Abstract
Tuberculosis is a chronic infectious disease caused by Mycobacterium tuberculosis (Mtb). Due to the extensive use of anti-tuberculosis drugs and the development of mutations, the emergence and spread of multidrug-resistant tuberculosis is recognized as one of the most dangerous threats to global tuberculosis control. Some single mutations have been identified to be significantly linked with drug resistance. However, the prior research did not take gene-gene interactions into account, and the emergence of transmissible drug resistance is connected with multiple genetic mutations. In this study we use the bioinformatics software GBOOST (The Hong Kong University, Clear Water Bay, Kowloon, Hong Kong, China) to calculate the interactions of Single Nucleotide Polymorphism (SNP) pairs and identify gene pairs associated with drug resistance. A large part of the non-synonymous mutations in the drug target genes that were included in the screened gene pairs were confirmed by previous reports, which lent sound solid credits to the effectiveness of our method. Notably, most of the identified gene pairs containing drug targets also comprise Pro-Pro-Glu (PPE) family proteins, suggesting that PPE family proteins play important roles in the drug resistance of Mtb. Therefore, this study provides deeper insights into the mechanisms underlying anti-tuberculosis drug resistance, and the present method is useful for exploring the drug resistance mechanisms for other microorganisms.
Keywords: Mycobacterium tuberculosis, drug resistance, gene pair, GBOOST
1. Introduction
Tuberculosis (TB) is one of most dangerous chronic infectious diseases and it is caused by Mycobacterium tuberculosis (Mtb) infection. Mtb is typically transmitted by aerosols and reaches the lungs, where macrophages and other immune cells are recruited during the early innate response to infection [1,2]. TB has become a significant threat to public health. The World Health Organization (WHO) Global Tuberculosis Report indicated that in 2014, approximate 9.6 million people were carriers of Mtb, and 1.5 million people died of TB [3]. In the early 20th century, with the development of effective antibiotics and vaccines, the deaths caused by Mtb were controlled. However, TB became an ongoing threat to global health in the late 20th century, and the current epidemic is fueled by Human Immunodeficiency Virus (HIV) coinfection and an increasing incidence of drug-resistant strains of Mtb [4]. The extensive treatment courses result in poor compliance and drug resistance, and the emergence of multidrug-resistant strains has become a serious public health threat and represents a new challenge in TB control.
The first TB therapy, streptomycin (STR), was discovered and used to combat TB in 1944 [5]. Soon afterwards, many more anti-TB agents were developed. Drugs employed in TB treatment are classified into five groups, as shown in Table 1 [6]. Group 1 represents the first-line oral agents, which include isoniazid (INH), rifampicin (RMP), ethambutol (EMB), pyrazinamide and rifabutin. Group 2 includes the injectable agents, which comprise aminoglycoside antibiotics (kanamycin (KAN); amikacin; STR) and peptide drugs (capreomycin (CPM)). Group 2 drugs can be used together with group 3 fluoroquinolone drugs, which include ofloxacin, levofloxacin, moxifloxacin and gatifloxacin. Group 4 drugs include the oral bacteriostatic second-line agents: p-aminosalicylic acid, cycloserine, terizidone, ethionamide (ETH) and prothionamide. Group 5 comprises agents with unclear efficacy and primarily includes clofazimine, linezolid and amoxicillin. The group 5 drugs can be used only when none of other groups of drugs can be used for TB treatment [7].
Table 1.
Anti-tuberculosis drug classification, with inclusion of new compounds under evaluation a.
| Group | Drug |
|---|---|
| Group 1: First-line oral agents | isoniazid; rifampicin; ethambutol; pyrazinamide |
| Group 2: Injectable agents | kanamycin; amikacin; capreomycin; streptomycin |
| Group 3: Fluoroquinolones | ofloxacin; levofloxacin; moxifloxacin; gatifloxacin |
| Group 4: Oral bacteriostatic second-line agents | p-aminosalicylic acid; cycloserine; terizidone; ethionamide; protionamide |
| Group 5: Agents with unclear efficacy | clofazimine; linezolid; amoxicillin; imipenem; clarithromycin; thioacetazone |
a According to Reference [6].
In general, the mechanism of anti-TB drugs can be divided into four types: (i) inhibition of RNA synthesis; (ii) inhibition of protein synthesis; (iii) inhibition of cell wall biosynthesis; and (iv) interference with the synthesis of cell membranes [8]. INH is a widely used first-line drug to treat TB and is a prodrug susceptible to oxidative reactions catalyzed by catalase-peroxidase (KatG). The formed activated isonicotinoyl-NAD adduct binds to InhA (enoyl-acyl carrier protein reductase) and inhibits the biosynthesis of mycolic acids, which are components of the mycobacterial cell wall, to induce cell lysis [9]. RMP is another first-line drug for the treatment of TB. RMP interferes with DNA-dependent RNA synthesis by inhibiting bacterial DNA-dependent RNA polymerase [8]. EMB is one of the first-line drugs used in treating TB, and the mechanism of EMB is the inhibition of the transfer of arabinogalactan [10]. The second-line anti-TB injectable drugs, which include STR, KAN and CPM, are aminoglycosides. These drugs exhibit similar mechanisms of antibacterial and metabolic activity by blocking the initiation of protein synthesis [11]. Ofloxacin (OFX) belongs to the fluoroquinolone class of drugs that inhibit DNA gyrase (topoisomerase II) and topoisomerase IV, resulting in microbial death [10]. ETH is an oral bacteriostatic second-line agent and is a structural analog of INH. The two drugs exhibit a similar mechanism of action by inhibiting mycolic acid biosynthesis [12].
Due to the extensive use and long-term treatment with the first-line and second-line anti-TB drugs, strains of Mtb resistant to anti-TB agents have emerged and increased rapidly. The general anti-TB drug resistance-associated genetic regions are summarized and presented in Table 2 [13]. Drug resistance patterns may vary widely from single-drug to multiple-drug resistance, and most transmissible drug resistance is associated with multiple genetic mutations. Therefore, it is critical to seek out and clarify the resistance mechanism of anti-TB agents and to develop a rapid and comprehensive approach to tackle the current ominous situation [14].
Table 2.
Anti-tuberculosis drug resistance-associated genes a.
| Drug | Genes Involved in Resistance |
|---|---|
| Capreomycin (CPM) | rrs, tlyA |
| Ethambutol (EMB) | embA, embB, embC |
| Ethionamide (ETH) | inhA, katG |
| Isoniazid (INH) | katG, inhA, ahpC, fabG, fadE24 |
| Kanamycin (KAN) | rpsL rrs |
| Ofloxacin (OFX) | gyrA, gyrB |
| Rifampicin (RMP) | rpoA, rpoB, rpoC |
| Streptomycin (STR) | gidB, rpsL rrs |
a According to reference [13].
Recently, many research groups have conducted whole genome sequencing on Mtb strains with a range of resistance profiles and have identified some new mutations in genes that are significantly linked with Mtb drug resistance [15,16]. However, the conventional method does not take gene-gene interactions into account. Accumulating evidence reveals that the emergence of transmissible drug-resistant Mtb is the result of a multitude of additional genetic mutations, many of which interact [17]. Therefore, in this paper we performed a bioinformatics analysis to identify gene pairs associated with Mtb drug resistance.
2. Results and Discussion
All of the interactions of Single Nucleotide Polymorphism (SNP) pairs obtained by GBOOST and the screened corresponding gene pairs from the two datasets are predicted (Tables S1 and S2). Because the mutations of target genes are of most significance in Mtb drug resistance [18], we primarily focused on the gene pairs that contained at least one target gene. The drug target information is from the DrugBank database and is listed in Table S3. In dataset 1, katG-PPE54, rpoB-PPE54 gene pairs associated with INH resistance and RMP resistance were identified, respectively (Table 3). In dataset 2, embA-PPE68, embB-PPE54 gene pairs associated with EMB resistance were detected, and katG-PPE54, katG-transcriptional regulator, katG-ccsA, katG-fdxB and katG-oxidoreductase gene pairs associated with ETH-resistance were discovered (Table 4).
Table 3.
The obtained gene pairs containing target genes in dataset 1.
| Gene ID (Target Name) | Gene ID (Gene Name) | SNP1 | SNP2 | Chi-Square Test | Permutation Test |
|---|---|---|---|---|---|
| Rv1908c (katG) a | Rv3343c (PPE54) a | rs_5245 | rs_8894 | 1.69 × 10−3 | <0.0001 |
| Rv0667 (rpoB) b | Rv3343c (PPE54) b | rs_2019 | rs_8894 | 1.28 × 10−4 | <0.0001 |
a associated with isoniazid resistance; b associated with rifampicin resistance.
Table 4.
The obtained gene pairs containing target genes in dataset 2.
| Gene ID (Target Name) | Gene ID (Gene Name) | SNP1 | SNP2 | Chi-Square Test | Permutation Test |
|---|---|---|---|---|---|
| Rv3794 (embA) a | Rv3873 (PPE68) a | rss_17400 | rss_17926 | 1.98 × 10−3 | 0.0016 |
| Rv3795 (embB) a | Rv3343c (PPE54) a | rss_17410 | rss_15182 | 2.29 × 10−3 | <0.0001 |
| Rv3795 (embB) a | Rv3343c (PPE54) a | rss_17411 | rss_15181 | 8.55 × 10−4 | 0.0004 |
| Rv3795 (embB) a | Rv3343c (PPE54) a | rss_17411 | rss_15182 | 2.19 × 10−3 | <0.0001 |
| Rv1908c (katG) b | Rv3343c (PPE54) b | rss_8886 | rss_15184 | 2.16 × 10−3 | 0.0023 |
| Rv1908c (katG) b | Rv0576 (transcriptional regulator) b | rss_8886 | rss_3040 | 1.86 × 10−3 | <0.0001 |
| Rv1908c (katG) b | Rv0529 (ccsA) b | rss_8886 | rss_2852 | 1.78 × 10−3 | 0.0038 |
| Rv1908c (katG) b | Rv3554 (fdxB) b | rss_8886 | rss_16297 | 1.20 × 10−3 | 0.0009 |
| Rv1908c (katG) b | Rv0575c (oxidoreductase) b | rss_8900 | rss_3037 | 1.80 × 10−3 | 0.0009 |
a associated with ethambutol resistance; b associated with ethionamide resistance.
To validate the effectiveness of the screening procedure, we sorted out all of the non-synonymous mutations in the drug targets. Overall, five non-synonymous mutations were found in drug targets; three mutations have experimental or theoretical evidence, and the other two are new discoveries (Table 5). One non-synonymous mutation R463L in the katG target gene associated with INH resistance was screened out in dataset 1 (Table 5). The R463L SNP site of katG was predicted to generate resistance for INH, which had been verified by previous experiments [19]. ETH resistance-associated mutations Y155C and R463L of the katG target gene were sorted from gene pairs of dataset 2, as shown in Table 5. The Y155C mutation was consistent with the results of Zhang et al. [16]. The R463L mutation was consistent with the results of INH resistance-associated mutations in dataset 1. ETH is a structural analog of INH (Figure 1). The structural similarity and the existence of cross-resistant phenotypes have suggested that these two drugs share common molecular targets, i.e., inhA and katG [20]. Therefore, we believe that the R463L mutation would lead to the Mtb resistance to ETH. Two EMB resistance-associated mutations, N399T and G406S, of the embB target gene were selected. The G406S mutation was consistent with the results of Zhang et al. and was verified by previous experiments [21]. Comparison with the results of Zhang et al. indicated that the N399T mutation was a new site identified by our method, possibly because the previous study did not consider the joint mutation of gene pairs, and the new mutation site is worth experimental verification.
Table 5.
The non-synonymous mutations in the target genes from the two datasets.
| Drug | SNP | SNP Loci | Gene ID | Target | Base Mutation | Amino Acid Mutation | Previously Reported |
|---|---|---|---|---|---|---|---|
| INH a | rs_5245 | 2154724 | Rv1908c | KatG | CGG > CTG | R463L | [19] |
| EMB b | rss_17410 | 4247709 | Rv3795 | embB | AAC > ACC | N399T | – |
| EMB b | rss_17411 | 4247729 | Rv3795 | embB | GGC > AGC | G406S | [16,21] |
| ETH c | rss_8886 | 2154724 | Rv1908c | KatG | CGG > CTG | R463L | – |
| ETH c | rss_8900 | 2155648 | Rv1908c | KatG | TAC > TGC | Y155C | [16] |
a isoniazid; b ethambutol; c ethionamide.
Figure 1.
The structures of isoniazid and ethionamide.
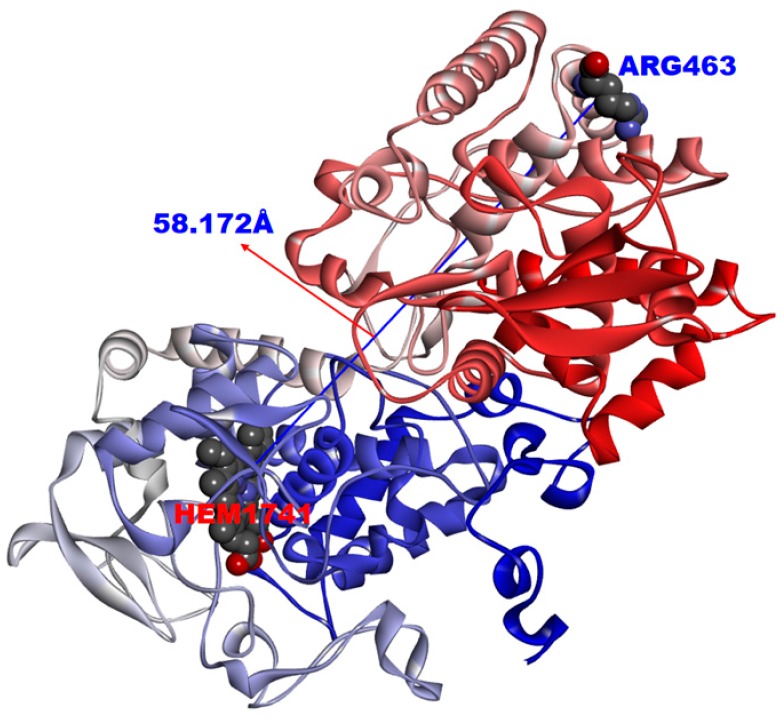
Interestingly, according to the research report of Phelan et al., there is a strong association between the (shorter) distance of the mutation to the ligand in the protein structure and INH resistance (greater minimum inhibitory concentrations (MIC) values), and the distance between the mutation location and drug binding site was not greater than 9.93 Å [15]. The R463L mutation was not reported in their results. The crystal structure of the KatG protein has been reported (PDB code: 2CCA) [22], and structural analysis revealed that the mutation of R463L is located far from the drug binding site (the distance was approximately 58.172 Å, much greater than 9.93 Å), as shown in Figure 2. This result also lends credit to the present method, which can identify the resistance mutations that far from the drug binding site.
Figure 2.
The distance of the active site of the KatG target and Arg463 residue (the distance was approximately 58.172 Å, far from drug binding site).
As shown in Table 3 and Table 4, more than 60% of the gene pairs contain one drug target and one PPE family protein (i.e., PPE68 and PPE54). PPE family proteins are unique to mycobacteria and are particularly abundant in pathogenic mycobacteria but are absent from the vaccine strains of Bacille Calmette-Guérin (BCG) [23]. Members of the PPE families are characterized by the presence of a conserved Pro-Pro-Glu (PPE) motif in their N-terminal region [24]. The literatures reported that most of the PPE proteins are located on the cell surface, and PPE proteins have been found to play important roles in generating antigenic variation and immune evasion [25,26,27]. The mutation of PPE protein results in failure to activate the body’s immune response [23]. PPE68 is a member of the PPE protein family and is a membrane protein expressed on the surface of Mtb [28,29]. PPE68 is primarily present in the cell wall of Mtb and is an immunogenic protein. Experiments confirmed that PPE68 cell surface proteins can induce a specific cellular immune response in mice [30]. Therefore, we infer that the changes in the protein structure caused by mutations would lead to the loss of function of PPE68. However, the biological functions for PPE54 remain elusive. Given that drug effects are associated with immunity and the fact that most of the gene pairs included one drug target and PPE family proteins, the present results suggest that PPE proteins play an important role in Mtb drug resistance, which is of apparent interest for future explorations.
In addition, katG-transcriptional regulator, katG-ccsA, katG-fdxB and katG-oxidoreductase gene pairs associated with drug resistance were also predicted. Limited by the existing information, the hidden mechanisms of these genes underlying drug resistance are unclear.
3. Materials and Methods
3.1. Data Source and SNP Calling
In this study, two sets of data were analyzed. The first set of data contained 127 samples of Mtb, which included 34 EMB-, 65 INH-, 53 RMP- and 45 STR-resistant strains. A total of 38 pan-susceptible strains of Mtb were involved in this dataset. All sequencing data of the first set were downloaded from TDR TB Strain Bank (ENA accession number: PRJEB11653) [15]. All the data of the first set were divided into four groups according to the different resistant strains for the following data analysis, as did by the previous study [15]. The second set of data contained 161 samples of Mtb, and all sequencing data were downloaded from the NCBI Sequence Read Archive (SRA) (SRA065095), which included 22 CPM-, 69 EMB-, 36 ETH-, 117 INH-, 21 KAN-, 54 OFX-, 117 RMP- and 83 STR-resistant strains [16]. And a total of 44 pan-susceptible strains of Mtb were involved in this set. Similarly, all the data of the second set were divided into eight groups according to the type of resistant strains for the following data analysis, as did by the prior research [16].
All of the downloaded sequencing data were treated with Trimmomatic v0.32 software [31] to remove or truncate reads of low quality (parameter: LEADING: 3; TRAILING: 3; MINLEN: 36; SLIDINGWINDOW: 4:20). High quality reads were then mapped to the Mtb H37Rv reference genome (GenBank accession: AL123456.3) using the Bowtie2 v2.2.6 [32] with default parameters. Sequencing filtering and SNP calling were performed by SAMtools v1.2 [33] and VarScan v2.4 [34] (parameters: min-coverage = 10; min-freq-for-hom = 0.95; min-avg-qual = 20), and SNPs with a minor allele frequency (MAF) <0.01 [35] were presumed to be sequencing errors and were discarded. Finally, we used PLINK v1.07 software [36] to filter phylogenetically related SNPs that are likely to influence the gene pair identification (parameter: indep-pairwise = 10, 5, 0.5) [37]. This procedure can exclude the gene pairs that could simply result from the strains genotype.
3.2. SNP Pair and Gene Pair Calculations
Gene-gene interactions have long been recognized to be fundamentally important in understanding the genetic causes of complex disease trait. GBOOST is a software package identifying gene-gene interactions in genome-wide case-control studies [38]. In this study, we used GBOOST software (parameters: wm = GPU; st = 12.838; tt = 12.838) to detect SNP-SNP interactions on risk of drug resistance. GBOOST adopts a chi-square test method to examine the interaction effect between two SNPs and phenotypes. We set the threshold to p < 0.005 (χ2 > 12.838).
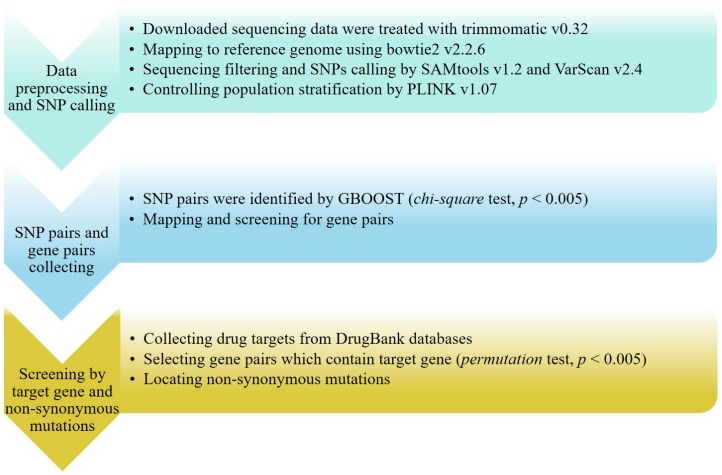
Taking into account that the mutation in a target gene is a key factor for drug resistance, the SNP pairs obtained by GBOOST were assigned to the corresponding genes. Subsequently the gene pairs that contained at least one target gene were screened for further analysis. We also used permutation tests to reduce the risk of false-positives (calculating 10,000 times in total). The results of the chi-square test can be further corrected using permutation tests (FDR < 0.005). The overall workflow of our method is presented as Figure 3.
Figure 3.
The workflow of the method used in this study.
4. Conclusions
The emergence and spread of resistant tuberculosis has become the most significant threat to global TB control. Drug resistance patterns may vary widely from single-drug to multiple-drug resistance, and the emergence of transmissible drug resistance is associated with multiple genetic mutations. To reveal the mechanisms of drug resistance and to develop better control strategies, we identified gene pairs associated with Mtb drug resistance.
All of the associations of SNP pairs were screened and assigned to the corresponding genes. Subsequently, to reveal the potential resistance mechanisms of the Mtb strains, the obtained gene pairs were filtered by drug targets and non-synonymous mutations. The obtained non-synonymous mutations in the drug target genes were compared with the reported experiments, and the results validated the effectiveness of our method. Many drug resistance-associated gene pairs that contained a target gene were identified: one for INH, one for RMP, four for EMB and five for ETH. Further gene pair analyses revealed that the detected katG-PPE54 and rpoB-PPE54 gene pairs are associated with INH and RMP resistance, respectively. The embA-PPE68 and embB-PPE54 gene pairs play possible roles in Mtb resistance to EMB. The joint mutations of katG-PPE54 gene pairs would bring Mtb resistance to ETH.
In summary, this study indicated the potential of bioinformatics methods in predicting drug resistance-associated gene pairs and clarifying the resistance mechanisms of anti-tuberculosis drugs. Moreover, the present approach is also useful for exploring the mechanisms of drug resistance for other microorganisms.
Acknowledgments
This work was supported by the Natural Science Foundation of Hubei Province (Grant 2013CFA016 and 2015CFB442), and the Natural Science Foundation of China (Grant 31402231).
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/17/9/1417/s1.
Author Contributions
Hong-Yu Zhang, Qing-Ye Zhang and Qiang Zhu conceived and designed the project; Ze-Jia Cui performed and analyzed the data; Qing-Ye Zhang and Ze-Jia Cui wrote the manuscript; Qing-Yong Yang and Qiang Zhu helped in preparing the manuscript and revised the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Cooper A.M. Cell-mediated immune responses in tuberculosis. Annu. Rev. Immunol. 2009;27:393–422. doi: 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.3rd Global GLC meeting World Health Organization, Geneva, Switzerland, 17–19 October 2012 Meeting Report. [(accessed on 11 January 2013)]. Available online: http://apps.who.int/iris/bitstream/10665/77948/1/WHO_HTM_TB_2012.13_eng.pdf.
- 3.World Health Organisation Global Tuberculosis Report. [(assessed on 28 October 2015)]. Available online: http://www.who.int/tb/Global_TB_Facts.pdf.
- 4.Narendran G., Swaminathan S. TB–HIV co-infection: A catastrophic comradeship. Oral Dis. 2016;22:46–52. doi: 10.1111/odi.12389. [DOI] [PubMed] [Google Scholar]
- 5.Bates J.H. Tuberculosis chemotherapy: The need for new antituberculosis drugs is urgent. Am. J. Respir. Crit. Care Med. 1995;151:942–943. doi: 10.1164/ajrccm/151.4.942. [DOI] [PubMed] [Google Scholar]
- 6.Pontali E., Matteelli A., Migliori G.B. Drug-resistant tuberculosis. Curr. Opin. Pulm. Med. 2013;19:266–272. doi: 10.1097/MCP.0b013e32835f1bf3. [DOI] [PubMed] [Google Scholar]
- 7.Falzon D., Jaramillo E., Schünemann H.J., Arentz M., Bauer M., Bayona J., Blanc L., Caminero J.A., Daley C.L., Duncombe C., et al. WHO guidelines for the programmatic management of drug-resistant tuberculosis: 2011 update. Eur. Respir. J. 2011;38:516–528. doi: 10.1183/09031936.00073611. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., Yew W.W. Mechanisms of drug resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung. Dis. 2015;19:1276–1289. doi: 10.5588/ijtld.15.0389. [DOI] [PubMed] [Google Scholar]
- 9.Wiseman B., Carpena X., Feliz M., Donald L.J., Pons M., Fita I., Loewen P.C. Isonicotinic acid hydrazide conversion to isonicotinyl-NAD by catalase-peroxidases. J. Biol. Chem. 2010;285:26662–26673. doi: 10.1074/jbc.M110.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sreevatsan S., Stockbauer K.E., Pan X. Ethambutol resistance in Mycobacterium tuberculosis: Critical role of embB mutations. Antimicrob. Agents Chemother. 1997;41:1677–1681. doi: 10.1128/aac.41.8.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caminero J.A., Sotgiu G., Zumla A., Migliori G.B. Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect. Dis. 2010;10:621–629. doi: 10.1016/S1473-3099(10)70139-0. [DOI] [PubMed] [Google Scholar]
- 12.Morlock G.P., Metchock B., Sikes D., Crawford J.T., Cooksey R.C. ethA, inhA, and katG Loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 2003;47:3799–3805. doi: 10.1128/AAC.47.12.3799-3805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller B., Borrell S., Rose G., Gagneux S. The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet. 2013;29:160–169. doi: 10.1016/j.tig.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gandhi N.R., Moll A., Sturm A.W., Pawinski R., Govender T., Lalloo U., Zeller K., Andrews J., Friedland G. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- 15.Phelan J., Coll F., McNerney R., Ascher D.B., Pires D.E., Furnham N., Coeck N., Hill-Cawthorne G.A., Nair M.B., Mallard K., et al. Mycobacterium tuberculosis whole genome sequencing and protein structure modelling provides insights into anti-tuberculosis drug resistance. BMC Med. 2016 doi: 10.1186/s12916-016-0575-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H., Li D., Zhao L., Fleming J., Lin N., Wang T., Liu Z., Li C., Galwey N., Deng J., et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet. 2013;45:1255–1260. doi: 10.1038/ng.2735. [DOI] [PubMed] [Google Scholar]
- 17.Trauner A., Borrell S., Reither K., Gagneux S. Evolution of drug resistance in tuberculosis: Recent progress and implications for diagnosis and therapy. Drugs. 2014;74:1063–1072. doi: 10.1007/s40265-014-0248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lou Z., Zhang X. Protein targets for structure-based anti-Mycobacterium tuberculosis drug discovery. Protein Cell. 2010;1:435–442. doi: 10.1007/s13238-010-0057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jessica N.T., Lynthia V.P., Timothy C.R., Victor T.C., Amallraja A.M., Elghraoui A., Goodmanson A.P., Ramirez-Busby S.M., Chawla A., Zadorozhny V., et al. Novel katG mutations causing isoniazid resistance in clinical M. tuberculosis isolates. Emerg. Microbes Infect. 2015 doi: 10.1038/emi.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee A., Dubnau E., Quemard A., Balasubramanian V., Um K.S., Wilson T., Collins D., Delisle G., Jacobs W.R. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 21.Ramaswamy S.V., Amin A.G., Goksel S., Stager C.E., Dou S.J., El Sahly H., Moghazeh S.L., Kreiswirth B.N., Musser J.M. Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2000;44:326–336. doi: 10.1128/AAC.44.2.326-336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X., Yu H., Yu S., Wang F., Sacchettini J.C., Magliozzo R.S. Hydrogen peroxide-mediated isoniazid activation catalyzed by Mycobacterium tuberculosis catalase-peroxidase (KatG) and its S315T mutant. Biochemistry. 2006;45:4131–4140. doi: 10.1021/bi051967o. [DOI] [PubMed] [Google Scholar]
- 23.Sampson S.L. Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin. Dev. Immunol. 2011 doi: 10.1155/2011/497203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed A., Das A., Mukhopadhyay S. Immunoregulatory functions and expression patterns of PE/PPE family members: Roles in pathogenicity and impact on anti-tuberculosis vaccine and drug design. IUBMB Life. 2015;67:414–427. doi: 10.1002/iub.1387. [DOI] [PubMed] [Google Scholar]
- 25.Riley R., Pellegrini M., Eisenberg D. Identifying cognate binding pairs among a large set of paralogs: The case of PE/PPE proteins of Mycobacterium tuberculosis. PLoS Comput. Biol. 2008 doi: 10.1371/journal.pcbi.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh P., Rao R.N., Reddy J.R., Prasad R., Kotturu S.K., Ghosh S., Mukhopadhyay S. PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence. Sci. Rep. 2016 doi: 10.1038/srep21624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukhopadhyay S., Balaji K.N. The PE and PPE proteins of Mycobacterium tuberculosis. Tuberculosis. 2011;91:441–447. doi: 10.1016/j.tube.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Duan Z.L., Li Q., Wang S., Chen X.Y., Liu H.F., Chen B.K., Li D.Z., Huang X., Wen J.S. Identification of Mycobacterium tuberculosis PPE68-specific HLA-A*0201-restricted epitopes for tuberculosis diagnosis. Curr. Microbiol. 2015;70:769–778. doi: 10.1007/s00284-015-0786-x. [DOI] [PubMed] [Google Scholar]
- 29.Demangel C., Brodin P., Cockle P.J., Brosch R., Majlessi L., Leclerc C., Cole S.T. Cell envelope protein PPE68 contributes to Mycobacterium tuberculosis RD1 immunogenicity independently of a 10-kilodalton culture filtrate protein and ESAT-6. Infect. Immun. 2004;72:2170–2176. doi: 10.1128/IAI.72.4.2170-2176.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Miltner E., Wu M., Petrofsky M., Bermudez L.E. A Mycobacterium avium PPE gene is associated with the ability of the bacterium to grow in macrophages and virulence in mice. Cell. Microbiol. 2005;7:539–548. doi: 10.1111/j.1462-5822.2004.00484.x. [DOI] [PubMed] [Google Scholar]
- 31.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavy N., Deschenes A., Blais S., Lavigne P., Beaulieu J., Isabel N., Mackay J., Bousquet J. The landscape of nucleotide polymorphism among 13,500 genes of the conifer picea glauca, relationships with functions, and comparison with Medicago truncatula. Genome Biol. Evol. 2013;5:1910–1925. doi: 10.1093/gbe/evt143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abraham G., Inouye M. Fast principal component analysis of large-scale genome-wide data. PLoS ONE. 2014;9:1417. doi: 10.1371/journal.pone.0093766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yung L.S., Yang C., Wan X., Yu W. GBOOST: A GPU-based tool for detecting gene-gene interactions in genome-wide case control studies. Bioinformatics. 2011;27:1309–1310. doi: 10.1093/bioinformatics/btr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.