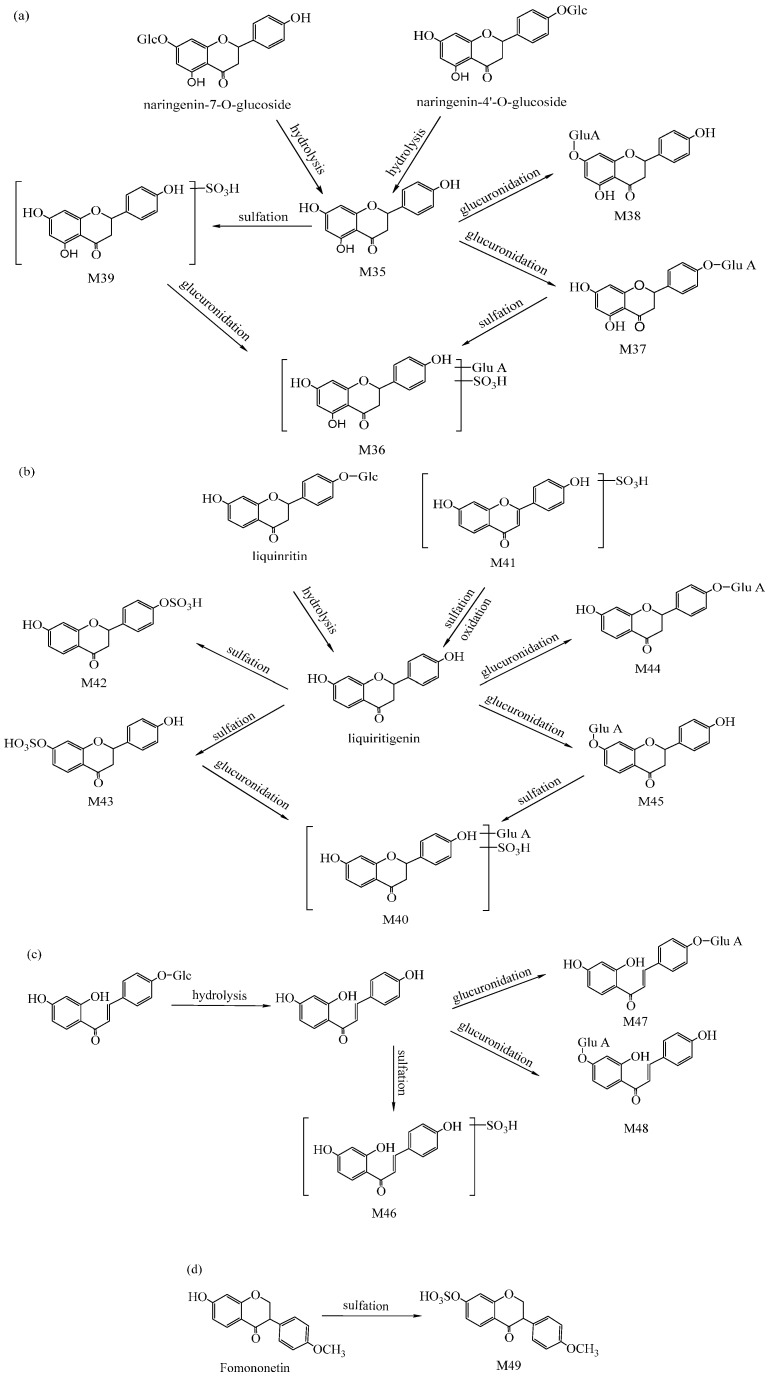
The metabolites of flavone-related are the major metabolites from Radix Glycyrrhizae. There are different kinds of complicated flavone components such as naringenin-O-glucoside, liquiritigenin and fomononetin existing in GZD. Sixteen compounds altogether were assigned as flavone-related metabolites. Among them, six originated from naringenin-O-glucoside-related metabolites, six from liquiritigenin-related metabolites, three from isoliquiritigenin-related metabolites and one from fomononetin-related metabolites. The potential metabolism profile of the main flavone-related metabolites was described as shown in Figure 6, and information about all the flavone-related metabolites is listed in Table 2.
Figure 6.
The potential metabolic profile of flavone-related metabolites. (a) Naringenin-O-glucoside-related metabolites; (b) Liquiritigenin-related metabolites; (c) Isoliquiritigenin-related metabolites; and (d) Formononetin-related metabolite.
2.3.4. Characterization of Other Metabolites
Paeoniflorin-related, cinnamic acid-related and gingerol-related compounds were also the most important metabolites of GZD. There are five metabolites belonging to them, among which three originated from paeoniflorin-related metabolites, one from cinnamic acid-related metabolites and one from gingerol-related metabolites. Further details can be seen in Table 2.
3. Discussion
In present study, we mainly discussed the identification and detection of (epi) catechin-related, gallic acid-related and flavone-related metabolites as they were found to be significant metabolites with high content in GZD. In addition, other metabolites such as gingerol-related metabolites are briefly introduced.
M1 (tR = 4.494 min) showed [M−H]− at m/z 465.0785 in a negative BPC model. The [aglycon−H]− at 289.0622 was observed, conforming to a neutral loss of 176 Da (C6H8O6). In addition, the identification of its major fragment ion at m/z 245.0724 was in line with the related literature [26]. Thus, M1 was preliminary identified as catechin-O-glucuronide and its molecular formula was C21H22O12. M2 and M3 displayed [M−H]− at m/z 479.1170 and 479.1208, respectively. Considering the appearance of the same ions at m/z 303.0860 and 175.0234, M2 and M3 are speculated to be isomers of each other. Furthermore, the loss of 176 Da indicated M2 and M3 were glucuronide conjugates. Compared to M1, catechin-O-glucuronide, M2 and M3 had an additional CH2 unit according to their high resolution mass spectrometer (HRMS) data, which determined their formula to be C22H24O12. Hence, M2 and M3 were identified as Methyl (epi) catechin conjugating with glucuronide. Following a previously research [27], M2 and M3 were tentatively identified as 3’-O-methyl (epi) catechin 5-O-glucuronide and 3’-O-methyl (epi) catechin 7- or 4’-O-glucuronide, respectively. In the same way, another three (epi) catechin-related metabolites conjugating with glucuronide, M4, M6 and M20 were tentatively detected and identified as 5-(3,4-dihydroxyphenyl)-γ-valerolactone glucuronide, 5-(3-methoxyl-4-hydroxyphenyl)-valerolactone glucuronide and benzoyl glucuronide, respectively. M21 (tR = 10.089 min) was determined to be C9H9NO4 according to the speculation of the HRMS data, and its displayed [M−H]− at m/z 194.0298. The main fragment ion at m/z 150.0562 showed a loss of 44 Da which indicated M21 might be an acid. According to a previous report [28], M21 was inferred to be 3-hydroxyhipuric acid. As for the (epi) catechin-related metabolites conjugating with sulfate, the neutral loss of molecular weight was 80 Da. According to this regulation, their HRMS data and related literature [29,30,31], we completely identified 14 compounds which were sulfate conjugates. Altogether, 21 constituents of (epi) catechin-related metabolites were identified in rat blood and urine samples.
M22 displayed [M−H]− at m/z 345.1354, and its ion fragmentation was predominant at m/z 169.1232 which made clear the elimination of glucuronide residue of M22. Combining with the HRMS data, M22 was considered as gallic acid glucuronide and its molecular formula was C13H14O11. Likewise, the identification of another eight metabolites was carried out, including two glucuronide conjugates (M24 and M28) and six sulfate conjugates (M25, M26, M27, M30 and M31). Isomeric compounds were also examined. For the sake of distinguishing between them, a comparison of their retention times and consultation with previous literature [25,32] was performed. M23 displayed [M−H]− at m/z 183.0887. Coincidently, one of the identified gallic acid-related glucuronide conjugates—M24 (4-O-methylgallic acid glucuronide)—showed a main ion fragment at m/z 183.0944. Moreover, the major ion fragment at m/z 183.0944 was the [aglycon−H]− of M24. Therefore, M23 might be 4-O-methylgallic acid. The main fragment ion of M23 at m/z 139.0268 and 168.0197 confirmed that M23 was 4-O-methylgallic acid. In total, there were nine compounds of gallic acid-related metabolites which were tentatively detected.
M36, M37, M38, M39 were chosen as instances to elaborate the process of flavone-related identification. M36 displayed a [M−H]− at m/z 527.0246. M37 and M38 showed the same [M−H]− at m/z 447.1025. M39 showed a [M−H]− at m/z 350.9982. M37 and M38 had the same ion fragments at m/z 271.0887 and 175.0160, which not only indicated M37 and M38 were isomeric but also confirmed that both of them conjugate with glucuronide. Based on HRMS data and related literature [25,33], M37 and M38 were tentatively identified respective as naringenin-4’-O-glucuronide and naringenin-7-O-glucuronide, and their molecular formula was C21H20O11. M39 had major MS/MS ions at m/z 271.0350 and 150.9829, which indicated M39 was a naringenin-related compound. The neutral loss of 80 Da showed that M39 was a sulfate conjugate. Therefore, M39 was tentatively assigned as naringenin-O-sulfate. M36 had its main fragment ions at m/z 447.0951, 351.0192 and 271.0628. This information suggested that M36 was conjugating with both sulfate and glucuronide. In addition, it might be a naringenin-related compound. Therefore, M36 was assigned as naringenin-O-glucuronide-O-sulfate. Altogether, there were 16 components of flavone-related metabolites which were tentatively identified.
M51 showed [M+H]+ at m/z 471.1015. Its major ion fragment was at m/z 295.0564 implying that M51 might be a compound conjugating glucuronide. Therefore, M51 was tentatively identified as 6-gingerol-O-glucuronide. Unfortunately, there are no other gingerol-related even if Rhizoma Zingiberis Recens-related metabolites were detected and identified. Considering there are many components of originated from Rhizoma Zingiberis Recens, the probable reason was that the main components of Rhizoma Zingiberis Recens were hard to metabolize. A previous report deemed that the major components of Rhizoma Zingiberis Recens are volatile oils which are not detectable and retainable with RRLC-Q-TOF-MS analysis [34]. The real reason actually remains unknown.
4. Materials and Methods
4.1. Reagents
Methanol and formic acid of HPLC grade were obtained from Merck (Darmstadt, Germany). Acetonitrile of HPLC grade was obtained from Fisher Scientific (Pittsburgh, PA, USA). Ultra-high purity water was prepared by Milli-Q system (Millipore, Billerica, MA, USA). All other chemicals were commercially available (Beijing Chemical Works, Beijing, China) and were of analytical reagent (AR) grade.
Radix Paeoniae Alba Ramulus Cinnamomi, Radix Glycyrrhizae, and Fructus Jujubae were obtained from Tong-Ren-Tang drugstore (Beijing, China). Rhizoma Zingiberis Recens was purchased from Dong-Zhi-Men vegetable market (Beijing, China). All of the crude drugs were identified by Chunsheng Liu (Academy of Traditional Chinese Medicine, Beijing University of Chinese Medicine, Beijing, China). The standards of Paeoniflorin, liquirituin, cinnamic acid, glycyrrhizic acid were got from National Institute for the Control of Pharmaceutical and Biological Products.
4.2. Rapid Resolution Liquid Chromatography with Quadrupole-Time-of-Flight Mass Spectrometry (RRLC-Q-TOF-MS) Analysis
RRLC-MS system is made up of an Agilent 1260 RRLC system coupled with Agilent 6520 Q-TOF mass spectrometer (6520, Aglient Technologies, Santa Clara, CA, USA). Both positive and negative ion modes were operated by an ESI source (6520, Aglient Technologies). Furthermore, full wavelength scanning analysis over an m/z range of 100–1500 was performed in positive or negative ionization mode. Data acquisition and processing were performed using Mass Hunter Qualitative Analysis B.04.00 software (Aglient Technologies).
The analytical column was operated by an Agilent ZORBAX SB-C18 (2.1 × 50 mm, 1.8 μm). The mobile phase is made up of 0.1% formic acid water (A) and acetonitrile (B). The linear elution gradient was as follows: 0–5 min, 5% B; 5–15 min, 5%–25% B; 15–25 min, 25%–40% B; 25–45 min, 40%–95% B. The injection volume was 2 mL. The flow rate was 0.3 mL/min and column temperature was at 35 °C. The conditions of the mass spectrometer under the ESI mode were as follows: ion spray voltage was 3500 V; N2 as drying gas and its flow rate was 10 L/min. The temperature of N2 350 °C; the pressure of nebulizer was 40 spi. The collision energy was set at 20 V in initial and then changed when necessary.
4.3. Preparation of Guizhi Decoction (GZD)
To prepare the GZD, the crude drugs—namely, Ramulus Cinnamomi (9 g), Radix Paeoniae Alba (9 g), Radix Glycyrrhizae (6 g), Rhizoma Zingiberis Recens (9 g) and Fructus Jujubae (12 pieces)—were immersed in an eight-fold mass of distilled water for 30 min. After that, the mixture decocted for 30 min and filtered. Subsequently, a six-fold mass of distilled water was added to the mixture and decocted for another 30 min. We were combining the two filtrates and making the concentration of crude drug to 1 g/L.
4.4. Animal Housing Environment and Experiments
Fourteen healthy male Sprague-Dawley (SD) rats (200 ± 20 g) used in the experiments were purchased from Vital River Laboratory Animal Co., Ltd. (Beijing, China). They were maintained under standard conditions with light cycles of 12 h on and 12 h off. The room temperature and relative humidity were 23 ± 3 °C and 50% ± 10%, respectively. Principles of laboratory animal care and all protocols were in accordance with the relevant national legislation and local guidelines and were approved by Animal Care and Use Committee of the Institute of Basic Theory for Chinese Medicine, China Academy of Chinese Medical Sciences (Date: 6 August 2014; No.: 201408-16).
After 7 days’ acclimation in the metabolism cages and 12 h fasting with only water ad libitum, animals were used in experiments and randomly divided into two groups. Rats were orally administrated with GZD (10 mL/kg) once a day for two consecutive days as experimental group in Group 1. Rats were orally administrated with an equivalent volume of distilled water as normal control in Group 2. Then, 24 h urine was collected on the 9th day.
On the 9th day after the final oral administration, all rats were anaesthetized with 4% chloral hydrate, and the blood samples were collected from the portal vein. The animal serum of the same group was combined. The serum samples were obtained by centrifugation of blood at 3500 rpm for 15 min and were kept frozen at −80 °C until necessary.
4.5. Sample Preparation
4.5.1. Preparation of GZD
To prepare the test products of GZD, GZD (prepared before, 1 g crude drug per milliliter) of 0.1 mL were placed in 10 mL volumetric flasks, diluted with methanol to volume and filtered with 0.22 µm PTFE membrane.
4.5.2. Preparation of Plasma Samples
Two milliliter plasma was spiked with 6 mL methanol by vortex mixing for 30 s. Then, the mixture was immediately centrifuged for 15 min at 3500 rpm and at 4 °C to obtain the supernatant. The supernatant was shifted and evaporated to dryness. Then the residue was dissolved in 1 mL methanol. After that, the reconstituted extraction was centrifuged again for 15 min at 12,000 rpm and at 4 °C. At last, the supernatants were stored at 4 °C until RRLC-Q-TOF-MS analysis.
4.5.3. Preparation of Urine Samples
Urine samples were evaporated at 65 °C to dryness. The residues were dissolved in 10 mL methanol, and the reconstituted extraction was centrifuged for 15 min at 3500 rpm and at 4 °C to obtain the supernatant. The supernatant was transferred and evaporated to dryness. Then, the residues were thoroughly dissolved in 5 mL methanol. After that, the mixture was filtered with 0.22 µm PTFE membrane. Finally, the filtrates were stored at 4 °C until RRLC-Q-TOF-MS analysis.
5. Conclusions
In this paper, an efficient RRLC-Q-TOF-MS method was used for separation and identification of absorbed constituents and metabolites in rat blood and urine after gavaging GZD. Altogether, 67 constituents comprising 15 prototype compounds and 52 metabolites were detected and tentatively identified in rat urine and plasma samples. In addition, 71 components altogether originating from GZD were detected or characterized. The results showed that phenolic compounds such as gingerol and shogaol were the main constituents of Rhizoma Zingiberis Recens in GZD. Cinnamic acid was the major compound of Ramulus Cinnamomi in GZD. Both phenolic compounds and cinnamic acid were found to be small polor compounds, which can be quickly absorbed into plasma. This feature indicated that GZD can be used for clinical treatment of exterior syndromes such as common colds and pyretic conditions. The major constituents of Radix Paeoniae Alba, Radix Glycyrrhizae and Fructus Jujubae were flavone and saponin. Most of them were absorbed into plasma and were metabolized by all kinds of metabolism enzymes in liver. After two-phase reaction in the liver, they were metabolized to secondary metabolites, which took effect in vivo. Analysis of metabolites showed that (epi) catechin, gallic acid and flavone were the major sources of metabolites, which originated from metabolism of GZD in vivo. Sulfation and glucuronidation were the main metabolic pathways in the metabolic process of GZD in vivo. This study systematically explored the plasma and urine metabolic profiles of GZD. The results of this study can offer essential data for deeper pharmacological and clinical studies in GZD.
Acknowledgments
This work was supported by the Youth Project of National Natural Science Foundation (No. 81403282) and the Natural Science Foundation of Beijing (No. 7133251).
Author Contributions
Hongjun Xiang, Haimin Lei and Dong Bai designed research; Yinglan Nie and Bin Fan performed RRLC-Q-TOF-MS experiment; Lishi Zhang performed animal experiment; Dong Bai, Lishi Zhang and Bin Fan analysis the data; Jiannan Song guided the experiment; Hongjun Xiang wrote the paper; All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Liu S.H., Chuang W.C., Lam W. Safety surveillance of traditional Chinese medicine: Current and future. Drug Saf. 2015;38:117–128. doi: 10.1007/s40264-014-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee T.Y., Chang H.H., Wu M.Y., Lin H.C. Yin-Chen-Hao-Tang ameliorates obstruction-induced hepatic apoptosis in rats. J. Pharm. Pharmacol. 2007;59:583–590. doi: 10.1211/jpp.59.4.0014. [DOI] [PubMed] [Google Scholar]
- 3.Wang H., Feng F. Identification of components in Zhi-Zi-Da-Huang decoction by HPLC coupled with electrospray ionization tandem mass spectrometry, photodiode array and fluorescence detectors. J. Pharm. Biomed. Anal. 2009;49:1157–1165. doi: 10.1016/j.jpba.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 4.Jiang W.Y. Therapeutic wisdom in traditional Chinese medicine: A perspective from modern science. Trends Pharmacol. Sci. 2005;26:558–563. doi: 10.1016/j.tips.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Zhou J., Zhou T.Y., Chen M.J., Jiang M., Wang X.X., Liu Q., Zhan Z., Zhang X. Research progress on synergistic anti-tumor mechanisms of compounds in traditional Chinese medicine. J. Tradit. Chin. Med. 2014;34:100–105. doi: 10.1016/S0254-6272(14)60062-5. [DOI] [PubMed] [Google Scholar]
- 6.Shen L., Cong W.J., Lin X., Hong Y.L., Hu R.W., Feng Y., Xu D.S., Ruan K.F. Characterization using LC/MS of the absorption compounds and metabolites in rat plasma after oral administration of a single or mixed decoction of Shaoyao and Gancao. Chem. Pharm. Bull. 2012;60:712–721. doi: 10.1248/cpb.60.712. [DOI] [PubMed] [Google Scholar]
- 7.Wang X.J. Progress and future developing of the serum pharmacochemistry of traditional Chinese medicine. China J. Chin. Mater. Med. 2006;31:789–792. [PubMed] [Google Scholar]
- 8.Yang X., Peng W.B., Yue X.Q. Syndrome differentiation and treatment of Taiyang disease in Shanghan Lun. J. Chin. Integr. Med. 2009;7:171–174. doi: 10.3736/jcim20090215. [DOI] [PubMed] [Google Scholar]
- 9.Zhang B.G., Liu Q.F. Clinic utilization of Guizhi decoction in modern times. China J. Chin. Mater. Med. 2007;32:757–760. [PubMed] [Google Scholar]
- 10.Katakai M., Tani T. A pair of crude drugs used in Shang-Han-Lun, especially the ways of using roasted licorice. Jpn. J. Hist. Pharm. 2003;38:151–160. [PubMed] [Google Scholar]
- 11.Zhang B.G., Liang X.X., Liu Q.F. Pharmacodynamics research of Guizhi decoction in modern times. China J. Chin. Mater. Med. 2007;32:557–561. [PubMed] [Google Scholar]
- 12.Li X., Jiang Y.H., Jiang P. Effect of Guizhi decoction on heart rate variability and regulation of cardiac autonomic nervous imbalance in diabetes mellitus rats. Chin. J. Integr. Med. 2014;20:524–533. doi: 10.1007/s11655-014-1861-z. [DOI] [PubMed] [Google Scholar]
- 13.Wang S., Chen L., Leng J., Chen P., Fan X., Cheng Y. Fragment ion diagnostic strategies for the comprehensive identification of chemical profile of Gui-Zhi-Tang by integrating high-resolution MS, multiple-stage MS and UV information. J. Pharm. Biomed. Anal. 2014;98:22–35. doi: 10.1016/j.jpba.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 14.Zhou S., Liang H., Cai S.Q., Zhao Y.-Y. Studies on the chemical constituents of the active fraction of GuiZhi decoction. J. Chin. Pharm. Sci. 2007;16:24–26. [Google Scholar]
- 15.Iwabu J., Watanabe J., Hirakura K., Ozaki Y., Hanazaki K. Profiling of the compounds absorbed in human plasma and urine after oral administration of a traditional Japanese (Kampo) medicine, Daikenchuto. Drug Metab. Dispos. 2010;38:2040–2048. doi: 10.1124/dmd.110.033589. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki H., Sugiyama Y. Role of metabolic enzymes and efflux transporters in the absorption of drugs from the small intestine. Eur. J. Pharm. Sci. 2000;12:3–12. doi: 10.1016/S0928-0987(00)00178-0. [DOI] [PubMed] [Google Scholar]
- 17.Moscovitz J.E., Aleksunes L.M. Establishment of metabolism and transport pathways in the rodent and human fetal liver. Int. J. Mol. Sci. 2013;14:23801–23827. doi: 10.3390/ijms141223801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herbert M. Forensic relevance of glucuronidation in phase-II-metabolism of alcohols and drugs. Leg. Med. 2009;11:S22–S26. doi: 10.1016/j.legalmed.2009.01.037. [DOI] [PubMed] [Google Scholar]
- 19.Kampe T., König A., Schroeder H., Hengstler J.G., Niemeyer C.M. Modular microfluidic system for emulation human phase I/phase II metabolism. Anal. Chem. 2014;86:3068–3074. doi: 10.1021/ac404128k. [DOI] [PubMed] [Google Scholar]
- 20.Mullen W., Boitier A., Stewart A.J., Crozier A. Flavonoid metabolites in human plasma and urine after the consumption of red onions: Analysis by liquid chromatography with photodiode array and full scan tandem mass spectrometric detection. J. Chromatogr. A. 2004;1058:163–168. doi: 10.1016/S0021-9673(04)01476-1. [DOI] [PubMed] [Google Scholar]
- 21.Miners J.O., Smith P.A., Sorich M.J., McKinnon R.A., Mackenzie P.I. Predicting human drug glucuronidation parameters: Application of in vitro and in silico modeling approaches. Annu. Rev. Pharmacol. Toxicol. 2004;44:1–25. doi: 10.1146/annurev.pharmtox.44.101802.121546. [DOI] [PubMed] [Google Scholar]
- 22.Lin Z.J., Qiu S.X., Wufuer A., Shum L. Simultaneous determination of glycyrrhizin, a marker component in radix Glycyrrhizae, and its major metabolite glycyrrhetic acid in human plasma by LC–MS/MS. J. Chromatogr. B. 2005;814:201–207. doi: 10.1016/j.jchromb.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y.R., Ma Y.M., Ma W. Pharmacokinetics and bioavailability of cinnamic acid after oral administration of Ramulus Cinnamomi in rats. Eur. J. Drug Metab. Pharmacokinet. 2009;34:51–56. doi: 10.1007/BF03191384. [DOI] [PubMed] [Google Scholar]
- 24.Liang J., Xu F., Zhang Y.Z., Huang S., Zang X.Y., Zhao X., Zhang L., Shang M.Y., Yang D.H., Wang X., Cai S.Q. The profiling and identification of the absorbed constituents and metabolites of Paeoniae Radix Rubra decoction in rat plasma and urine by the HPLC-DAD-ESI-IT-TOF-MSn technique: A novel strategy for the systematic screening and identification of absorbed constituents and metabolites from traditional Chinese medicines. J. Pharm. Biomed. Anal. 2013;83:108–121. doi: 10.1016/j.jpba.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 25.Yan Z., Chen Y., Li T., Zhang J., Yang X. Identification of metabolites of Si-Ni-San, a traditional Chinese medicine formula, in rat plasma and urine using liquid chromatography/diode array detection/triple–quadrupole spectrometry. J. Chromatogr. B. 2012;885:73–82. doi: 10.1016/j.jchromb.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 26.Jack J.W., Rendan B.W., Ferruzzi M.G., Reuhs B.L., Cooper B.R., Harwood J.S., Shulaev V., Pasinetti G., Dixon R.A. Synthesis and Quantitative Analysis of Plasma-Targeted Metabolites of Catechin and Epicatechin. J. Agric. Food Chem. 2015;63:2233–2240. doi: 10.1021/jf505922b. [DOI] [PubMed] [Google Scholar]
- 27.Shali N.A., Curtis G.M., Powell A.B. Sulfation of the flavonoids quercetin and catechin by rat liver. Xenobiotica. 1991;21:881–893. doi: 10.3109/00498259109039528. [DOI] [PubMed] [Google Scholar]
- 28.Gonthier M.P., Donovan J.L., Texier O., Felgines C., Remesy C., Scalbert A. Metabolism of dietary procyanidins in rats. Free Radic. Biol. Med. 2003;35:837–844. doi: 10.1016/S0891-5849(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 29.Tourino S., Perez-Jimenez J., Mateos-Martin M.L., Fuguet E., Vinardell M.P., Cascante M., Torres J.L. Metabolites in contact with rat digestive tract after ingestion of a phenolic-rich dietary fiber matrix. J. Agric. Food Chem. 2011;59:5955–5963. doi: 10.1021/jf200159f. [DOI] [PubMed] [Google Scholar]
- 30.Urpi-Sarda M., Garrido I., Monagas M., Gómez-Cordovés C., Medina-Remón A., Andres-Lacueva C., Bartolomé B. Profile of plasma and urine metabolites after the intake of almond [Prunus dulcis (Mill.) DA Webb] polyphenols in humans. J. Agric. Food Chem. 2009;57:10134–10142. doi: 10.1021/jf901450z. [DOI] [PubMed] [Google Scholar]
- 31.Liang J., Xu F., Zhang Y.Z., Zang X.-Y., Wang D., Shang M.-Y., Wang X., Chui D.-H., Cai S.-Q. The profiling and identification of the metabolites of (+)-catechin and study on their distribution in rats by HPLC-DAD-ESI-IT-TOF-MSn technique. Biomed. Chromatogr. 2014;28:401–411. doi: 10.1002/bmc.3034. [DOI] [PubMed] [Google Scholar]
- 32.Yasuda T., Inaba A., Ohmori M., Endo T., Kubo S., Ohsawa K. Urinary metabolites of gallic acid in rats and their radical-scavenging effects on 1, 1-diphenyl-2-picrylhydrazyl radical. J. Nat. Prod. 2000;63:1444–1446. doi: 10.1021/np0000421. [DOI] [PubMed] [Google Scholar]
- 33.Bredsdorff L., Nielsen I.L.F., Rasmussen S.E., Cornett C., Barron D., Bouisset F., Offord E., Williamson G. Absorption, conjugation and excretion of the flavanones, naringenin and hesperetin from α-rhamnosidase-treated orange juice in human subjects. Br. J. Nutr. 2010;103:1602–1609. doi: 10.1017/S0007114509993679. [DOI] [PubMed] [Google Scholar]
- 34.Tan G., Liu M., Dong X., Wu S., Fan L., Qiao Y., Chai Y., Wu H. A strategy for rapid analysis of xenobiotic metabolome of Sini decoction in vivo using ultra-performance liquid chromatography-electrospray ionization quadrupole-time-of-flight mass spectrometry combined with pattern recognition approach. J. Pharm. Biomed. Anal. 2014;96:187–196. doi: 10.1016/j.jpba.2014.03.028. [DOI] [PubMed] [Google Scholar]