Abstract
The regulation of insulin secretion by pancreatic β cells is perturbed in several diseases, including adult-onset (type 2) diabetes and persistent hyperinsulinemic hypoglycemia of infancy (PHHI). The first mouse model for PHHI has a conditional deletion of the gene encoding the winged-helix transcription factor Foxa2 (Forkhead box a2, formerly Hepatocyte nuclear factor 3β) in pancreatic β cells. Using isolated islets, we found that Foxa2 deficiency resulted in excessive insulin release in response to amino acids and complete loss of glucose-stimulated insulin secretion. Most PHHI cases are associated with mutations in SUR1 (Sulfonylurea receptor 1) or KIR6.2 (Inward rectifier K+ channel member 6.2), which encode the subunits of the ATP-sensitive K+ channel, and RNA in situ hybridization of mutant mouse islets revealed that expression of both genes is Foxa2 dependent. We utilized expression profiling to identify additional targets of Foxa2. Strikingly, one of these genes, Hadhsc, encodes short-chain L-3-hydroxyacyl-coenzyme A dehydrogenase, deficiency of which has been shown to cause PHHI in humans. Hadhsc is a direct target of Foxa2, as demonstrated by cotransfection as well as in vivo chromatin immunoprecipitation experiments using isolated islets. Thus, we have established Foxa2 as an essential activator of genes that function in multiple pathways governing insulin secretion.
Introduction
The pancreatic β cell plays a central role in the regulation of glucose homeostasis. Insufficient insulin release in response to elevated glucose levels is a hallmark of both type 1 and type 2 diabetes. Given the importance of the β cell, great effort has been focused on understanding the molecular pathways that govern glucose-stimulated insulin secretion. Likewise, many laboratories are attempting to elucidate the transcription factor hierarchy that controls the development of the β cell in the hope of enabling the controlled expansion and differentiation of β cell populations for use in widespread cell replacement therapy.
Foxa2 encodes a transcription factor (forkhead box a2) that has been postulated to play a central role in β cell development due to its ability to bind to and transactivate pancreatic duodenal homeobox 1 (Pdx1) cis-regulatory elements in vitro (1–3). Because embryos homozygous for a Foxa2-null allele die in midgestation (4, 5), genetic analysis of the role of Foxa2 in the pancreatic β cell required the development of cell type–specific gene ablation using the Cre-loxP recombination system.
We have previously reported the derivation of mice with a pancreatic β cell–specific Foxa2 deletion (Foxa2loxP/loxP;Ins.Cre), where Ins.Cre is a transgene with Cre recombinase under control of the rat insulin 2 promoter (6). Foxa2loxP/loxP;Ins.Cre mice, which lack Foxa2 in 85% of their β cells by postnatal day 8 (P8), exhibit growth retardation, disorganized islet architecture, severe hypoglycemia, and relative hyperinsulinemia, and usually die between P9 and P12. Blood glucose levels of Foxa2loxP/loxP;Ins.Cre mice are lower than those of euglycemic littermate controls (∼25 mg/dl versus ∼100 mg/dl), but plasma insulin levels are unchanged and the ratio of plasma insulin to glucagon is elevated (7). This aberrant release of insulin despite low glucose levels suggests a misfiring of the glucose-stimulated pathway of insulin secretion or the activation of an alternate insulin-release mechanism.
Insulin secretion from the pancreatic β cell is a tightly regulated process that couples the uptake of glucose with depolarization of the cell membrane to stimulate the exocytosis of insulin granules (8, 9). Expression analysis of key components of the glucose-sensing insulin secretory mechanism (glucose transporter 2 [Glut2], glucokinase, and glutamate dehydrogenase) revealed no significant changes in their mRNA levels in Foxa2loxP/loxP;Ins.Cre mice (7). However, steady-state mRNA levels of both subunits of the ATP-dependent K+ channel (KATP), sulfonylurea receptor 1 (Sur1) and the inward rectifier potassium channel member 6.2 (Kir6.2), are reduced by approximately 75% (7). This requirement of Foxa2 for maintenance of Sur1 and Kir6.2 expression was subsequently confirmed by Northern blot analysis in insulinoma-1 cells overexpressing a dominant negative mutant of Foxa2 (10).
KATP channels are heteromultimers composed of four Sur1 subunits surrounding an inner core of four Kir6.2 subunits. These ATP-sensitive K+ channels are found in both α and β cells of the islet and play an important role in the regulation of insulin and glucagon secretion (11–14). In humans, mutations in either the SUR1 or KIR6.2 genes can lead to persistent hyperinsulinemic hypoglycemia of infancy (PHHI), also referred to as congenital hyperinsulinism (15). In this disease, the lack of functional KATP channels “translates” into perpetual depolarization of β cells (16, 17), resulting in the activation of voltage-gated calcium channels and the subsequent release of insulin.
Foxa2loxP/loxP;Ins.Cre mice share many metabolic characteristics with PHHI patients, including β-cell dysfunction in the form of uncontrolled insulin secretion despite severe hypoglycemia (7, 18). The majority of familial PHHI patients inherit a recessive mutation in either the SUR1 or KIR6.2 gene (19). Interestingly, however, mice homozygous for null alleles in either Sur1 or Kir6.2 reproduce only some aspects of the phenotypes of either PHHI patients or Foxa2loxP/loxP;Ins.Cre mice. Therefore, we hypothesized that Foxa2 regulates additional targets in the pancreatic β cell that contribute to the phenotype. Here we characterize the genetic and physiological consequences of the β cell–specific ablation of Foxa2 in mice in detail and identify novel Foxa2 targets that explain its role in glucose homeostasis in the β cell.
Results
A prerequisite for understanding the functional role of Foxa2 in the pancreatic β cell is the determination of its transcriptional targets. We have shown previously that Sur1 and Kir6.2 mRNAs were reduced by approximately 75% in islets from Foxa2loxP/loxP;Ins.Cre mice (7). In order to interpret the physiological response of Foxa2-deficient β cells, it was essential to determine if the remaining Sur1 and Kir6.2 mRNA was present in the Foxa2-deficient β cells or if it was due to the contribution of α cells in our islet RNA preparations. To this end, we performed RNA in situ hybridization of pancreas sections from control and mutant P8 mice using digoxigenin-labeled antisense probes (Figure 1). Glucagon mRNA was abundantly expressed in pancreatic α cells, which were localized to the outer edges of control islets (Figure 1, A and E). Although Foxa2loxP/loxP;Ins.Cre islets also contained glucagon-secreting α cells, they were not confined to the islet perimeter but were also found in the core, confirming the perturbed islet architecture described previously in these mice (7) (Figure 1, C and G). Sur1 and Kir6.2 mRNAs were expressed in both α and β cells of control islets at approximately equal levels (Figure 1, B and F). The remaining mRNA expression of both KATP channel subunits in the mutant islets is confined to α cells (Figure 1, D and H), which comprise 15–20% of the islet (20). Therefore, β cells lacking Foxa2 are essentially deficient for both subunits of the KATP channel.
Figure 1.

Sur1 and Kir6.2 mRNAs are undetected in Foxa2-deficient β cells. (A–H) RNA in situ hybridization using paraffin-embedded pancreatic sections and digoxigenin probes for Glucagon, Sur1, and Kir6.2. Glucagon is expressed in the α cells on the perimeter of both control islets (A and E) and mutant Foxa2loxP/loxP; Ins.Cre islets (C and G). Sur1 (B) and Kir6.2 (F) are expressed throughout control islets, but are confined to the α cells of Foxa2loxP/loxP;Ins.Cre islets (D and H). Note the similar staining pattern between Glucagon and both KATP subunits in mutant islets (C versus D and G versus H). Magnification, ×40 for all images.
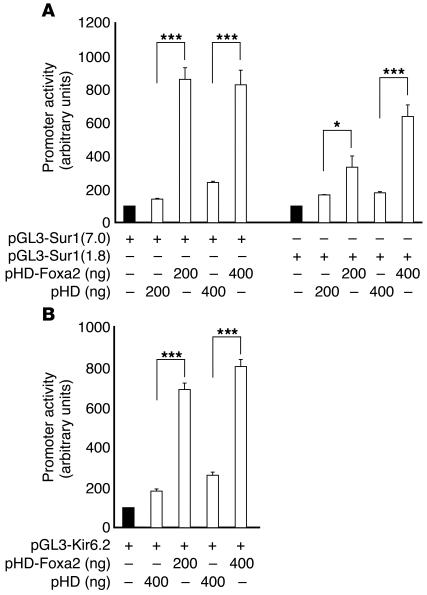
These findings suggested that Foxa2 acts as a transcriptional activator of Sur1 and Kir6.2 in β cells. To test this possibility, we transfected baby hamster kidney (BHK) cells with Sur1 or Kir6.2 promoter/luciferase reporter constructs along with a Foxa2 expression plasmid. Over 3-fold activation was observed with 1.8 kb of the Sur1 promoter and more than 6-fold activation was measured with the 7.0-kb promoter fragment construct (Figure 2A). Similarly, the addition of Foxa2 resulted in nearly 4-fold activation of Kir6.2 (Figure 2B), confirming that Foxa2 is a potent transactivator of both genes.
Figure 2.
Foxa2 activates the promoters of Sur1 and Kir6.2. (A) Cotransfection of a Foxa2 expression plasmid (pHD-Foxa2) results in stimulation of luciferase activity from Sur1 promoter constructs containing 1.8 kb [pGL3-Sur1(1.8)] and 7.0 kb [pGL3-Sur1(7.0)] of promoter sequence in BHK cells. (B) Cotransfection with pGL3-Kir6.2, containing the entire Kir6.2 promoter region, reveals Foxa2-dependent activation. For each condition, n = 3. *P ≤ 0.05 and ***P ≤ 0.0001 by ANOVA.
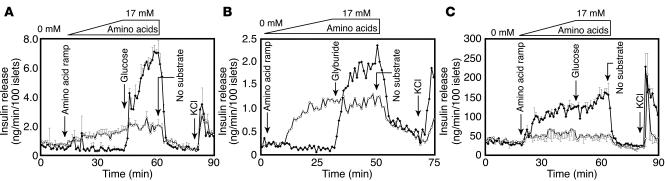
KATP channels in the pancreatic β cell couple glucose metabolism to insulin secretion (13). The absence of Sur1 and Kir6.2 mRNA in Foxa2-deficient β cells prompted us to investigate further the physiological consequences of this downregulation. Circulating insulin levels in Foxa2loxP/loxP;Ins.Cre mice are inappropriately high given their low blood sugar levels, suggesting abnormal regulation of insulin secretion. We had previously investigated the secretory responses of these mice to a variety of stimuli using minced pancreas pieces (7). Newly optimized isolation techniques now allowed us to perform perifusion assays using isolated islets without any potential impairment of stimulus-secretion coupling by surrounding exocrine tissue or the presence of digestive enzymes. After being exposed to a ramp of increasing concentrations of all 20 amino acids, control islets had a negligible insulin secretion response (Figure 3A). Mutant islets, however, displayed a dose-dependent insulin secretion response to amino acids, with peak secretion at 17 mM (Figure 3A). With the addition of 25 mM glucose, control islets responded with robust insulin secretion, while mutant islets showed no additional response (Figure 3A).
Figure 3.
Foxa2loxP/loxP;Ins.Cre islets exhibit misregulated hormone secretion in response to glucose and amino acids. (A) Control islets (filled symbols) immediately secrete insulin in response to 25 mM glucose but do not respond to amino acids alone. In contrast, Foxa2loxP/loxP;Ins.Cre islets (open symbols) secrete insulin in a dose-dependent manner upon exposure to increasing concentrations of a mixture of all 20 amino acids (0.55 mM/min for 30 minutes up to 17 mM) followed by 20 minutes at 17 mM, with no additional response to 25 mM glucose. Data shown are mean ± SEM of 2 identical experiments. Similar responses were seen in 8 additional trials (data not shown). (B) Control islets do not respond to amino acid stimulation, but 300 nM glyburide closes KATP channels and leads to insulin secretion. As in A, Foxa2loxP/loxP;Ins.Cre islets respond to the identical amino acid ramp, but glyburide has no additional effect. Trace shown is representative of 2 similar experiments. (C) Control islets secrete glucagon in response to low concentrations of an amino acid ramp (0.55 mM/min for 30 minutes up to 17 mM), but Foxa2loxP/loxP;Ins.Cre islets do not. While the amino acid concentration is held constant at 17 mM, additional exposure to 25 mM glucose for 20 minutes has no effect on secretion from either control or mutant islets. Mean ± SEM of two identical experiments is shown. Downward arrows indicate times of reagent administration.
In a similar islet perifusion, an identical amino acid ramp was applied to control or Foxa2loxP/loxP;Ins.Cre islets (Figure 3B). Again, control islets did not respond to amino acid stimulation, but mutant islets showed the same steady increase in insulin secretion, which correlated with amino acid concentration in a dose-dependent manner (Figure 3B). The perifusate was then held at the highest amino acid concentration while the islets were exposed to 300 nM glyburide for 20 minutes. Glyburide is a sulfonylurea that binds to KATP channels in both α and β cells (21, 22) and prevents their opening. This prolonged closure of the KATP channel prevents the outflow of K+ ions from β cells, causing depolarization of their cell membranes and the subsequent opening of voltage-gated calcium channels and insulin secretion. The addition of glyburide to Foxa2loxP/loxP;Ins.Cre islets neither blunted nor enhanced the insulin secretion response to amino acids, while control islets exhibited a robust secretory response to glyburide as expected (Figure 3B), confirming that mutant β cells lack functional KATP channels.
We previously reported that glucagon expression is unchanged at the level of both pancreatic content and steady-state mRNA in Foxa2loxP/loxP;Ins.Cre mice. The significant decrease in circulating plasma glucagon, therefore, suggests a defect not in the biosynthesis of this hormone but in secretion (7). To investigate the glucagon secretion defect, we exposed islets to an amino acid ramp and determined glucagon release. Even at very low concentrations of the amino acid mixture, control islets were stimulated to secrete glucagon, but Foxa2loxP/loxP;Ins.Cre were not. The addition of glucose in conjunction with the maximal concentration of amino acids (17 mM) for 20 minutes did not alter glucagon secretion in control or mutant islets (Figure 3C).
The severely hyperinsulinemic, hypoglycemic phenotype of Foxa2loxP/loxP;Ins.Cre mice is usually lethal between P9 and P12. The downregulation of Sur1 and Kir6.2 described above cannot be the sole effect of the β cell–specific deletion of Foxa2, as the phenotype of mice homozygous for a null mutation in either Sur1 or Kir6.2 is mild compared with that of Foxa2loxP/loxP;Ins.Cre mice (23–25). Although all three mouse models, Sur1–/–, Kir6.2–/–, and Foxa2loxP/loxP;Ins.Cre, demonstrate KATP channel deficiency in their pancreatic β cells, only mice lacking Foxa2 succumb to severe hyperinsulinemic hypoglycemia by P9–P12. Because these mice do not simply recapitulate the phenotype of the Sur1–/– or Kir6.2–/– mouse models, we initiated a systematic search for additional pancreas-specific target genes that may contribute to lethality and the abnormal insulin secretion response to amino acids in Foxa2loxP/loxP;Ins.Cre mice.
Total RNA was purified from isolated islets pooled from 5–7 P8 mice of like genotype for control and Foxa2loxP/loxP;Ins.Cre samples. For quantitative expression studies, it was imperative to use only the highest quality RNA of equal purity. In the representative trace of RNA from isolated islets shown in Figure 4A, the rRNA fluorescence ratio (28S/18S) is about 2.5, which is the theoretical limit, with no evidence of degradation. Although minimal contamination with exocrine cells does not affect assays such as islet perifusions, RNA contributed by non-endocrine cells has the potential to significantly skew microarray or real-time PCR results. We developed a quantitative method for the evaluation of exocrine impurities in islet preparations (for details, see Methods). Amplified islet RNA was quantified, reverse-transcribed, and used in real-time PCR analysis with primers for insulin and amylase (Figure 4C). Using total pancreas RNA for comparison, we were able to calculate the “fold enrichment” of endocrine mRNA for each of our samples and select only the purest for microarray hybridization and real-time PCR. For the representative samples shown, the difference in cycle threshold (Ct) values for insulin and amylase between total pancreas cDNA (Figure 4B) and islet cDNA (Figure 4C) was 12.24 cycles, which “translates” into a 4,837-fold enrichment in endocrine tissue, resulting in about 99% endocrine purity.
Figure 4.
Quality and purity assessment of islet RNA for gene expression analysis. (A) RNA Nano 6000 Assay of P8 islet RNA pooled from 5 mutant Foxa2loxP/loxP;Ins.Cre mice using the Agilent 2100 Bioanalyzer. Islet RNA was quantified and evaluated for evidence of degradation using the ratio of 28S rRNA peaks to 18S rRNA peaks. In the representative sample shown, the 28S/18S ratio is approximately 2.5 with a concentration of 79 ng/μl and no evidence of degradation. (B) Real-time PCR analysis of total pancreas cDNA for endocrine and exocrine content using insulin (open circles) and amylase (filled squares) as markers and TBP, TATA-box binding protein (filled triangles), a “housekeeping” gene, as an internal control. (C) Representative real-time PCR analysis of cDNA from isolated islets. In relation to the total pancreas cDNA shown in B, the insulin/amylase ratio has been enriched for 12.24 cycles to achieve a purity of about 99%. dRn, raw fluorescence normalized for baseline and reference dye intensities by MX4000 software.
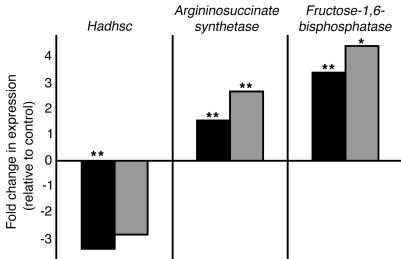
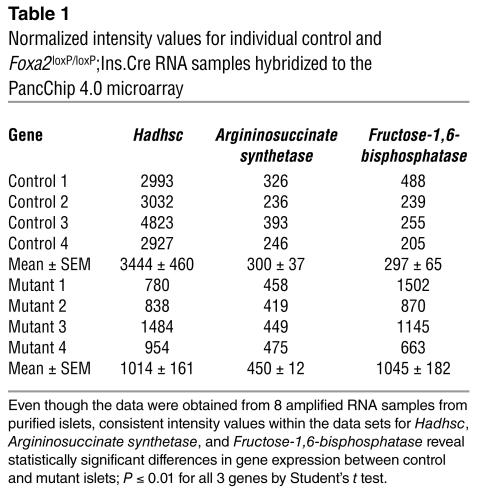
For the microarray analysis, we used the PancChip 4.0, which contains nearly 14,000 elements representing over 10,000 unique genes, most of which are expressed in the endocrine pancreas (26, 27). To analyze gene expression differences between control and Foxa2loxP/loxP;Ins.Cre P8 islets, we chose four amplified RNA samples of matched quality and endocrine purity from control and mutant genotypes. Overall, the gene expression profiles were very similar between mutant and control islets, demonstrating that the absence of Foxa2 does not cause a global defect in differentiation of β cells. Only approximately 0.3% of the genes were significantly changed, and for most of them their biochemical function is still unknown. However, three metabolic genes were identified in this screen and confirmed by quantitative real-time PCR analysis as being dependent on Foxa2 (Figure 5 and Table 1): Argininosuccinate synthetase, Fructose-1,6-bisphosphatase, and Hadhsc, the gene that encodes short-chain L-3-hydroxyacyl coenzyme A dehydrogenase (schad). Additional real-time PCR analysis with primers for known regulators of β-cell development and function revealed a significant decrease in Pdx1 mRNA in Foxa2loxP/loxP;Ins.Cre islets (data not shown), further confirming Foxa2 as an upstream transcriptional regulator of Pdx1 (28). We also confirmed our earlier experiments that reported unchanged levels of other genes involved in glucose metabolism, namely Glut2 and Glucokinase (ref. 7 and data not shown).
Figure 5.
Gene expression changes discovered by microarray analysis are confirmed by real-time quantitative PCR. Fold changes in gene expression calculated from microarray data (black bars) are similar in value to those calculated from quantitative real-time PCR results (gray bars). All fold changes refer to deviations from gene expression in control islets. *P ≤ 0.05 and **P ≤ 0.01 by Student’s t test.
Table 1.
Normalized intensity values for individual control and Foxa2loxP/loxP;Ins.Cre RNA samples hybridized to the PancChip 4.0 microarray
Our findings from the microarray experiment prompted us to investigate the mechanism that could explain the abnormal physiology of Foxa2loxP/loxP;Ins.Cre islets. We first considered how upregulation of Argininosuccinate synthetase could contribute to the aberrant islet secretion responses seen in our islet perifusions. In pancreatic β cells, this enzyme converts L-citrulline to L-argininosuccinate, which is then metabolized by argininosuccinate lyase to L-arginine. The third member of this biochemical cycle, nitric oxide synthase, metabolizes L-arginine back to L-citrulline, with an additional release of nitric oxide. Overactivation of this citrulline-argininosuccinate-arginine recycling pathway, which produces elevated levels of nitric oxide, has been associated with increases in intracellular calcium concentration in β cells (29). We hypothesized that upregulation of this biochemical pathway may trigger elevated basal intracellular calcium levels and the subsequent high levels of insulin secretion seen in Foxa2loxP/loxP;Ins.Cre islets. Real-time PCR analysis (data not shown) revealed modest upregulation of argininosuccinate lyase (P < 0.05) and a trend toward an increase in nitric oxide synthase transcript abundance in mutant islet RNA, suggesting that this pathway is indeed upregulated in Foxa2loxP/loxP;Ins.Cre mice.
We further investigated the Foxa2-dependent mRNA expression of schad, encoded by Hadhsc, that was identified through our microarray screen (Figure 5 and Table 1). Schad is a soluble mitochondrial matrix protein that plays an essential role in the β-oxidation of short chain fatty acids (30, 31). This enzyme has reportedly high levels of activity in pancreatic islets, suggesting a crucial influence on insulin secretion (32, 33). The strongest evidence for the importance of schad in glucose metabolism came from the discovery that mutations in the corresponding human gene cause PHHI (34–36). Thus, the approximately 3-fold reduction in Hadhsc expression in Foxa2loxP/loxP;Ins.Cre mice probably contributes to the severity of the hypoglycemic, hyperinsulinemic phenotype.
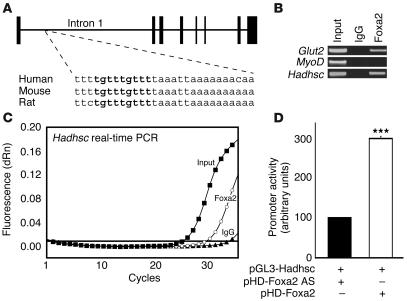
Analysis of steady-state mRNA levels does not address the question whether Hadhsc is a direct transcriptional target of Foxa2 in the β cell or whether the reduction in its expression is a secondary consequence of the altered metabolic state of Foxa2loxP/loxP;Ins.Cre mice. We analyzed potential cis-regulatory elements of the Hadhsc gene for Foxa2-binding sites. Sequence alignments from mouse, rat, and human revealed no areas of conservation within the promoter region of Hadhsc, but detected a span of 28 bases within intron 1 with 93% identity (26 of 28 bases) among all three species (Figure 6A). A transcriptional element search algorithm revealed a putative Foxa2-binding site (TGTTTGTTT) within this region. Although there is no strict consensus Foxa-binding site, this exact sequence had previously demonstrated a high affinity for Foxa2 (hepatocyte nuclear factor 3β [HNF3β]) protein in vitro (37). To investigate whether this site is occupied by Foxa2 in β cells, we performed chromatin immunoprecipitation (ChIP) with a Foxa2-specific antibody on isolated islets. As shown in Figure 6, B and C, the intron of Hadhsc was indeed bound by Foxa2 in vivo. Cotransfection of a luciferase construct containing the Foxa2-binding site within intron 1 of Hadhsc demonstrated that Foxa2 not only bound to Hadhsc but also activated transcription up to 3-fold (Figure 6D). The magnitude of this transactivation is in good agreement with the magnitude of the downregulation of Hadhsc we observed in islets deficient for Foxa2 shown in Figure 5 and Table 1. Thus, we have shown that Hadhsc is a direct target of Foxa2 in pancreatic β cells.
Figure 6.
Identification of a conserved Foxa2-binding site in intron 1 of the Hadhsc gene, encoding schad. (A) Mouse Hadhsc is located on chromosome 3 and contains 8 exons. Exons are represented as vertical black bars, with bar width indicative of exon size. The 28-base region of the 1st Hadhsc intron shown is 93% conserved (26 of 28 bases) among mouse, rat, and human and includes the identical Foxa2-binding site shown in bold. (B) ChIP using mouse islets and a Foxa2 antibody followed by PCR confirmed the binding site on Hadhsc shown in A. Glut2 (a known Foxa2 target) served as a positive PCR control and MyoD is the negative PCR control. (C) Real-time PCR of purified DNA from ChIP eluates using primers for Hadhsc confirms the occupancy of this intron enhancer by Foxa2. Input (filled squares) and Foxa2 (open circles) Ct values were approximately 25 and 30, respectively. IgG (filled triangles) served as the ChIP control. (D) Cotransfection experiments with a luciferase construct containing 100 bp of Hadhsc intron 1 (pGL3-Hadhsc) and a Foxa2 expression plasmid (pHD-Foxa2) result in 3-fold activation compared with transfections with antisense Foxa2 (pHD-Foxa2 AS). n = 3 and ***P ≤ 0.0001 by Student’s t test.
Discussion
In this study, we have characterized the functional consequences of a β cell–specific deletion of Foxa2 in order to better define its role in glucose metabolism. We had previously shown that the mRNA levels of both subunits of the KATP channel were reduced in Foxa2-deficient islets by about 70% (7). Here, we have demonstrated that the reduction in Sur1 and Kir6.2 expression in β cells is even more than previously published, as these transcripts are below the detection limit of in situ hybridization. This, together with the demonstration that Foxa2-deficient β cells are unresponsive to glyburide, shows that Foxa2 is absolutely required for the activation of Sur1 and Kir6.2 expression in vivo. This raises the question of whether Foxa2 is a direct or indirect activator of the Sur1 and Kir6.2 genes.
The mouse Sur1 gene contains 39 exons and spans about 80 kb on chromosome 7. Only about 5 kb separates the 3′ end of Sur1 from the 5′ start of the Kir6.2 gene, which is composed of a single 1.5-kb exon on the same chromosome (25). This close linkage of the two genes is conserved between mouse and human, as the human genes both map to chromosome 11p15.1. The proximity of these genes combined with RNA expression data from this study suggests an elegant hypothesis by which Foxa2 could regulate both genes coordinately. Indeed, our cotransfection experiments revealed that both Sur1 and Kir6.2 promoters are activated by Foxa2.
A striking finding is that Foxa2-deficient β cells are completely refractory to glucose, yet respond robustly to stimulation with amino acids. This situation is reminiscent of that of fetal rodent islets, whose secretory properties differ tremendously from those of adult islets (38, 39). Before birth, serum glucose levels are low (40) and amino acid concentrations are elevated (41). As Foxa2loxP/loxP;Ins.Cre mice are hypoglycemic, demonstrate a lack of glucose-stimulated insulin secretion, and also respond to amino acids (ref. 7 and our data here), we considered that Foxa2 mutant islets may be simply developmentally delayed. However, when we compared the expression profile of P8 mutant islets with that of controls, we found that the transcriptional programs were very similar, excluding the possibility of a pervasive developmental delay. In addition, fetal islets have a 50% reduction in protein and mRNA levels of Glut2 (42), which conceivably contributes to the lack of glucose sensitivity in fetal islets. However, both our previous studies (7) and real-time PCR from our study here revealed no difference in Glut2 mRNA expression between Foxa2loxP/loxP;Ins.Cre and control mice at P8. Furthermore, fetal islets have intact and fully functional KATP channels (43). Foxa2loxP/loxP;Ins.Cre β cells have no detectable Sur1 or Kir6.2 message (Figure 1) and demonstrate no KATP channel activity (Figure 3), strongly suggesting that the mutant Foxa2 phenotype is not due to delayed islet maturation.
In our initial characterization of Foxa2loxP/loxP;Ins.Cre mice, we found that both glucagon protein content and steady-state mRNA levels were similar to those of control littermates; however, plasma glucagon levels were nearly 5-fold lower (7). In the present study, mutant islets did not secrete glucagon in response to amino acids (Figure 3C) and secreted nearly 3-fold less glucagon than did control islets after exposure to pyruvate (data not shown). Taken together, these studies suggest a defect not in Glucagon transcription or protein biosynthesis, but in secretion of this hormone. As the glucagon-secreting α cells in our mouse model retain Foxa2, the decreased glucagon secretion is a secondary effect of the metabolic dysfunction of the β cells, which comprise the majority of the islet cell population. A number of mechanisms have been implicated in the suppression of glucagon secretion, including improper KATP or voltage-gated Na+ channel activity (14), insulin (44), and, most recently, zinc (45), suggesting that the nonphysiological insulin secretion by Foxa2-deficient β cells in response to amino acids is responsible for the suppression of glucagon release.
We utilized the power of expression profiling using the NIH-sponsored PancChip cDNA array to screen for additional Foxa2 targets that could contribute to the severe β cell defect in Foxa2loxP/loxP;Ins.Cre mice. This screen revealed schad, which uses NAD+ to convert L-3-hydroxyacyl-CoA to 3-ketoacyl-CoA to complete the second to last step of the fatty acid oxidation spiral, was differentially expressed. A deficiency in this mitochondrial enzyme results in an accumulation of short chain acyl-CoA esters. Such a buildup inhibits the outer mitochondrial enzyme carnitine palmitoyltransferase I and prevents the transport of long-chain fatty acyl-CoAs (LCFA-CoAs) into the mitochondria. LCFA-CoAs can be converted to triglycerides, diacylglycerol, fatty acids, and acylated proteins, all of which are postulated to enhance insulin secretion by amplifying mechanisms that circumvent the β-cell KATP channel (34, 46).
The greater than 3-fold downregulation of Hadhsc transcripts in Foxa2-deficient β cells along with the ability of Foxa2 to bind to and activate this gene suggests an intriguing mechanism by which to explain the severe hypoglycemia and hyperinsulinemia seen in Foxa2loxP/loxP;Ins.Cre mice. This particular phenotype is reminiscent of human PHHI, which has an incidence of nearly 1:40,000 live births (47). Although many clinical cases of PHHI can be attributed to mutations in either SUR1 or KIR6.2, there are recent reports of hypoglycemic hyperinsulinemic infants with no determinable mutations in either subunit of the KATP channel. Instead, these patients harbor various mutations in the gene encoding SCHAD (34–36). Cases like these, along with additional SCHAD deficiencies that result in abnormal glucose metabolism (48), reveal the influence of fatty acid β-oxidation, in addition to glucose, on nutrient-stimulated insulin secretion. Although much remains to be learned about the cooperation between the triggering (i.e., KATP channel–dependent) mechanism of insulin secretion (49) and the amplifying metabolic pathways that augment insulin release, our discovery that Foxa2 regulates both aspects of secretion demonstrates a universal role for this transcription factor in the pancreatic β cell.
In summary, we have utilized both molecular and physiological approaches to define the β cell defect induced by Foxa2 deficiency. The complete absence of KATP channels on the β-cell membrane provides an explanation for the lack of glucose responsiveness of Foxa2-deficient β cells. In addition, increased activation of the citrulline-argininosuccinate-arginine cycle may contribute to the relative hyperinsulinemia seen in Foxa2loxP/loxP;Ins.Cre mice through possible hyper-responsiveness of mutant β cells to amino acids. We have discovered a novel and crucial dependence of amplifying (i.e., KATP channel–independent) mechanisms of insulin secretion on Foxa2 through direct binding to and transcriptional activation of Hadhsc, the gene encoding one of the key enzymes in fatty acid metabolism. Thus, we have demonstrated that Foxa2 is an essential transcriptional regulator of β cell function.
Methods
Animals and genotype analysis.
The derivation of the Ins.Cre transgenic line (RipCre) and Foxa2loxP/loxP mice has been described previously (6, 7). Genotyping was performed by PCR analysis using genomic DNA isolated from the tail tips of newborn mice. All studies were performed on P8 mice because mutants succumb to severe hypoglycemia at P9–P12 (7). Foxa2loxP/+ littermates without the Ins.Cre transgene served as controls in all experiments. All procedures involving mice were conducted in accordance with approved Institutional Animal Care and Use Committee protocols.
Riboprobe production and RNA in situ hybridization.
A 288-bp mouse Glucagon cDNA fragment (obtained by PCR with the following primers: 5′-CCGTGCCCAAGATTTTGTGCA-3′ and 5′-CGCCAGTGTGCTGGAATTCAT-3′) was cloned into the EcoRV site of pBluescript II KS– (Stratagene, La Jolla, California, USA). The resulting plasmid was digested with BamHI and was transcribed in the presence of Digoxigenin RNA Labeling Mix (Roche, Indianapolis, Indiana, USA) with T3 RNA polymerase (Promega, Madison, Wisconsin, USA) for synthesis of an antisense riboprobe. Similarly, a 1,730-bp mouse Kir6.2 cDNA fragment (obtained by PCR with the following primers: 5′-GGACAAGGGCTAGAGAAGGA-3′ and 5′-GAGGAACTGCAACTCAGGACA-3′) was cloned into pCRII using the TA Cloning kit (Invitrogen, Carlsbad, California, USA). The resulting plasmid was digested with XbaI and was transcribed with SP6 RNA Polymerase (Promega). A clone containing the mouse Sur1 sequence (I.M.A.G.E. clone 5663177) was digested with NotI and was transcribed with T7 RNA polymerase (Promega).
Pancreata from P8 mice were fixed overnight at 4°C in Bouin’s fixative (Poly Scientific, Warrington, Pennsylvania, USA), embedded in paraffin, and cut into sections 6 μm in thickness, which were applied to Superfrost/Plus Microscope slides (Fisher Scientific, Pittsburgh, Pennsylvania, USA). In situ hybridization was performed on deparaffinized and rehydrated slides as described (50), except that the proteinase K treatment was shortened to 4 minutes and the ×0.2 SSC wash was performed at 55°C. Anti-digoxigenin (Roche) was used at a dilution of 1:1,500. Slides were exposed to 5-bromo-4-chloro-3-indolyl-phosphate and 4-nitro blue tetrazolium chloride (Roche) until the color reaction was fully developed. Images were captured with Nomarski optics on a Nikon Eclipse E600 microscope.
Cotransfections.
A clone containing Sur1 promoter sequence was identified in a bacterial artificial chromosome (BAC) library screen (Invitrogen). PCR products of 1.8 kb (primers used: 5′-GAGGAGCAGGAGAAAAAGGA-3′ and 5′-GCCTCCCGCTCCTCTGTT-3’) and 7.0 kb (primers used: 5′-CGCGCGTAATACGACTCACTAT-3′ and 5′-CTCTGTTCCTCGCAGCACC-3’), containing the Sur1 transcriptional start site and upstream sequence, respectively, were cloned into the SmaI site and the BamHI/SmaI sites, respectively, of the promoterless luciferase reporter plasmid pGL3-Basic (Promega). A 6.0-kb fragment from a BAC (Invitrogen) containing the entire Kir6.2 promoter sequence was cloned into the BglII site of pGL3-Basic (Promega). A 150-bp PCR product from intron 1 of Hadhsc containing a putative Foxa2-binding site (5′-GGTCCAGGGTTAGTTACATTTAGCC-3′ and 5′-CTGCAGGCTAAGAGGTTTGGTT-3′) was cloned into the Asp718 and BglII sites of the pGL3-Promoter luciferase vector (Promega). Foxa2 or antisense Foxa2 was expressed from the pHD plasmid (51). The day before transfection, 5 × 105 BHK cells were seeded on 60-mm dishes and were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, L-glutamine, and antibiotics. Effectene reagent (Qiagen, Valencia, California, USA) was used in transient transfections according to manufacturer’s instructions. Cells were harvested 24 hours after transfection and luciferase was measured with the Dual-Luciferase Reporter Assay (Promega) in an Orion Microplate Luminometer (Berthold, Bad Wildbad, Germany). Firefly luciferase activity was normalized to Renilla luciferase expression from pRL-SV40 (Promega).
Islet perifusions.
For each experiment, islets were isolated from 5–7 P8 mice of like genotype using standard collagenase digestion followed by purification through a Ficoll gradient (52). One hundred islets were “hand-picked” under a light microscope and perifusions were performed as described previously (7). The amino acid mixture consisted of 19 amino acids (15 mM) plus 2 mM glutamine, and additional secretagogues were as reported in the figure legends. At the end of all perifusion experiments, islet viability was verified through stimulation with 30 mM KCl, causing full depolarization of β- and α-cell membranes and the subsequent release of insulin or glucagon.
RNA isolation and amplification.
Isolated islets from 5–7 P8 mice of like genotype were homogenized in 1 ml of TRIzol reagent (Invitrogen). Glycogen (20 μg; Roche) was added to each sample as a carrier, followed by chloroform extraction and isopropanol precipitation. After being washed with 70% ethanol, RNA pellets were resuspended in 300 μl of 10 mM Tris, pH 7.5, 1 mM EDTA, and 0.1% SDS. RNA was re-extracted with 600 μl phenol/chloroform/isoamyl alcohol (25:24:1, vol/vol/vol) and was precipitated with 1/10 volume 3 M sodium acetate and three volumes of ethanol. RNA was quantified with the RNA 6000 Nano Assay program of the Agilent 2100 Bioanalyzer (Agilent Technologies, Wilmington, Delaware, USA).
Islet RNA was amplified once with the MessageAmp antisense RNA (aRNA) Kit (Ambion, Austin, Texas, USA). Briefly, 200 ng of total RNA was reverse-transcribed at 42°C for 2 hours with T7 Oligo(dT) Primer and reverse transcriptase, both of which are included in the MessageAmp kit. Second-strand synthesis was performed using the MessageAmp DNA Polymerase, and the resulting cDNA was purified with a filter cartridge. Double-stranded cDNA was concentrated and transcribed in vitro at 37°C to make aRNA. The aRNA was treated with DNase, purified with a filter cartridge, eluted with nuclease-free water, and stored at –80°C until use.
Determination of islet purity.
Islet purity was determined by real-time PCR analysis. Because insulin mRNA is synthesized only in β cells and amylase is produced by acinar cells, we used these two genes as markers representing the two different cell types in our samples. Our assessment of islet purity encompasses a number of assumptions: (a) β cells represent 2.4% of all cells in the newborn mouse pancreas (53); (b) amylase-secreting acinar cells, in our estimation, compose roughly 80% of the pancreas, with the remaining percentages being α, duct, nerve, adipose, and blood cells, among others; and (c) endocrine and exocrine cells contain the same amount of RNA per cell. By comparing the Ct values of total pancreas to isolated islet preparations, we can estimate the “fold enrichment” and relative purity of islet RNA.
Islet aRNA labeling.
Control and Foxa2loxP/loxP; Ins.Cre aRNA samples (2 μg each) were reverse-transcribed for incorporation of amino-allyl dUTP as described previously (26), except that 2 μg of random hexamers were used to prime the reactions. The resulting cDNAs were dried with a Microcon YM-30 Concentrator (Millipore, Bedford, Massachusetts, USA) and were labeled with Cy5-bis-OSU, N,N′-biscarboxypentyl-5,5′-disulfonatoindodicarbocyanine (Cy5) or Cy3-bis-OSU, N,N′-biscarboxypentyl-5,5′-disulfonatoindodicarbocyanine (Cy3) (Amersham Pharmacia, Piscataway, New Jersey, USA) following a modified indirect labeling protocol (27). Coupled samples were combined, incubated with T7 Oligo(dT) to block nonspecific binding to the microarray, purified using the Qiaquick PCR Purification Kit (Qiagen), and precipitated at –20°C overnight with 1 μl polyacyl carrier (Molecular Research Center Inc., Cincinnati, Ohio, USA), 1/10 volume 1 M sodium acetate (pH 5.2), and three volumes ethanol. Pellets were air-dried after precipitation.
Microarray hybridization and analysis.
Construction of the PancChip 4.0 microarray and a source description of its more than 10,000 nonredundant elements has been reported recently (27). Control and Foxa2loxP/loxP; Ins.Cre samples were hybridized using a “dye-swap” design in which two coupling reactions were set up for each pair of cDNAs and Cy3 and Cy5 labeling was alternated to control for variability between dyes. Four independent biological replicates were prepared for both control and mutant islets. Hybridization of the labeled samples to the PancChip 4.0 and subsequent scanning and image analysis using Genepix Pro 3.0 were identical to those in our previous report (27). Microarray data have been deposited in the “minimal information about a microarray experiment–compliant” RNA abundance database (54) and can be accessed at http://www.cbil.upenn.edu/RAD/php/queryStudy.php?study_id=550.
Islet RNA reverse-transcription and real-time PCR.
Two pools each of control and Foxa2loxP/loxP; Ins.Cre islet RNA were assembled, with each pool consisting of four high-quality total islet RNA samples and each sample representing 5–7 P8 animals of like genotype. Islet RNA (500 ng) from each pool was reverse-transcribed at 42°C for 1 hour with 1 μg Oligo(dT) Primer (Invitrogen), 5x First Strand Buffer (Invitrogen), 100 mM DTT (Invitrogen), 10 mM dNTP, RNasin (Promega), and Superscript II Reverse Transcriptase (Invitrogen). Islet cDNA was used at a dilution of 1:10 or 1:20 in water in subsequent real-time PCR reactions.
Primers for real-time PCR were designed for all chosen genes using Primer3 software (55). PCR reaction mixtures included the Brilliant SYBR Green QPCR Master Mix (Stratagene), 10 μM primers, and the included reference dye at a dilution of 1:200 according to manufacturer’s instructions, except that the total reaction volume was “scaled down” from 50 μl to 25 μl. Reactions were performed with the SYBR Green (with Dissociation Curve) program on the Mx4000 Multiplex Quantitative PCR System (Stratagene). Cycling parameters were 95°C for 10 minutes and then 40 cycles of 95°C (30 s), 58°C or 60°C (1 min), and 72°C (30 s), followed by a melting curve analysis. All reactions were performed in triplicate with reference dye normalization, and the median Ct value was used for analysis. Primer sequences are available upon request.
Computational identification of Foxa2-binding sites in the Hadhsc gene.
In order to identify potential Foxa2-binding sites in the Hadhsc gene, we first performed an alignment of all available mammalian sequences (mouse, rat, human) using the University of California Santa Cruz Genome Browser (www.genome.ucsc.edu). The strongest conserved area was located in intron 1, while the promoter and immediate upstream sequence showed no conservation. Manual inspection of the aligned sequence in intron 1 identified a block of 28 bases, of which 26 were identical among all three species. Using the transcription factor–binding site–identification algorithm Transcriptional Element Search Schema (http://www.cbil.upenn.edu/tess), we identified a high-scoring Foxa2 (HNF3β)–binding site within this 28 bp sequence.
Formaldehyde cross-linking and ChIP.
Four hundred isolated islets from adult female CD1 mice were suspended in 1% formaldehyde in PBS and were incubated for 10 minutes at room temperature while being rotated. Cross-linking and chromatin sonication were performed as described (56).
For each immunoprecipitation, 100 μl of cross-linked chromatin was precleared by incubation for 1 hour at 4°C with 150 μl of protein G–agarose (Upstate Biotechnology, Lake Placid, New York, USA) in a total volume of 1 ml ChIP dilution buffer (20 mM Tris-HCl, pH 8.1, 1% Triton X-100, 2 mM EDTA, and 150 mM NaCl). After this preclearing, the supernatant was incubated overnight with 4 μg Foxa2 antibody (Santa Cruz Biotechnology, Santa Cruz, California, USA) or control IgG. Immunoprecipitation was performed as described (56). The precipitated and noncrosslinked DNA was purified on a Qiaquick PCR purification column (Qiagen) and eluted in 50 μl 10 mM Tris, pH 8.5. PCR reactions were performed for 35–40 cycles, as described above, in Stratagene’s Mx4000 Multiplex Quantitative PCR System. Purified input DNA template was used at a dilution 1:3 relative to IgG or Foxa2 ChIP templates. Primer sequences are available upon request.
Acknowledgments
The authors wish to acknowledge J. Fulmer for taking care of the mouse colony, as well as the University of Pennsylvania Morphology Core and the Radioimmunoassay Core of the Penn Diabetes Center for sample processing. This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases grants R01-DK55342 and U01-DK56947 to K.H. Kaestner.
Footnotes
Nonstandard abbreviations used: antisense RNA (aRNA); ATP-dependent potassium channel (KATP); baby hamster kidney (BHK); bacterial artificial chromosome (BAC); chromatin immunoprecipitation (ChIP); Cy3-bis-OSU, N,N′-biscarboxypentyl-5,5′-disulfonatoindodicarbocyanine (Cy3); Cy5-bis-OSU, N,N′-biscarboxypentyl-5,5′-disulfonatoindodicarbocyanine (Cy5); cycle threshold (Ct); forkhead box a2 (Foxa2); glucose transporter 2 (Glut2); hepatocyte nuclear factor 3β (HNF3β); inward rectifier potassium channel member 6.2 (KIR6.2); long-chain fatty acyl-CoAs (LCFA-CoAs); pancreatic duodenal homeobox 1 (Pdx1); persistent hyperinsulinemic hypoglycemia of infancy (PHHI); postnatal day 8 (P8); short-chain l-3-hydroxyacyl-coenzyme A dehydrogenase (schad); short-chain l-3-hydroxyacyl-coenzyme A dehydrogenase gene (Hadhsc); sulfonylurea receptor 1 (SUR1); transgenic mouse with Cre recombinase under control of the rat insulin 2 promoter (Ins.Cre).
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Ben-Shushan E, Marshak S, Shoshkes M, Cerasi E, Melloul D. A pancreatic beta-cell-specific enhancer in the human PDX-1 gene is regulated by hepatocyte nuclear factor 3beta (HNF-3beta), HNF-1alpha, and SPs transcription factors. J. Biol. Chem. 2001;276:17533–17540. doi: 10.1074/jbc.M009088200. [DOI] [PubMed] [Google Scholar]
- 2.Sharma S, et al. Hormonal regulation of an islet-specific enhancer in the pancreatic homeobox gene STF-1. Mol. Cell. Biol. 1997;17:2598–2604. doi: 10.1128/mcb.17.5.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu KL, et al. Hepatocyte nuclear factor 3beta is involved in pancreatic beta-cell-specific transcription of the pdx-1 gene. Mol. Cell. Biol. 1997;17:6002–6013. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinstein DC, et al. The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- 5.Ang SL, Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- 6.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 7.Sund NJ, et al. Tissue-specific deletion of Foxa2 in pancreatic beta cells results in hyperinsulinemic hypoglycemia. Genes Dev. 2001;15:1706–1715. doi: 10.1101/gad.901601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguilar-Bryan L, et al. Toward understanding the assembly and structure of KATP channels. Physiol. Rev. 1998;78:227–245. doi: 10.1152/physrev.1998.78.1.227. [DOI] [PubMed] [Google Scholar]
- 9.Ashcroft FM, Gribble FM. Correlating structure and function in ATP-sensitive K+ channels. Trends Neurosci. 1998;21:288–294. doi: 10.1016/s0166-2236(98)01225-9. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Gauthier BR, Hagenfeldt-Johansson KA, Iezzi M, Wollheim CB. Foxa2 (HNF3beta) controls multiple genes implicated in metabolism-secretion coupling of glucose-induced insulin release. J. Biol. Chem. 2002;277:17564–17570. doi: 10.1074/jbc.M111037200. [DOI] [PubMed] [Google Scholar]
- 11.Babenko AP, Aguilar-Bryan L, Bryan J. A view of sur/KIR6.X, KATP channels. Annu. Rev. Physiol. 1998;60:667–687. doi: 10.1146/annurev.physiol.60.1.667. [DOI] [PubMed] [Google Scholar]
- 12.Miki T, Nagashima K, Seino S. The structure and function of the ATP-sensitive K+ channel in insulin-secreting pancreatic beta-cells. J. Mol. Endocrinol. 1999;22:113–123. doi: 10.1677/jme.0.0220113. [DOI] [PubMed] [Google Scholar]
- 13.Cook DL, Satin LS, Ashford ML, Hales CN. ATP-sensitive K+ channels in pancreatic beta-cells. Spare-channel hypothesis. Diabetes. 1988;37:495–498. doi: 10.2337/diab.37.5.495. [DOI] [PubMed] [Google Scholar]
- 14.Gopel SO, et al. Regulation of glucagon release in mouse α-cells by KATP channels and inactivation of TTX-sensitive Na+ channels. J. Physiol. 2000;528:509–520. doi: 10.1111/j.1469-7793.2000.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meissner T, Beinbrech B, Mayatepek E. Congenital hyperinsulinism: molecular basis of a heterogeneous disease. Hum. Mutat. 1999;13:351–361. doi: 10.1002/(SICI)1098-1004(1999)13:5<351::AID-HUMU3>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 16.Thomas PM, et al. Mutations in the sulfonylurea receptor gene in familial persistent hyperinsulinemic hypoglycemia of infancy. Science. 1995;268:426–429. doi: 10.1126/science.7716548. [DOI] [PubMed] [Google Scholar]
- 17.Kane C, et al. Loss of functional KATP channels in pancreatic beta-cells causes persistent hyperinsulinemic hypoglycemia of infancy. Nat. Med. 1996;2:1344–1347. doi: 10.1038/nm1296-1344. [DOI] [PubMed] [Google Scholar]
- 18.Dunne MJ, Cosgrove KE, Shepherd RM, Ammala C. Potassium channels, sulphonylurea receptors and control of insulin release. Trends Endocrinol. Metab. 1999;10:146–152. doi: 10.1016/s1043-2760(98)00135-0. [DOI] [PubMed] [Google Scholar]
- 19.Kukuvitis A, Deal C, Arbour L, Polychronakos C. An autosomal dominant form of familial persistent hyperinsulinemic hypoglycemia of infancy, not linked to the sulfonylurea receptor locus. J. Clin. Endocrinol. Metab. 1997;82:1192–1194. doi: 10.1210/jcem.82.4.3904. [DOI] [PubMed] [Google Scholar]
- 20.Edlund H. Pancreatic organogenesis--developmental mechanisms and implications for therapy. Nat. Rev. Genet. 2002;3:524–532. doi: 10.1038/nrg841. [DOI] [PubMed] [Google Scholar]
- 21.Cherksey B, Altszuler N. Tolbutamide and glyburide differ in effectiveness to displace alpha- and beta-adrenergic radioligands in pancreatic islet cells and membranes. Diabetes. 1984;33:499–503. doi: 10.2337/diab.33.5.499. [DOI] [PubMed] [Google Scholar]
- 22.Gorus FK, Schuit FC, In’t Veld PA, Gepts W, Pipeleers DG. Interaction of sulfonylureas with pancreatic beta-cells. A study with glyburide. Diabetes. 1988;37:1090–1095. doi: 10.2337/diab.37.8.1090. [DOI] [PubMed] [Google Scholar]
- 23.Miki T, et al. Defective insulin secretion and enhanced insulin action in KATP channel-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 1998;95:10402–10406. doi: 10.1073/pnas.95.18.10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seghers V, Nakazaki M, DeMayo F, Aguilar-Bryan L, Bryan J. Sur1 knockout mice. A model for KATP channel-independent regulation of insulin secretion. J. Biol. Chem. 2000;275:9270–9277. doi: 10.1074/jbc.275.13.9270. [DOI] [PubMed] [Google Scholar]
- 25.Shiota C, et al. Sulfonylurea receptor type 1 knock-out mice have intact feeding-stimulated insulin secretion despite marked impairment in their response to glucose. J. Biol. Chem. 2002;277:37176–37183. doi: 10.1074/jbc.M206757200. [DOI] [PubMed] [Google Scholar]
- 26.Scearce LM, et al. Functional genomics of the endocrine pancreas: the pancreas clone set and PancChip, new resources for diabetes research. Diabetes. 2002;51:1997–2004. doi: 10.2337/diabetes.51.7.1997. [DOI] [PubMed] [Google Scholar]
- 27.Kaestner KH, et al. Transcriptional program of the endocrine pancreas in mice and humans. Diabetes. 2003;52:1604–1610. doi: 10.2337/diabetes.52.7.1604. [DOI] [PubMed] [Google Scholar]
- 28.Lee CS, et al. Foxa2 controls Pdx1 gene expression in pancreatic beta-cells in vivo. Diabetes. 2002;51:2546–2551. doi: 10.2337/diabetes.51.8.2546. [DOI] [PubMed] [Google Scholar]
- 29.Nakata M, Yada T, Nakagawa S, Kobayashi K, Maruyama I. Citrulline-argininosuccinate-arginine cycle coupled to Ca2+-signaling in rat pancreatic beta-cells. Biochem. Biophys. Res. Commun. 1997;235:619–624. doi: 10.1006/bbrc.1997.6854. [DOI] [PubMed] [Google Scholar]
- 30.Barycki JJ, et al. Biochemical characterization and crystal structure determination of human heart short chain L-3-hydroxyacyl-CoA dehydrogenase provide insights into catalytic mechanism. Biochemistry. 1999;38:5786–5798. doi: 10.1021/bi9829027. [DOI] [PubMed] [Google Scholar]
- 31.Vredendaal PJ, et al. Human short-chain L-3-hydroxyacyl-CoA dehydrogenase: cloning and characterization of the coding sequence. Biochem. Biophys. Res. Commun. 1996;223:718–723. doi: 10.1006/bbrc.1996.0961. [DOI] [PubMed] [Google Scholar]
- 32.Agren A, Borg K, Brolin SE, Carlman J, Lundqvist G. Hydroxyacyl CoA dehydrogenase, an enzyme important in fat metabolism in different cell types in the islets of Langerhans. Diabete Metab. 1977;3:169–172. [PubMed] [Google Scholar]
- 33.Hammar H, Berne C. The activity of beta-hydroxyacyl-CoA dehydrogenase in the pancreatic islets of hyperglycaemic mice. Diabetologia. 1970;6:526–528. doi: 10.1007/BF01211896. [DOI] [PubMed] [Google Scholar]
- 34.Clayton PT, et al. Hyperinsulinism in short-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency reveals the importance of beta-oxidation in insulin secretion. J. Clin. Invest. 2001;108:457–465. doi:10.1172/JCI200111294. doi: 10.1172/JCI11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eaton S, et al. Short-chain 3-hydroxyacyl-CoA dehydrogenase deficiency associated with hyperinsulinism: a novel glucose-fatty acid cycle? Biochem. Soc. Trans. 2003;31:1137–1139. doi: 10.1042/bst0311137. [DOI] [PubMed] [Google Scholar]
- 36.Molven A, et al. Familial hyperinsulinemic hypoglycemia caused by a defect in the SCHAD enzyme of mitochondrial fatty acid oxidation. Diabetes. 2004;53:221–227. doi: 10.2337/diabetes.53.1.221. [DOI] [PubMed] [Google Scholar]
- 37.Overdier DG, Porcella A, Costa RH. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol. Cell. Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hellerstrom C, Swenne I. Functional maturation and proliferation of fetal pancreatic beta-cells. Diabetes. 1991;40:89–93. doi: 10.2337/diab.40.2.s89. [DOI] [PubMed] [Google Scholar]
- 39.Freinkel N, et al. Differential effects of age versus glycemic stimulation on the maturation of insulin stimulus-secretion coupling during culture of fetal rat islets. Diabetes. 1984;33:1028–1038. doi: 10.2337/diab.33.11.1028. [DOI] [PubMed] [Google Scholar]
- 40.Eriksson U, Andersson A, Efendic S, Elde R, Hellerstrom C. Diabetes in pregnancy: effects on the foetal and newborn rat with particular regard to body weight, serum insulin concentration and pancreatic contents of insulin, glucagon and somatostatin. Acta Endocrinol. 1980;94:354–364. doi: 10.1530/acta.0.0940354. [DOI] [PubMed] [Google Scholar]
- 41.Girard JR, et al. Fuels, hormones, and liver metabolism at term and during the early postnatal period in the rat. J. Clin. Invest. 1973;52:3190–3200. doi: 10.1172/JCI107519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes SJ. The role of reduced glucose transporter content and glucose metabolism in the immature secretory responses of fetal rat pancreatic islets. Diabetologia. 1994;37:134–140. doi: 10.1007/s001250050083. [DOI] [PubMed] [Google Scholar]
- 43.Rorsman P, et al. Failure of glucose to elicit a normal secretory response in fetal pancreatic beta cells results from glucose insensitivity of the ATP-regulated K+ channels. Proc. Natl. Acad. Sci. U. S. A. 1989;86:4505–4509. doi: 10.1073/pnas.86.12.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenbaum CJ, Havel PJ, Taborsky GJ, Jr, Klaff LJ. Intra-islet insulin permits glucose to directly suppress pancreatic A cell function. J. Clin. Invest. 1991;88:767–773. doi: 10.1172/JCI115375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishihara H, Maechler P, Gjinovci A, Herrera PL, Wollheim CB. Islet beta-cell secretion determines glucagon release from neighbouring alpha-cells. Nat. Cell Biol. 2003;5:330–335. doi: 10.1038/ncb951. [DOI] [PubMed] [Google Scholar]
- 46.Prentki M. New insights into pancreatic beta-cell metabolic signaling in insulin secretion. Eur. J. Endocrinol. 1996;134:272–286. doi: 10.1530/eje.0.1340272. [DOI] [PubMed] [Google Scholar]
- 47.Glaser B, Thornton P, Otonkoski T, Junien C. Genetics of neonatal hyperinsulinism. Arch. Dis. Child. Fetal Neonatal Ed. 2000;82:F79–F86. doi: 10.1136/fn.82.2.F79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennett MJ, et al. Fatal hepatic short-chain L-3-hydroxyacyl-coenzyme A dehydrogenase deficiency: clinical, biochemical, and pathological studies on three subjects with this recently identified disorder of mitochondrial beta-oxidation. Pediatr. Dev. Pathol. 1999;2:337–345. doi: 10.1007/s100249900132. [DOI] [PubMed] [Google Scholar]
- 49.Henquin JC. Triggering and amplifying pathways of regulation of insulin secretion by glucose. Diabetes. 2000;49:1751–1760. doi: 10.2337/diabetes.49.11.1751. [DOI] [PubMed] [Google Scholar]
- 50.Wilkinson, D.G. 1992. In situ hybridization: a practical approach. IRL Press at Oxford University Press. New York, New York, USA. 163 pp.
- 51.Kaestner KH, Katz J, Liu Y, Drucker DJ, Schutz G. Inactivation of the winged helix transcription factor HNF3alpha affects glucose homeostasis and islet glucagon gene expression in vivo. Genes Dev. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scharp DW, Kemp CB, Knight MJ, Ballinger WF, Lacy PE. The use of Ficoll in the preparation of viable islets of Langerhans from the rat pancreas. Transplantation. 1973;16:686–689. doi: 10.1097/00007890-197312000-00028. [DOI] [PubMed] [Google Scholar]
- 53.Hara M, et al. Transgenic mice with green fluorescent protein-labeled pancreatic beta-cells. Am. J. Physiol. Endocrinol. Metab. 2003;284:E177–E183. doi: 10.1152/ajpendo.00321.2002. [DOI] [PubMed] [Google Scholar]
- 54.Manduchi E, et al. RAD and the RAD Study-Annotator: an approach to collection, organization, and exchange of all relevant information for high-throughput gene expression studies. Bioinformatics. 2004;20:452–459. doi: 10.1093/bioinformatics/btg428. [DOI] [PubMed] [Google Scholar]
- 55.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 56.Wells J, Farnham PJ. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. . Methods. 2002; 26:48–56. doi: 10.1016/S1046-2023(02)00007-5. [DOI] [PubMed] [Google Scholar]