Abstract
Naturally occurring resistance-associated substitutions (RASs) can negatively impact the response to direct-acting antivirals (DAAs) agents-based therapies for hepatitis C virus (HCV) infection. Herein, we set out to characterize the RASs in the HCV1b genome from serum samples of DAA-naïve patients in the context of the SINERGIE (South Italian Network for Rational Guidelines and International Epidemiology, 2014) project. We deep-sequenced the NS3/4A protease region of the viral population using the Ion Torrent Personal Genome Machine, and patient-specific majority rule consensus sequence summaries were constructed with a combination of freely available next generation sequencing data analysis software. We detected NS3/4A protease major and minor variants associated with resistance to boceprevir (V36L), telaprevir (V36L, I132V), simeprevir (V36L), and grazoprevir (V36L, V170I). Furthermore, we sequenced part of HCV NS5B polymerase using Sanger-sequencing and detected a natural RAS for dasabuvir (C316N). This mutation could be important for treatment strategies in cases of previous therapy failure.
Keywords: hepatitis C virus 1b, resistance-associated substitutions, direct-acting antiviral agents, deep-sequencing
1. Introduction
Treatment of hepatitis C virus (HCV) infection has significantly improved in the past few years after the introduction of direct-acting antivirals (DAAs) agents [1]. While patients treated with pegylated interferon-α (pegIFN-α) plus ribavirin (RBV) reached sustained virological response (SVR) in only 42% of cases [2], clinical trials have demonstrated that combinations including a first-generation DAA and pegIFN-α + RBV are effective in obtaining a SVR in up to 75% of patients infected with HCV genotype 1 (1a and 1b subtypes) [3]. Lately, new interferon-free regimens are able to eradicate HCV infection in more than 90% of patients after 12 weeks of treatment, irrespective of the HCV genotype [4].
However, the targets of these DAAs are viral proteins, and due to the rapid evolution of the virus, viral variants resistant to drugs can emerge and lead to therapy failure [5,6]. For this reason, it is sometimes advised to consider the presence of resistance-associated substitutions (RASs) at therapy initiation, and now importantly also at virological failure, to guide drug selection and for the rational management of HCV infection [1,7,8,9].
In order to prescribe the appropriate treatment with regards to the drug type and length of treatment, determining the correct HCV subtype is crucial [1]. Likewise, the genetic background of different subtypes is also important for the interpretation of RASs [7]. In clinical practice, the infecting subtype is determined by a reverse hybridization line probe assay (LiPA) that distinguishes subtypes based on the heterogeneity in the 5′UTR and core regions, with a misclassification rate of around 11% [10]. Indeed, it has become clear that more accurate subtyping results can be obtained through the analysis of more diverse regions, such as the NS5B gene [11,12,13]. Recently, Di Maio et al. [14] reported that HCV NS3 sequencing, followed by phylogenetic analysis, also improves the specificity and sensitivity of subtype assignments.
The large diversity of HCV [15] complicates the bulk sequencing of viral populations with Sanger-sequencing. Furthermore, population sequencing can only reliably detect variants that make up around 20% of the viral population [16], which makes this a suboptimal approach for assessing the impact of lower-frequency RASs on therapy outcome [9]. These limitations make massively parallel sequencing methods an attractive alternative for the study of complex virus populations [17]. Indeed, next-generation sequencing (NGS) has been successfully applied in a wide range of HCV analyses, including characterization of virus transmission [18] and positions of mutation (hot spot) in NS3/4A protease [19].
The vast amount of data produced by deep sequencing platforms, each with its particular characteristics, stimulated bioinformatics developments to efficiently translate the raw sequence data into interpretable information [20,21]. The choice of NGS platform and post-processing largely depend on the research purpose. The short read platforms (e.g., the Illumina® sequencers) are currently the most widely used, but the advantages of longer read lengths in reads assembly and the preservation of mutational linkage over longer distances [22,23,24] ensure a place for longer read technologies, such as the Ion Torrent platforms for virus sequencing. As a case in point, the Ion Personal Genome Machine (PGM) Sequencer (Life Technologies, Carlsbad, CA, USA) has recently been used to detect low-level drug resistance variants in HCV and HIV quasispecies [25,26,27,28,29].
Herein, we used the PGM sequencer in concert with a number of freely available analysis software packages [30,31,32,33,34] to generate a baseline resistance profile for eight samples from DAA-naïve patients chronically infected with HCV1b, targeting NS3 protease. Also, the NS5B Sanger sequences, used to genotype/subtype virus, were screened for RASs to polymerase inhibitors.
2. Results
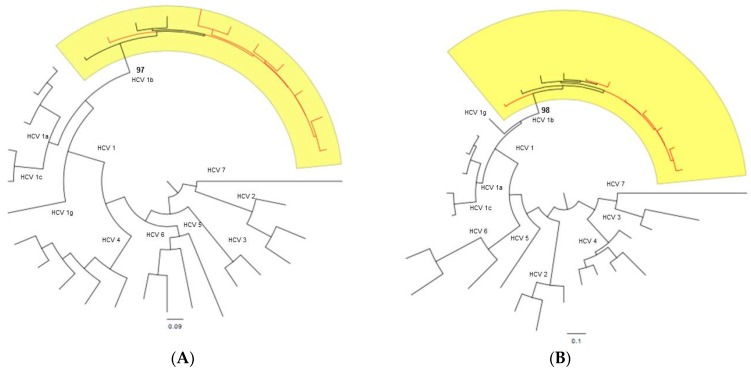
The Versant HCV genotype 2.0 assay classified HCV isolates from all patients as HCV subtype 1b, except for one patient whose genotype was characterized as a mixed genotype 1b/4 (Table 1). In contrast, all samples were classified as genotype 1b by both the Oxford and COMET subtyping tools, and by phylogenetic analysis of the NS3 (Figure 1A) and NS5B (Figure 1B) regions.
Table 1.
Patients’ characteristics.
| Patient ID | Gender | Age | LiPA Genotyping | Liver Stiffness (KPa) * | Risk Factors # | Response Previous Therapy |
|---|---|---|---|---|---|---|
| HCV04 | female | 47 | 1b | 6.0 | Surgery and Cohabitation | Partial responder |
| HCV06 | male | 68 | 1b | 6.5 | Surgery and Cohabitation | Partial responder |
| HCV08 | male | 46 | 1b | 6.0 | Surgery | Relapser |
| HCV09 | male | 62 | 1b/4 | 6.1 | Surgery and Cohabitation | Relapser |
| HCV17 | female | 69 | 1b | 14.0 | Surgery | Naïve |
| HCV19 | male | 45 | 1b | 6.9 | Surgery and Tattoo | Relapser |
| HCV20 | male | 51 | 1b | 22.0 | Surgery | Relapser |
| HCV21 | male | 66 | 1b | 25.0 | Not Available | Partial responder |
* According to Castéra et al. [35], 2005, KPa ≤ 7.1 = F0−F1 (minimal fibrosis), 7.1 < KPa ≤ 9.5 = F2 (moderate fibrosis), 9.5 < KPa ≤ 14.5 = F3 (severe fibrosis), and KPa > 14.5 = F4 (cirrhosis); # Surgery and cohabitation with HCV-antibodies (Ab) positive individuals or tattoo are transmission risks reported by the same patient.
Figure 1.
RAxML phylogenetic trees were estimated using 24 hepatitis C virus (HCV) reference sequences (black) downloaded from Los Alamos HCV Sequence Database and 8 HCV isolates (red) in this study for NS3 (A) and NS5B (B) regions, respectively. The reliability of the phylogenetic clustering was evaluated using bootstrap analysis with 1000 replicates. Bootstrap support values are only shown for the HCV1b clades (yellow area). The scale bars at the bottom of the figure represent genetic distance.
Seven patients were previously treated with pegIFN-α/RBV and were classified as partial responders (3/7) or relapsers (4/7). The eighth patient was naïve to any prior HCV treatment. The median age of the patients was 55 years (interquartile range (IQR): 46.8–66.5), and six out of eight patients were males. Concerning transmission route, surgery was the most frequently reported risk factor (Table 1).
HCV RNA levels during follow-up (scheduled according to official guidelines EASL 2015) [1] and adverse events for each patient are listed in Table 2. The median baseline RNA viral load was 2,130,000 IU/mL. At week 4 of treatment, no HCV RNA was detected for 3/8 samples (HCV04, HCV08, HCV20 patients) while for 4 samples a viremia of <15 IU/mL was measured, and patient HCV21, who was treated with boceprevir (BOC), had an HCV RNA level of 11,900 IU/mL. Viremia was <15 IU/mL at 8 weeks in patient HCV21. All patients had undetectable RNA from week 12 to the end of therapy. Patients were asked to report any adverse events; three patients (HCV09, HCV17, HCV21) reported anemia and one patient (HCV20) reported anemia and neutropenia. The initial liver stiffness was tested by FibroScan® (Table 1) for its association with SVR and the occurrence of adverse events (Table 2). In particular, among our patients: 5/8 (HCV04, HCV06, HCV08, HCV09, HCV19) were classified as Metavir F0−F1, 1 patient (HCV17) was classified as F3, and 2/8 (HCV20, HCV21) were classified as F4 (Table 1).
Table 2.
Direct-acting antiviral (DAA) therapy, HCV RNA viral load, adverse events, and treatment response.
| Patient ID | DAA Therapy * | HCV RNA (IU/mL) | Adverse Events | SVR 12 | |||||
|---|---|---|---|---|---|---|---|---|---|
| – | – | Baseline | 4 Weeks | 12 Weeks | 24 Weeks | 36 Weeks | 48 Weeks | – | – |
| HCV04 | TVR | 7,410,000 | TND | TND | TND | TND | TND | None | Yes |
| HCV06 | TVR | 1,580,000 | <15 | TND | TND | TND | TND | None | Yes |
| HCV08 | TVR | 2,090,000 | TND | TND | TND | TND | TND | None | Yes |
| HCV09 | TVR | 7,930,000 | <15 | TND | TND | TND | TND | Anemia | Yes |
| HCV17 | TVR | 2,170,000 | <15 | TND | TND | TND | TND | Anemia | Yes |
| HCV19 | TVR | 4,290,000 | <15 | TND | TND | TND | TND | None | Yes |
| HCV20 | TVR | 1,520,000 | TND | TND | TND | TND | TND | Anemia and Neutropenia | Yes |
| – | – | Baseline | 4 Weeks | 8 Weeks | 12 Weeks | 24 Weeks | 36 Weeks | – | – |
| HCV21 | BOC | 1,030,000 | 11,900 | <15 | TND | TND | TND | Anemia | Yes |
SVR = Sustained Virological Response; TND = Target Not Detected; * Patients HCV04, HCV06, HCV09, HCV17, HCV19 and HCV20 were treated with pegylated interferon-α plus ribavirin (PegIFN-α/RBV) + telaprevir (TVR) for 12 weeks, afterward with PegIFN-α/RBV for 36 weeks; Patient HCV08 was treated with PegIFN-α/RBV + TVR for 12 weeks, afterward with PegIFN-α/RBV for 12 weeks; Patient HCV21 was treated with PegIFN-α/RBV for 4 weeks, afterward with PegIFN-α/RBV + boceprevir (BOC) for 44 weeks.
We identified several nonsynonymous substitutions in the majority rule NS3 consensus sequences (Table 3), and in minor viral populations. Substitution NS3V36L—conferring resistance to BOC and possibly to telaprevir (TVR) or simeprevir (SMV) [5] or grazoprevir (GZR) [36]—was detected in a treatment-naïve patient’s (HCV17) HCV isolate. We also identified substitution NS3I132V [29], which is associated with possible resistance to TVR, in 4 patients: HCV04, HCV06, HCV08, HCV19. The NS3V170I GZR RAS was found in HCV isolate from patient HCV20 [37] (Table 3). However, for minor substitutions in the NS3 region, nucleotide substitutions were detected at amino acid (AA) positions 55 (10.9%) and 132 (0.05%) in isolates from HCV17 and HCV21 patients, respectively, but these changes did not modify the corresponding AA (i.e., synonymous substitutions). The NS5BC316N mutation associated with resistance to the NS5B polymerase inhibitor, dasabuvir (DSV) [38], was found in two patients (HCV06, HCV19), who also harbored NS3I132V and NS5BC316N (Table 3).
Table 3.
Nonsynonymous substitutions detected for NS3 and NS5B target region in HCV isolates from SVR patients.
| Patient ID | NS3 Protease | NS5B Polymerase | ||
|---|---|---|---|---|
| – | RASs | Polymorphisms | RASs | Polymorphisms |
| HCV04 | I132V | S7A | none | V338A |
| D30E | ||||
| P86Q | ||||
| M94L | ||||
| V114I | ||||
| S122N | ||||
| I170V | ||||
| HCV06 | I132V | L14F | C316N | R254K |
| D30E | – | S300T | ||
| V48I | V338A | |||
| P86Q | – | |||
| P89S | ||||
| M94L | ||||
| V114I | ||||
| S122N | ||||
| V150A | ||||
| I170V | ||||
| HCV08 | I132V | D30E | none | V338A |
| T72I | – | |||
| P86Q | ||||
| M94L | ||||
| V114I | ||||
| V150A | ||||
| I170V | ||||
| HCV09 | none | D30E | none | L266M |
| V48I | V338A | |||
| T72I | – | |||
| P89S | ||||
| M94L | ||||
| H110Y | ||||
| S122N | ||||
| V150A | ||||
| I170V | ||||
| HCV17 | V36L | L14I | none | A252V |
| D30E | Q309R | |||
| V48I | V338A | |||
| S61A | – | |||
| P86Q | ||||
| A87S | ||||
| M94L | ||||
| V114I | ||||
| V150A | ||||
| I170V | ||||
| HCV19 | I132V | L14F | C316N | Q309R |
| D30E | – | S335N | ||
| S61T | V338A | |||
| P86Q | – | |||
| M94L | ||||
| V114I | ||||
| V150A | ||||
| I170V | ||||
| HCV20 | V170I | D30E | none | Q309R |
| S61A | S335N | |||
| P86Q | V338A | |||
| M94L | – | |||
| V114I | ||||
| V150A | ||||
| HCV21 | none | L14F | none | V338A |
| D30E | – | |||
| V48I | ||||
| I71V | ||||
| T95A | ||||
| V114I | ||||
| V150A | ||||
| I170V | ||||
We also found genotype 1b polymorphisms [39], NS5BV338A and NS3D30E + NS3I170V, in all HCV isolates (Table 3). A mutation analysis for the last generation DAAs showed that none of the isolates harbor RASs for NS3/4A protease inhibitor paritaprevir or NS5B polymerase inhibitor sofosbuvir [5] (Table 3).
3. Discussion
In this work we capitalized on the sequence output of the Ion Torrent PGM Sequencer to generate a baseline NS3 resistance profile of eight DAA-naïve patients infected by HCV1b subtype. The NS5B coding region of the same isolates was sequenced with conventional Sanger technology for genotyping purposes, and was also screened for RASs to polymerase inhibitors.
We performed bulk sequencing of virus populations and were able to construct a consensus representation of the intra-host viral population so that secondary variants could also be detected. However, the threshold for clinical relevance of RASs in population sequencing is set at 15%–25% [40]. This contrasts sharply with the excellent in-depth view of the viral population composition that can be achieved with NGS methods, even though replicate analysis is advised to average the effects of random variations in the sample pre-processing procedures [23,41]. We have applied this technique for mutation analysis to a sample of clinical isolates. Although the predictive potential of baseline resistance substitutions remains controversial, RASs have been associated with reduced response to DAA regimens in several studies [5,6]. Using deep sequencing with PGM, we conducted a thorough assessment on HCV NS3 protease major and minor variants in HCV1b isolates from samples of chronic DAA-naïve patients; samples were gathered prior to the telaprevir- or boceprevir-based triple therapy. In addition, once NS5B sequences were obtained by conventional Sanger-sequencing method for subtyping analysis, we also looked for RASs to polymerase nucleotide inhibitors within the dominant viral population [42]. A potential useful application of deep sequencing is that this technique could detect minor variants, whose impact on clinical response to treatment is significant.
We found a dual resistance profile, NS3I132V and NS5BC316N, against TVR (a protease inhibitor) and DSV (a polymerase inhibitor), respectively, in two patients (Table 3). This finding underlines the value of a wider genomic view on resistance testing when multiple viral proteins are targeted [39]. The fact that all patients (including those with RASs) reached SVR with the TVR- or BOC-based triple therapy seems to suggest that other factors, besides the presence of RASs, contribute to the eventual treatment response. The present study adds to the available information regarding the prevalence of baseline HCV RASs and polymorphisms. Furthermore, we recognize that the studied drug combinations remain acceptable in selected patients for whom the new DAAs regimens are not available [1] and further studies involving patients treated with current regimens are needed. Lastly, the sequenced NS5B region, used for subtyping analysis, allowed us to identify NS5BC316N RAS even though a number of important AA positions for resistance against NS5B non-nucleoside inhibitors (i.e., 368, 411, 414, 448, 553, 554, 556) were not amplified by the used primers [43,44].
In the NS3 region, we found two RASs, NS3V36L [45] and NS3I132V [29], in isolates from one patient and four patients, respectively. The NS3V36L variant carried by patient HCV17 confers resistance to BOC and possibly to TVR or SMV and is reported to be present in 0.5% of all previously studied worldwide HCV1b strains [46]. An Italian study of 326 HCV-infected patients treated with TVR/BOC triple therapy, showed this NS3V36L variant in only two HCV1b patients with SVR [14], in line with our observations. The two isolates from HCV06 and HCV19 patients carried mutations for TVR (NS3I132V) and DSV (NS5BC316N). In phase 3 clinical trials, the NS3I132V was reported as RAS only for the HCV1a subtype [45], although NS3132V has recently been described as an HCV1b-resistant variant with a high prevalence at baseline [29]. In particular, NS3I132V variant is present at 0.5% and 73.7% in HCV1a versus HCV1b strains [46]. The NS3V170I GZR RAS, detected at baseline in patient HCV20, has also been reported at treatment failure during a randomized trial of HCV1b-infected patients [37].
As reported in the results section, in some cases (HCV isolates from patients HCV04, HCV06, HCV08, HCV17, and HCV19) RASs were detected but SVR was achieved. This may reflect an incomplete knowledge of the mutation patterns that have a clinical meaningfulness (i.e., predictive value for the risk of virological failure). Unfortunately, our sample size was too small to address these issues with an acceptable statistical power. Moreover, it is possible that host variability (including pharmacogenomical features, such as IL28B single nucleotide polymorphisms) may have favored virological response notwithstanding the presence of resistant virus quasispecies. Unfortunately, however, we did not include any pharmacogenomical evaluations and, again, our sample size was limited. Further studies are needed in this respect.
High-throughput deep sequencing allowed us to determine minor nucleotide variations in the viral population of each patient. The analysis highlighted nucleotide mutations in hot spot positions, ranging 0.05%–10.9%, that were not associated with HCV resistance to DAAs.
Our combination of the PGM and software packages was able to identify low frequency RASs in the viral quasispecies population, which could provide important insights when the methodology used in this study is applied in a larger cohort of patients.
The NS5B variant C316N, known to confer resistance to non-nucleoside inhibitor DSV, is observed in 31.4% of the worldwide HCV1b strains [46], and assessment of this mutation could be important for the treatment strategy in case of TVR failure. Indeed, DSV is approved by the FDA (Food and Drug Administration) in combination with other DAA agents in interferon-free regimens for HCV1, achieving high cure rates with few adverse effects [42]. Therefore, in case of TVR treatment failure, DSV could be contraindicated in the presence of the NS5BC316N variant. Furthermore, we found a substantial number of polymorphisms (Table 3) in the NS3 protease and NS5B polymerase regions, which could be characterized as RASs in the near future. These substitutions could prove to be useful for drug resistance studies [46], and our report adds to the knowledge regarding their general prevalence in Italian HCV isolates. For instance, Chen et al. [9] described the NS3I170V variant, observed in 7/8 of our patients, as BOC RASs in HCV1a, HCV2, HCV3, HCV4 and HCV6 genotypes, while the Geno2pheno team describes them as mutations in a hot spot position not yet related to resistance against BOC in HCV1b strains. We also highlight the polymorphism NS3S122N found in a hot spot position of the NS3 protease region. Lontok et al. recently reported A/G/I/T substitutions in the same amino acid residue at the time of virological failure [5]. Analogous to what has been observed in HIV studies, AA substitutions could be crucial for future analyses and possible correlations with the clinical outcome [47].
4. Materials and Methods
4.1. Ethic Statement
The study was approved by the Ethical Committee (#2012.58.E; 19 June 2013) of the Mater Domini University Hospital of Catanzaro, Italy. Written informed consent was obtained from each patient in accordance with the principles of the Helsinki Declaration (World Medical Association General Assembly, Seoul, Korea, 59 October 2008).
4.2. Study Population
Eight DAA-naïve patients chronically infected with HCV subtype 1b, who were not co-infected with HIV or HBV, were recruited between January and December 2014 at the “Mater Domini” University Hospital of Catanzaro, Italy, as part of the SINERGIE (South Italian Network for Rational Guidelines and International Epidemiology) project [48]. We provide an overview of the patients’ clinical information in Table 1. After baseline sampling and evaluation of initial fibrosis stage, a triple therapy protocol (pegIFN-α and RBV supplemented with telaprevir or boceprevir) was started for each patient [1].
4.3. HCV RNA Viral Load Determination
HCV RNA viral load was determined with the Cobas AmpliPrep/Cobas TaqMan HCV test (Roche Diagnostics, Milan, Italy) (quantification range of 15 to 100 million IU/mL).
4.4. Liver Stiffness
Fibrosis stage was estimated by transient elastometry (FibroScan®), interpreted as follows: KPa ≤ 7.1 = F0−F1 (minimal fibrosis), 7.1 < KPa ≤ 9.5 = F2 (moderate fibrosis), 9.5 < KPa ≤ 14.5 = F3 (severe fibrosis), and KPa > 14.5 = F4 (cirrhosis) [35].
4.5. HCV NS3 Protease Deep Sequencing
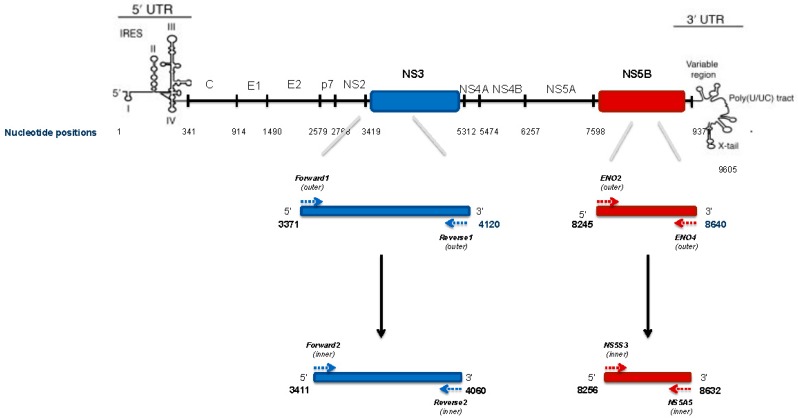
Viral RNA was extracted from 140 µL serum using the QIAmp viral RNA extraction kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s protocol. RNA was reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kits protocol (Applied Biosystems, Foster City, CA, USA). The 2X reverse transcription Master Mix (2 µL 10× RT Buffer, 0.8 µL 25× dNTP Mix (100 mM), 2 µL 10× RT Random Primers, 1 µL MultiScribe™ Reverse Transcriptase, 1 µL RNase Inhibitor and RNase-free water) was added at 10 µL RNA to make up a final volume of 20 µL. Synthesized cDNA was amplified by PCR technique using in-house-developed primers specific for NS3 genomic region (650 bp), covering all NS3/4A positions involved in drug resistance using GoTaq® DNA Polymerase (Promega, Madison, WI, USA). The nested PCR amplification for both rounds was performed under the following conditions: 1.25 units of GoTaq® DNA Polymerase, 50 mM KCl, 30 mM Tris-HCl, 1.5 mM Mg2+, 200 µM of each dNTP, 0.2 µM sense primer, 0.2 µM antisense primer. Six microliters of cDNA was used for the first round of PCR. NS3 primers were the following: Forward1 (outer) 5′-GGAGGGAGATACATCTGG-3′; Reverse1 (outer): 5′-GTTCAGGACAAGCACCTTAT-3′. Thermocycling conditions consisted in a denaturation step at 95 °C for 5 min, followed by 35 cycle at 95 °C for 30 s; 62 °C for 30 s; 72 °C for 1 min; a final elongation cycle of 7 min at 72 °C and finally a hold at 4 °C. Five microliters of the first round of PCR products was used for nested-PCR, using the same thermocycling conditions. NS3 nested primers were the following: Forward2 (inner) 5′-ACTCCTCGCGCCTATTACG-3′; Reverse2 (inner) 5′-TTAGTGCTCTTGCCGCTACC-3′ (Figure 2). In order to obtain enough material for sequencing, the second PCR was performed in duplicate. These PCR products were quantified by semi-fluidic electrophoresis (Agilent® Bioanalyzer®, Santa Clara, CA, USA), using Agilent High Sensitivity DNA Kit and diluted to in order to obtain 100 ng of HCV cDNA in a final volume of 40 µL.
Figure 2.
PCR primers positions and amplicons lenght for NS3 and NS5B HCV genomic regions, according to the HCV1b reference sequence (Con1 AJ238799).
High-throughput sequencing was performed using the Ion Torrent Personal Genome Machine (PGM) Sequencer [49]. The adapter- and barcoded-ligated library was prepared according to manufacturer’s protocol using the Ion Xpress Plus Fragment Library kit (Life Technologies, Foster, CA, USA). Briefly, amplicons were enzymatically fragmented for 5 min, using Ion Shear Reagents. Fragments were then purified by Agencourt® AMPure® XP Reagent and the fragmentation pattern was analyzed by semi-fluidic electrophoresis (Agilent® Bioanalyzer®, Santa Clara, CA, USA) Barcodes and adapters were ligated to the amplicons and library fragments of 300–400 nucleotides long were size selected using E-gel® SizeSelect Agarose Gel (Life Technologies, Carlsbad, CA, USA). Emulsion PCR and Ion sphere particle (ISP) enrichment was performed by using Ion OneTouch 400 Template Kit (Life Technologies). The enrichment of template on Ion Spheres particles (ISPs) was quantified using Qubit System (Life Technologies) and loaded into a 314 chip, according to Ion PGM Hi-Q sequencing kit protocol.
4.6. HCV NS3 NGS Read Analysis
We provide an overview of the sequence read post-processing steps in Figure 3. Briefly, Cutadapt [31] and Sickle tools [30] were used to remove low quality reads, low complexity reads, adapters and primers from fastq raw sequence data. After quality control of raw data using FastQC (Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and pre-processing with the Pollux error correction software [32], a de novo assembly was performed using software packages VICUNA [33] and V-FAT [34]. The sample-specific majority rule consensus sequences were used to infer the variant frequencies with V-phaser 2 [50], and we only considered variants that had no strand bias (p-value ≤ 0.05).
Figure 3.
Workflow of sample-specific consensus sequence of the NS3 region generated by an in-house-developed script.
4.7. Subtyping Analysis
The Versant HCV genotype v2.0 assay (LiPA, Siemens, Healthcare Diagnostic Inc., Tarrytown, NY, USA) was used for initial subtyping. We sought to confirm the subtype assignment by three methods using the NS3 majority rule consensus sequences (cfr. above) and the NS5B population consensus sequence. The NS5B region was amplified as previously described (Figure 2) [43,44] and sequenced by the traditional dideoxy chain termination method using ABI PRISM 3500 genetic analyzer (Applied Biosystems). These were submitted to the Oxford HCV Automated Subtyping Tool v.2.0 [51] and the COMET HCV typing tool [52], and were also used in a maximum likelihood phylogenetic inference incorporating reference sequences available from the Los Alamos National Laboratory HCV Sequence Database (Table S1) [53]. Sequences were aligned with Clustal W [54] and manually edited in MEGA v.5.2.2 [55]. Trees were estimated, inferring with the generalized time reversible (GTR) nucleotide substitution model with gamma (Г)-distribution by RAxML v.8.1.17 [56] and the reliability of phylogenetic clustering was evaluated using 1000 bootstrap replicates. Phylogenetic trees were visualized with FigTree v1.4.2 (Available online: http://tree.bio.ed.ac.uk/software/figtree/).
4.8. Genetic Variability Analysis
We compiled a list of positions known to possibly encode for RASs through a literature search [5,9,29] and with the Geno2pheno[HCV] 0.92 tool [57] using the rules as updated on 21 October 2015. Additionally, the same amino acid (AA) substitutions, aligning generated sequences to HCV1b (accession number AJ238799) wild-type reference by MUSCLE [58], were confirmed. This resulted in the following list of NS3 AA positions: 36, 43, 54, 55, 80, 107, 122, 132, 155, 156, 168, 170, 174, 175; for NS5B we evaluated variation at positions 282, 316, 321.
4.9. Public Availability of the Sequencing Data
The newly generated NS3 and NS5B sequences were submitted to the SINERGIE database (available from ARCA (dbARCA)-Antiviral Response Cohort Analysis (Available online: https://www.dbarca.net/)) and to GenBank® database [59]. All sequences can be retrieved from GenBank® under accession numbers: KX373275-KX373290.
5. Conclusions
In conclusion, we found pretreatment RASs in 6/8 patients. In spite of the presence of RASs, all patients achieved SVR at 12 weeks under TVR or BOC plus pegIFN-α/RBV triple therapy regimens. However, further studies focusing on current treatments are necessary to establish the impact of RASs and other polymorphisms on treatment response. We have determined that the combination of the PGM Sequencer in conjunction with an analysis pipeline consisting of freely available software could be a useful method to analyze genetic variability in the NS3 coding region. Our workflow is readily extensible to a wider genome perspective, but requires benchmarking on simulated data sets and/or larger patient cohorts. This piece of evidence will be extended through implementation of an ongoing international collaboration.
Acknowledgments
Members of the South Italian Network for Rational Guidelines and International Epidemiology (SINERGIE) Study Group at Magna Graecia University (UMG) are: Aida Giancotti and Emilia Zicca (Unit of Clinical Microbiology, School of Medicine, University of Magna Graecia, Catanzaro); Chiara Costa, Vincenzo Pisani, Maria Concetta Postorino and Maria Mazzitelli (Unit of Infectious and Tropical Diseases, School of Medicine, University of Magna Graecia, Catanzaro). Nadia Marascio was supported by a postdoctoral grant of POR (Regional Operating Program) CALABRIA European Social Founding (FSE) 2007/2013. Lize Cuypers was supported by a PhD grant of the FWO (Fonds Wetenschappelijk Onderzoek-Vlaanderen, Asp/12). Tim Dierckx was supported by the Institute for the Promotion of Innovation through Science and Technology Flanders (IWT-Vlaanderen), project 141614. The VIROGENESIS project receives funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 634650.
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/17/9/1416/s1.
Author Contributions
Nadia Marascio corresponding author, performed phylogenetic and mutations analysis, wrote subsequent drafts of the manuscript. Grazia Pavia performed Ion Torrent PGM sequences, and assisted in writing the manuscript. Alessio Strazzulla collected clinical data. Tim Dierckx, Lize Cuypers and Bram Vrancken designed Ion Torrent PGM data analysis methods, and assisted in writing the manuscript. Giorgio Settimo Barreca carried out laboratory analysis. Teresa Mirante, Donatella Malanga, and Duarte Mendes Oliveira performed Ion Torrent PGM sequences, and assisted in writing the manuscript. Anne-Mieke Vandamme assisted in the study design and assisted in writing the manuscript. Carlo Torti, Maria Carla Liberto and Alfredo Focà supervised the design of the study, and assisted extensively in writing the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.European Association for the Study of the Liver EASL recommendation on treatment of hepatitis C 2015. J. Hepatol. 2015;63:199–236. doi: 10.1016/j.jhep.2022.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Manns M.P., McHutchison J.G., Gordon S.C., Rustgi V.K., Shiffman M., Reindollar R., Goodman Z.D., Koury K., Ling M., Albrecht J.K. Peginterferon α-2b plus ribavirin compared with interferon α-2b plus ribavirin for initial treatment of chronic hepatitis C: A randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/S0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 3.Feeney E.R., Chung R.T. Antiviral treatment of hepatitis C. BMJ. 2014 doi: 10.1136/bmj.g3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asselah T., Boyer N., Saadoun D., Martinot-Peignoux M., Marcellin P. Direct-acting antivirals for the treatment of hepatitis C virus infection: Optimizing current IFN-free treatment and future perspectives. Liver Int. 2016;36:47–57. doi: 10.1111/liv.13027. [DOI] [PubMed] [Google Scholar]
- 5.Lontok E., Harrington P., Howe A., Kieffer T., Lennerstrand J., Lenz O., McPhee F., Mo H., Parkin N., Pilot-Matias T., Miller V. Hepatitis C virus drug resistance-associated substitutions: State of the art summary. Hepatology. 2015;61:1623–1632. doi: 10.1002/hep.27934. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed A., Felmlee D.J. Mechanisms of hepatitis c viral resistance to direct acting antivirals. Viruses. 2015;7:6716–6729. doi: 10.3390/v7122968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cento V., Chevaliez S., Perno C.F. Resistance to direct-acting antiviral agents: Clinical utility and significance. Curr. Opin. HIV AIDS. 2015;10:381–389. doi: 10.1097/COH.0000000000000177. [DOI] [PubMed] [Google Scholar]
- 8.Cuypers L., Ceccherini Silberstein F., Van Laethem K., Li G., Vandamme A.M., Rockstroh J.K. Impact of HCV genotype on treatment regimens and drug resistance: A snapshot in time. Rev. Med. Virol. 2016 doi: 10.1002/rmv.1895. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z., Li H., Ren H., Hu P. Global prevalence of pre-existing HCV variants resistant to direct-acting antiviral agents (DAAs): Mining the GenBank HCV genome data. Sci. Rep. 2016;6:20310–20319. doi: 10.1038/srep20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guelfo J.R., Macias J., Neukam K., di Lello F.A., Mira J.A., Merchante N., Mancebo M., Nunez-Torres R., Pineda J.A., Real L.M. Reassessment of genotype 1 hepatitis C virus subtype misclassification by LiPA 2.0: Implications for direct-acting antiviral treatment. J. Clin. Microbiol. 2014;52:4027–4029. doi: 10.1128/JCM.02209-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richter S.S. Laboratory assays for diagnosis and management of hepatitis C virus infection. J. Clin. Microbiol. 2002;40:4407–4412. doi: 10.1128/JCM.40.12.4407-4412.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmonds P. The origin of hepatitis C virus. Curr. Top. Microbiol. Immunol. 2013;369:1–15. doi: 10.1007/978-3-642-27340-7_1. [DOI] [PubMed] [Google Scholar]
- 13.Tong Y.Q., Liu B., Liu H., Zheng H.Y., Gu J., Liu H., Song E.J., Song C., Li Y. Accurate genotyping of hepatitis C virus through nucleotide sequencing and identification of new HCV subtypes in China population. Clin. Microbiol. Infect. 2015;21:9–21. doi: 10.1016/j.cmi.2015.05.034. [DOI] [PubMed] [Google Scholar]
- 14.Di Maio V.C., Cento V., di Paolo D., Aragri M., De Leonardis F., Tontodonati M., Micheli V., Bellocchi M.C., Antonucci F.P., Bertoli A., et al. HCV NS3 sequencing as a reliable and clinically useful tool for the assessment of genotype and resistance mutations for clinical samples with different HCV-RNA levels. J. Antimicrob. Chemother. 2016;71:739–750. doi: 10.1093/jac/dkv403. [DOI] [PubMed] [Google Scholar]
- 15.Smith D.B., Bukh J., Kuiken C., Muerhoff A.S., Rice C.M., Stapleton J.T., Simmonds P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology. 2014;59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergfors A., Leenheer D., Bergqvist A., Ameur A., Lennerstrand J. Analysis of hepatitis C NS5A resistance associated polymorphisms using ultra deep single molecule real time (SMRT) sequencing. Antivir. Res. 2016;126:81–89. doi: 10.1016/j.antiviral.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Vrancken B., Lequime S., Theys K., Lemey P. Covering all bases in HIV research: unveiling a hidden world of viral evolution. AIDS Rev. 2010;12:89–102. [PubMed] [Google Scholar]
- 18.Zhao Q., Wen Y., Jiang Y., Zhang C., Li Y., Zhang G., Zhang L., Qiu M. Next generation sequencing-based investigation of potential patient-to-patient hepatitis C virus transmission during hemodialytic treatment. PLoS ONE. 2016;11:1416. doi: 10.1371/journal.pone.0147566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogishi M., Yotsuyanagi H., Tsutsumi T., Gatanaga H., Ode H., Sugiura W., Moriya K., Oka S., Kimura S., Koike K. Deconvoluting the composition of low-frequency hepatitis C viral quasispecies: Comparison of genotypes and NS3 resistance-associated variants between HCV/HIV coinfected hemophiliacs and HCV monoinfected patients in Japan. PLoS ONE. 2015;10:1416. doi: 10.1371/journal.pone.0119145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barzon L., Lavezzo E., Militello V., Toppo S., Palu G. Applications of next-generation sequencing technologies to diagnostic virology. Int. J. Mol. Sci. 2011;12:7861–7884. doi: 10.3390/ijms12117861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brister J.R., Ako-Adjei D., Bao Y., Blinkova O. NCBI Viral Genomes Resource. Nucleic Acids Res. 2014;43:571–577. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capobianchi M.R., Giombini E., Rozera G. Next-generation sequencing technology in clinical virology. Clin. Microbiol. Infect. 2013;19:15–22. doi: 10.1111/1469-0691.12056. [DOI] [PubMed] [Google Scholar]
- 23.Poon A.F., Swenson L.C., Dong W.W., Deng W., Kosakovsky Pond S.L., Brumme Z.L., Mullins J.I., Richman D.D., Harrigan P.R., Frost S.D. Phylogenetic analysis of population-based and deep sequencing data to identify coevolving sites in the nef gene of HIV-1. Mol. Biol. Evol. 2010;27:819–832. doi: 10.1093/molbev/msp289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zagordi O., Däumer M., Beisel C., Beerenwinkel N. Read length versus depth of coverage for viral quasispecies reconstruction. PLoS ONE. 2012;7:1416. doi: 10.1371/journal.pone.0047046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang M.W., Oliveira G., Yuan J., Okulicz J.F., Levy S., Torbett B.E. Rapid deep sequencing of patient-derived HIV with ion semiconductor technology. J. Virol. Methods. 2013;189:232–234. doi: 10.1016/j.jviromet.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akuta N., Suzuki F., Seko Y., Kawamura Y., Sezaki H., Suzuki Y., Hosaka T., Kobayashi M., Hara T., Kobayashi M., et al. Emergence of telaprevir-resistant variants detected by ultra-deep sequencing after triple therapy in patients infected with HCV genotype 1. J. Med. Virol. 2013;85:1028–1036. doi: 10.1002/jmv.23579. [DOI] [PubMed] [Google Scholar]
- 27.Gibson R.M., Weber J., Winner D., Miller M.D., Quinones-Mateu M.E. Contribution of human immunodeficiency virus type 1 minority variants to reduced drug susceptibility in patients on an integrase strand transfer inhibitor-based therapy. PLoS ONE. 2014;9:1416. doi: 10.1371/journal.pone.0104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akuta N., Suzuki F., Fukushima T., Kawamura Y., Sezaki H., Suzuki Y., Hosaka T., Kobayashi M., Hara T., Kobayashi M., et al. Utility of detection of telaprevir-resistant variants for prediction of efficacy of treatment of hepatitis C virus genotype 1 infection. J. Clin. Microbiol. 2014;52:193–200. doi: 10.1128/JCM.02371-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann L., Faffe D.S., Lima J.F.C., Capitanio T.A., Cabral B.C.A., Urmenyi T.P., Coelho H.S., Rondinelli E., Villela-Nogueira C.A., Silva R. No correspondence between resistance mutations in the HCV-NS3 protease at baseline and early telaprevir-based triple therapy. BBA Clin. 2015;3:146–151. doi: 10.1016/j.bbacli.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joshi N.A., Fass J.N. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) [(accessed on 1 June 2016)]. Available online: http://www.oalib.com/references/15483271.
- 31.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 32.Marinier E., Brown D.G., McConkey B.J. Pollux: Platform independent error correction of single and mixed genomes. BMC Bioinform. 2015 doi: 10.1186/s12859-014-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X., Charlebois P., Gnerre S., Coole M.G., Lennon N.J., Levin J.Z., Qu J., Ryan E.M., Zody M.C., Henn M.R. De novo assembly of highly diverse viral populations. BMC Genomics. 2012;13:1416. doi: 10.1186/1471-2164-13-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charlebois P., Yang X., Newman R.M., Henn M.R., Zody M.C. V-FAT: A post-assembly pipeline for the finishing and annotation of viral genomes. [(accessed on 1 June 2016)]. Available online: http://www.broadinstitute.org/scientific-community/science/projects/viral-genomics/v-fat.
- 35.Castera L., Vergniol J., Foucher J., Le Bail B., Chanteloup E., Haaser M., Darriet M., Couzigou P., de Ledinghen V. Prospective comparison of transient elastography, Fibrotest, APRI, and liver biopsy for the assessment of fibrosis in chronic hepatitis C. Gastroenterology. 2005;128:343–350. doi: 10.1053/j.gastro.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 36.Howe A.Y., Black S., Curry S., Ludmerer S.W., Liu R., Barnard R.J., Newhard W., Hwang P.M., Nickle D., Gilbert C. Virologic resistance analysis from a phase 2 study of MK-5172 combined with pegylated interferon/ribavirin in treatment-naive patients with hepatitis C virus genotype 1 infection. Clin. Infect. Dis. 2014;59:1657–1665. doi: 10.1093/cid/ciu696. [DOI] [PubMed] [Google Scholar]
- 37.Zeuzem S., Ghalib R., Reddy K.R., Pockros P.J., Ben Ari Z., Zhao Y., Brown D.D., Wan S., di Nubile M.J., Nguyen B.Y. Grazoprevir-Elbasvir Combination therapy for treatment-naive cirrhotic and noncirrhotic patients with chronic hepatitis C virus genotype 1, 4, or 6 infection: A randomized trial. Ann. Intern. Med. 2015;163:1–13. doi: 10.7326/M15-0785. [DOI] [PubMed] [Google Scholar]
- 38.Di Maio V.C., Cento V., Mirabelli C., Artese A., Costa G., Alcaro S., Perno C.F., Ceccherini-Silberstein F. Hepatitis C virus genetic variability and the presence of NS5B resistance-associated mutations as natural polymorphisms in selected genotypes could affect the response to NS5B inhibitors. Antimicrob. Agents Chemother. 2014;58:2781–2797. doi: 10.1128/AAC.02386-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuypers L., Snoeck J., Vrancken B., Kerremans L., Vuagniaux G., Verbeeck J., Nevens F., Camacho R.J., Vandamme A.M., Van Dooren S. A near-full length genotypic assay for HCV1b. J. Virol. Methods. 2014;209:126–135. doi: 10.1016/j.jviromet.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Zeuzem S. What do We Need to Know about RAV’s Clinically, Oral presentation; Presented at 14th European Meeting on HIV & Hepatitis-Treatment Strategies & Antiviral Drug Resistance; Rome, Italy. May 2016. [Google Scholar]
- 41.Vrancken B., Trovao N.S., Baele G., van Wijngaerden E., Vandamme A.M., van Laethem K., Lemey P. Quantifying next generation sequencing sample pre-processing bias in HIV-1 complete genome sequencing. Viruses. 2016;8:12–26. doi: 10.3390/v8010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mantry P.S., Pathak L. Dasabuvir (ABT333) for the treatment of chronic HCV genotype I: A new face of cure, an expert review. Expert Rev. Anti Infect. Ther. 2016;14:157–165. doi: 10.1586/14787210.2016.1120668. [DOI] [PubMed] [Google Scholar]
- 43.Ciccozzi M., Equestre M., Costantino A., Marascio N., Quirino A., Lo Presti A., Cella E., Bruni R., Liberto M.C., Foca A., et al. Hepatitis C virus genotype 4d in Southern Italy: Reconstruction of its origin and spread by a phylodynamic analysis. J. Med. Virol. 2012;84:1613–1619. doi: 10.1002/jmv.23384. [DOI] [PubMed] [Google Scholar]
- 44.Marascio N., Ciccozzi M., Equestre M., Lo Presti A., Costantino A., Cella E., Bruni R., Liberto M.C., Pisani G., Zicca E., et al. Back to the origin of HCV2c subtype and spreading to the Calabria region (Southern Italy) over the last two centuries: A phylogenetic study. Infect. Genet. Evol. 2014;26:352–358. doi: 10.1016/j.meegid.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Sullivan J.C., De Meyer S., Bartels D.J., Dierynck I., Zhang E.Z., Spanks J., Tigges A.M., Ghys A., Dorrian J., Adda N., et al. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. Clin. Infect. Dis. 2013;57:221–229. doi: 10.1093/cid/cit226. [DOI] [PubMed] [Google Scholar]
- 46.Cuypers L., Li G., Libin P., Piampongsant S., Vandamme A.M., Theys K. Genetic diversity and selective pressure in hepatitis C virus genotypes 1–6: Significance for direct-acting antiviral treatment and drug resistance. Viruses. 2015;7:5018–5039. doi: 10.3390/v7092857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clavel F., Hance A.J. HIV drug resistance. N. Engl. J. Med. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 48.Torti C., Zazzi M., Abenavoli L., Trapasso F., Cesario F., Corigliano D., Cosco L., Costa C., Curia R.L., De Rosa M., et al. Future research and collaboration: The “SINERGIE” project on HCV (South Italian Network for Rational Guidelines and International Epidemiology) BMC Infect. Dis. 2012 doi: 10.1186/1471-2334-12-S2-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rothberg J.M., Hinz W., Rearick T.M., Schultz J., Mileski W., Davey M., Leamon J.H., Johnson K., Milgrew M.J., Edwards M., et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 50.Yang X., Charlebois P., Macalalad A., Henn M.R., Zody M.C. V-Phaser 2: Variant inference for viral populations. BMC Genom. 2013 doi: 10.1186/1471-2164-14-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Oliveira T., Deforche K., Cassol S., Salminen M., Paraskevis D., Seebregts C., Snoeck J., van Rensburg E.J., Wensing A.M., van de Vijver D.A., et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21:3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 52.Struck D., Lawyer G., Ternes A.M., Schmit J.C., Bercoff D.P. COMET: Adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuiken C., Hraber P., Thurmond J., Yusim K. The hepatitis C sequence database in Los Alamos. Nucleic Acids Res. 2008;36:512–516. doi: 10.1093/nar/gkm962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 57.Kalaghatgi P., Sikorski A.M., Knops E., Rupp D., Sierra S., Heger E., Neumann-Fraune M., Beggel B., Walker A., Timm J., Walter H., et al. Geno2pheno[HCV]—A web-based interpretation system to support hepatitis C treatment decisions in the era of direct-acting antiviral agents. PLoS ONE. 2016;11:1416. doi: 10.1371/journal.pone.0155869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113–132. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benson D.A., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2014;42:32–37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.