Abstract
Mammary gland tumors are one of the most common neoplasms in female dogs, and certain breeds are prone to develop the disease. The use of biomarkers in canines is still restricted to research purposes. Therefore, the necessity to analyze gene profiles in different mammary entities in large sample sets is evident in order to evaluate the strength of potential markers serving as future prognostic factors. The aim of the present study was to analyze the gene expression of 16 target genes (BRCA1, BRCA2, FOXO3, GATA4, HER2, HMGA1, HMGA2, HMGB1, MAPK1, MAPK3, MCL1, MYC, PFDN5, PIK3CA, PTEN, and TP53) known to be involved in human and canine mammary neoplasm development. Expression was analyzed in 111 fresh frozen (FF) and in 170 formalin-fixed, paraffin-embedded (FFPE) specimens of neoplastic and non-neoplastic canine mammary tissues using a multiplexed branched-DNA (b-DNA) assay. TP53, FOXO3, PTEN, and PFDN5 expression revealed consistent results with significant low expression in malignant tumors. The possibility of utilizing them as predictive factors as well as for assisting in the choice of an adequate gene therapy may help in the development of new and improved approaches in canine mammary tumors.
Keywords: canine mammary tumor, gene expression, RNA, multiplexed branched-DNA (b-DNA) assay, formalin-fixed, paraffin-embedded samples, fresh frozen tissue
1. Introduction
Approximately 50% of canine mammary tumors appear to be malignant [1,2]. Certain breeds are reported to be predisposed with influence of the geographic location [3]. The highest relative risk ratio of developing benign mammary tumors was found in Yorkshire terriers followed by poodles, dachshunds, and cocker spaniels in Germany. In malignant mammary tumors, the prevalence was higher in poodles followed by dachshunds, cocker spaniels, and Yorkshire terriers [4].
The postoperative median survival time can be shorter than two years depending on the clinical stage and histologic grade of the tumor [5]. Therefore, the clinical stage at mammary tumor diagnosis is one of the most important prognostic factors in dogs [6]. However, it is only possible to be assessed in symptomatic patients and is thus not suitable as a mammary tumor predictive parameter.
Immunohistochemistry (IHC), alongside conventional histopathology, plays an important role as a diagnostic tool in classifying tumors in humans and dogs [7,8]. Most studies on mammary tumors in dogs are made through IHC and determine biomarkers on the protein level [9]. This technique has proven to be suitable when performing large multicenter studies. However, the limitations rely on the semiquantitative and subjective interpretation of its results [10]. Therefore, the present study utilized branched DNA (b-DNA) assay which, in combination with xMAP® magnetic beads technology, permits the concomitant expression analysis of several target genes within a single sample [11]. It also presents a simpler and more rapid workflow, providing highly sensitive quantitative results when compared to other gene expression techniques such as real-time polymerase chain reaction (qPCR) [12], which is commonly the method of choice when analyzing gene expression [13]. The possibility of analyzing a vast number of samples at the same time is also an important advantage of the proposed technique.
The need to comprehensively analyze gene profiles in different canine mammary tumor (CMT) entities to find potential candidate genes which could serve as predictive factors is evident, considering little is known in this regard to date [14]. Thus, the detection of suitable biomarkers could assist in the neoplasms′ early detection and support in providing a therapeutic approach.
In contrast to the human counterpart, the use of mammary tumor biomarkers in canines is still restricted to research purposes [14,15]. Expression of BRCA1 and BRCA2 in CMT was analyzed and it was found to be associated with an increased mammary tumor risk in English Springer Spaniels (ESS) [16]. A possible influence of those genes in tumor development is also suggested in other studies [17,18,19,20]. Further targets of those analyzed herein have also been investigated in CMT such as TP53 [21,22], PTEN [23,24,25,26,27], PFDN5 [15,26], MYC [27], HER2 [28], and MCL1 [29] associating their expression patterns with the development of CMT.
The present study proposes the concomitant analysis of 16 onco- and suppressor-genes (BRCA1, BRCA2, FOXO3, GATA4, HER2, HMGA1, HMGA2, HMGB1, MAPK1, MAPK3, MCL1, MYC, PFDN5, PIK3CA, PTEN, and TP53) regarded as being involved in neoplasm development using multiplex branched-DNA technology in fresh frozen (FF) and formalin-fixed, paraffin-embedded (FFPE) tissues. A previous study demonstrated that the analysis of canine mammary FF and FFPE samples via b-DNA assay is feasible [30]. Therefore, the expression analysis in 111 FF and 170 FFPE specimens of canine mammary tissues aims to compare the expression patterns of the candidate genes in neoplastic (benign and malignant) and non-neoplastic tissues with the intention of finding new potential tumor markers. This characterization might serve to gain a better understanding of the tumor pathogenesis of different canine mammary tumor types in further studies. Moreover, potential target genes might contribute to determining whether certain breeds are at risk of developing mammary tumors and are able to contribute to the therapeutic approach.
2. Results
Of the 180 FFPE specimens, 15 of them (7 benign tumors and 6 malignant tumors) were entirely excluded due to values below the LOD (limit of detection). Ninety-eight of FFPE samples of GATA4 were excluded because of the LOD, whereas 84 FFPE samples of HMGA2 were excluded. BRCA1 also showed a high number of exclusions with 63 FFPE samples. The remaining samples of GATA4, HMGA2, and BRCA1 as well as all specimens of MYC, BRCA2, and MAPK1 in both FF and FFPE samples revealed low expression (below 0.1) and no significant differences were found between the histologic groups.
All genes from FFPE samples except for PTEN showed lower expression levels when compared to FF specimens (exemplary box plots for the gene PFDN5 are shown in Figure 1A and Figure 2A, where different expression levels between FF and FFPE specimens are noted). The samples of FF and FFPE origin were analyzed separately.
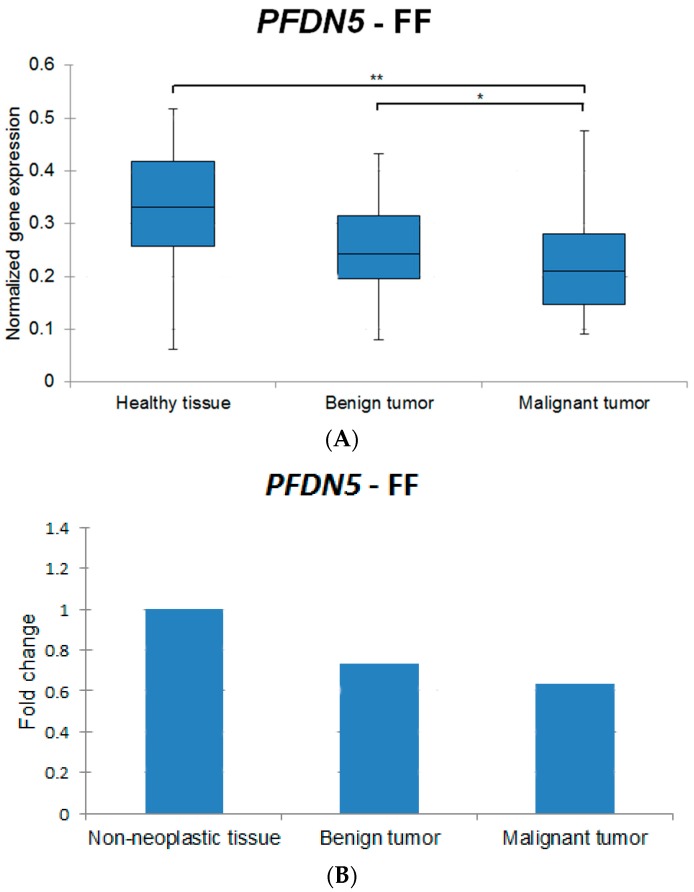
Figure 1.
(A) Exemplary box plot indicating the normalized gene expression of PFDN5 in fresh frozen samples in the different histologic groups. Statistically significant differences are indicated by an asterisk (*). Results are classified as significant (* p < 0.05) and very significant (** p < 0.01). The box includes cases from the 25th to the 75th percentile. The horizontal line within the box represents the median and the upper and lower bars are the largest and lowest observed values. Significant differences are revealed between the groups healthy tissue vs. malignant tumors and benign tumors vs. malignant tumors; (B) Normalized expression fold changes (mean) indicating the percentage change of PFDN5 in fresh frozen samples in the different histologic groups.
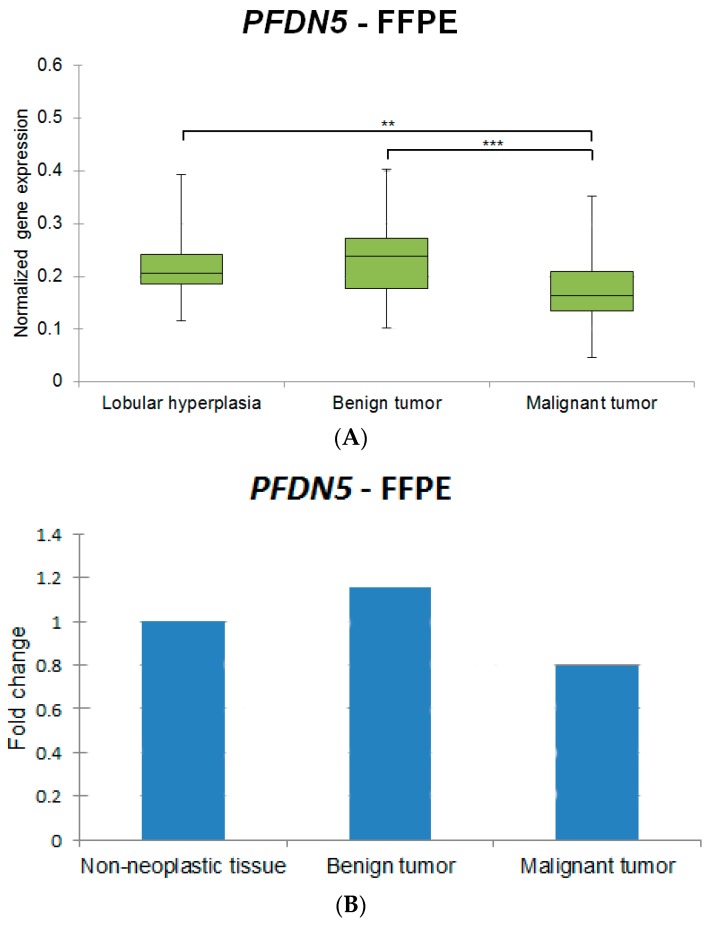
Figure 2.
(A) Exemplary box plot indicating the normalized gene expression of PFDN5 in formalin-fixed, paraffin-embedded samples in the different histologic groups. Statistically significant differences are indicated by an asterisk (*). Results are classified as very significant (** p < 0.01) and extremely significant (*** p < 0.001). The box includes cases from the 25th to the 75th percentile. The horizontal line within the box represents the median and the upper and lower bars are the largest and lowest observed values. Significant differences are revealed between the groups lobular hyperplasias vs. malignant tumors and benign tumors vs. malignant tumors. A lower level of expression can be clearly identified when comparing the expression of PFDN5 in fresh frozen samples (Figure 1); (B). Normalized expression fold changes (mean) indicating the percentage change of PFDN5 in formalin-fixed, paraffin-embedded samples in the different histologic groups.
Exemplary graphs representing the percentage change of PFDN5 between the histologic groups in FF and FFPE are represented in Figure 1B and Figure 2B, respectively.
The expression of the housekeeping genes (HKG) varied, and the mean expression of the three housekeeping genes was calculated to achieve a more accurate normalization.
The number of samples in each group are listed in Table 1.
Table 1.
Number of specimens of fresh frozen (FF) and formalin-fixed, paraffin-embedded (FFPE) per group.
| Group | FF (n) | FFPE (n) |
|---|---|---|
| 1. Healthy mammary tissue (non-neoplastic tissue) | 15 | 4 |
| 2. Lobular hyperplasias (non-neoplastic tissue) | 3 | 20 |
| 3. Benign tumors | 33 | 47 |
| 4. Malignant tumors | 60 | 84 |
| Total | 111 | 155 |
2.1. Gene Expression
2.1.1. Fresh Frozen Tissues
Table 2 shows the results of the target gene expression in FF samples when comparing the different histologic groups. The group of lobular hyperplasias was excluded due to the small number of samples (n = 3). The non-neoplastic tissue for FF specimens was considered to be healthy mammary tissue.
Table 2.
Gene expression of the target genes when comparing the different canine mammary tissues from FF samples. Arrows show a higher or lower expression of the first group in relation to the second one (example: first group vs. second group).
| Target Gene | Healthy Tissue vs. Benign Tumors | Healthy Tissue vs. Malignant Tumors | Benign Tumors vs. Malignant Tumors |
|---|---|---|---|
| BRCA1 | - | - | - |
| BRCA2 | - | - | - |
| FOXO3 | - | ↑ * | ↑ * |
| GATA4 | - | - | - |
| HER2 | - | ↑ * | ↑ * |
| HMGA1 | ↑ ** | ↑ * | - |
| HMGA2 | - | - | |
| HMGB1 | ↑ * | - | - |
| MAPK1 | - | - | - |
| MAPK3 | - | ↑ * | - |
| MCL1 | ↑ * | ↑ ** | ↑ * |
| MYC | - | - | - |
| PFDN5 | - | ↑ ** | ↑ * |
| PIK3CA | - | - | - |
| PTEN | - | - | - |
| TP53 | - | ↑ * | - |
Results are classified as significant (* p < 0.05) and very significant (** p < 0.01). Cells with hyphen (-) revealed no significant differences.
In the groups healthy tissue vs. benign tumors and healthy tissue vs. malignant tumors, all target genes with statistically significant differences showed higher expression in the healthy mammary tissue. In benign tumors vs. malignant tumors, all genes were more highly expressed in benign tumors (Table 2).
2.1.2. Formalin-Fixed, Paraffin-Embedded Tissues
Table 3 summarizes the results of the target gene expression in FFPE samples when comparing the different histologic groups. The group of healthy mammary tissue was excluded due to the insufficient number of samples (n = 4). The group lobular hyperplasia was considered as the non-neoplastic tissue for FFPE specimens. Results of lobular hyperplasias vs. benign tumors are not displayed in the table due to the lack of statistically significant differences.
Table 3.
Gene expression of the target genes when comparing the different canine mammary tissues from FFPE samples. Arrows show a higher or lower expression of the first group in relation to the second one (example: first group vs. second group).
| Target Gene | Lobular Hyperplasias vs. Malignant Tumors | Benign Tumors vs. Malignant Tumors |
|---|---|---|
| BRCA1 | - | - |
| BRCA2 | - | - |
| FOXO3 | ↑ ** | ↑ *** |
| GATA4 | - | - |
| HER2 | ↑ *** | ↑ *** |
| HMGA1 | ↑ * | - |
| HMGA2 | - | - |
| HMGB1 | ↑ *** | ↑ * |
| MAPK1 | - | - |
| MAPK3 | ↑ ** | ↑ *** |
| MCL1 | ↑ *** | ↑ *** |
| MYC | - | - |
| PFDN5 | ↑ ** | ↑ *** |
| PIK3CA | ↑ * | ↑ ** |
| PTEN | - | ↑ ** |
| TP53 | ↑ * | ↑ * |
Results are classified as significant (* p < 0.05), very significant (** p < 0.01), and extremely significant (*** p < 0.001). Cells with hyphen (-) revealed no significant differences.
All genes showing statistically significant differences in lobular hyperplasias vs. malignant tumors were more highly expressed in lobular hyperplasias (non-neoplastic tissue). Between the groups benign tumors vs. malignant tumors, all genes revealing statistically significant differences were more highly expressed in the benign tumors.
3. Discussion
The gene expression profile of the mammary tumor in dogs is, to date, not well characterized and the use of tumor markers is still for research purposes only [15]. The analysis and differentiation of the mammary tumors is the basis for pathogenesis, diagnosis, prognosis, new therapy options, and good breed hygiene practice. Until now, most mammary studies in dogs have utilized IHC and analyzed proteins [9]. IHC has proved to be an excellent technique when performing large multicenter studies. Nonetheless, it is laborious and time-consuming when analyzing several target genes [31]. The present study proposes the analysis of 16 target genes using branched-DNA assay which, when allied with xMAP® magnetic beads, enables the analysis of several genes within a single sample (multiplexing).
The main aim of this study was to compare the expression of target genes known to be involved in tumor growth in dogs and humans [32,33,34] in different FF and FFPE canine mammary samples, as well as to find new potential tumor markers.
The significant reduction in the expression of the tumor suppressors TP53, FOXO3, PTEN, and PFDN5 in malignant tumors found in the present study confirms previous reports [35,36,37,38] and underlines their role in cellular growth. Hence, these genes could represent potential markers to predict CMT.
To the best of our knowledge, no specific reports on the expression of FOXO3 in canine mammary tissues have been elucidated so far. The results of the present study corroborate what has already been reported in humans, demonstrating its higher expression in non-neoplastic tissues. The inactivation of FOXO subfamily controls several functions, including cell differentiation, proliferation, cell death, metabolism, and longevity [35]. A study from 2013 revealed that a high expression level of FOXO3 was significantly correlated with long-term survival, indicating that FOXO3 expression is a favorable prognostic marker in breast cancer [39] and this may also be applicable for dogs.
In the present study, TP53 was revealed to be more highly expressed in non-neoplastic tissue. However, most of the existing studies on humans and canines analyzed point mutations instead of gene expression levels [21,22,36,40]. Muto and colleagues revealed that mutations were detected not only in mammary carcinomas but also in benign tumors [21]. To the best of our knowledge, no gene expression analysis has been performed with TP53 in dogs to date. Thus, it is suggested that the results herein might correlate with existing reports [36,41] where the described presence of mutations could correspond to the lower levels of TP53 in neoplastic tissues found in our study. Moreover, TP53 is known to be a tumor suppressor gene [42] and, therefore, its lower expression in neoplastic tissues might lead to uncontrolled cellular growth, a fact which corroborates the findings herein. Further studies at mRNA level are worth carrying out to deepen existing knowledge concerning the role of this gene in CMT.
The present study revealed higher expression of PTEN in non-neoplastic tissues and benign tumors in FFPE samples, thus supporting existing studies. PTEN is known as a tumor suppressor gene [37] and the role of its loss has been largely investigated in human breast carcinomas [43,44,45]. Known for its importance in humans, the role of PTEN has also been investigated in CMT and its behavior in dogs resembles what has been seen in humans, associating low expression of PTEN with malignancy [23,24,25,26]. A recent study revealed frequent loss of the region harboring PTEN on the canine chromosome 26, indicating an important event in CMT development [27].
Rare reports mention the role of PFDN5 in breast cancer. However, it has been shown to be a tumor suppressor candidate in leukemia and tongue cancer [38]. The outcomes herein revealed higher expression in non-neoplastic tissue, suggesting it as a potential tumor marker just as demonstrated by Hennecke and colleagues [15]. Moreover, a previous study demonstrated that PFDN5 was recurrently deleted in CMT [26].
The genes MAPK3, HMGB1, HMGA1, PIK3CA, and MCL1 are genes known to stimulate cellular growth; however, contrary to what is normally reported in humans [46,47,48,49,50], in the present study these genes were shown to be consistently expressed at lower levels in malignant tumors. Therefore, it is hypothesized that those genes play a different role in canine mammary tumors.
The role of HER2 is well characterized in humans and it is widely used as a prognostic factor [51,52]. Interestingly, in contrast to a previous study on CMT which revealed different percentages of HER2 being overexpressed in carcinomas [28], the present study demonstrated lower expression of the referred gene in malignant tumors. The relevance of its overexpression as a prognostic factor has, however, still not been clearly determined [28]. Therefore, further analyses are necessary for a better understanding of the interaction of HER2 in CMT.
Intriguingly, the present study did not show conclusive results for BRCA1 and BRCA2 despite their well-characterized role and importance in human [53] and canine [16] mammary tumors. The reason for this could be the extremely low expression of both genes in the FF and FFPE samples investigated here.
4. Materials and Methods
4.1. Tissue Samples
4.1.1. Formalin-Fixed, Paraffin-Embedded Blocks
The samples utilized in this work were retrospectively retrieved from the Department of Pathology, University of Veterinary Medicine Hannover, Hannover, Germany, between the years 1993 and 2000. The mammary specimens were fixed in 10% neutral buffered formalin as routinely performed and embedded in paraffin wax. The blocks were stored in the archive at room temperature. A total number of 170 samples were selected for the analysis.
4.1.2. Fresh Frozen Tumor Samples
The samples were obtained from patients which had undergone surgery in the Small Animal Clinic, University of Veterinary Medicine Hannover, Hannover, Germany, between 2003 and 2011 (identification number: 96A697; 19 January 2005), as well as from patients from the Clinic of Small Animals, Institute of Veterinary Medicine, Georg-August-University Göttingen, Göttingen, Germany, from 2013 and 2014. All utilized tissue samples were removed with consent of the owner. The specimens were frozen in liquid nitrogen directly after surgical removal and maintained at −80 °C until RNA isolation. A total number of 111 samples were selected for the analysis.
4.2. Nucleic Acid Isolation and Quantification
4.2.1. Formalin-Fixed, Paraffin-Embedded Samples
Prior to RNA isolation, two 20 µm thick paraffin sections of the FFPE blocks were prepared using a microtome (pfm Slide 2003, pfm medical AG, Cologne, Germany). Samples were deparaffinized using the Deparaffinization Solution (Qiagen GmbH, Hilden, Germany) and the RNA was isolated as previously described [30]. Samples were stored at −80 °C until the experiment required further usage.
4.2.2. Fresh Frozen Samples
Prior to the nucleic acid isolation, frozen tissue was homogenized using a Tissue Lyser II and 5 mm stainless steel beads (Qiagen GmbH). Nucleic acid isolation was performed with the RNeasy Mini kit (Qiagen GmbH) following the manufacturer’s instructions, with an additional step of digestion using the RNase-free DNase Set (Qiagen GmbH) according to the manufacturer’s protocol. Additionally, genomic DNA was digested using RQ1 RNase-free DNase (Promega GmbH, Mannheim, Germany). RNA yield was determined with the Take3 Micro-Volume Plate (BioTek, Bad Friedrichshall, Heilbronn, Germany). Samples were stored at −80 °C until use.
4.2.3. Target Genes
The following 16 canine genes were analyzed: BRCA1, BRCA2, FOXO3, GATA4, HER2, HMGA1, HMGA2, HMGB1, MAPK1, MAPK3, MCL1, MYC, PFDN5, PIK3CA, PTEN, and TP53 (Table 4).
Table 4.
Target and housekeeping genes with their approved name and respective accession numbers.
| Target Gene | Approved Name | Accession Number |
|---|---|---|
| BRCA1 | BRCA1, DNA repair associated | NM_001013416 |
| BRCA2 | BRCA2, DNA repair associated | NM_001006653 |
| FOXO3 | Forkhead box O3 | NM_003639400 |
| GATA4 | GATA binding protein 4 | NM_001048112 |
| HER2 | Erb-b2 receptor tyrosine kinase 2 | NM_001003217 |
| HMGA1 | High mobility group AT-hook 1 | NM_001003387 |
| HMGA2 | High mobility group AT-hook 2 | XM_005625590 |
| HMGB1 | High mobility group box 1 | NM_001002937 |
| MAPK1 | Mitogen-activated protein kinase 1 | NM_001110800 |
| MAPK3 | Mitogen-activated protein kinase 3 | NM_001252035 |
| MCL1 | Myeloid cell leukemia 1 | NM_001003016 |
| MYC | V-myc avian myelocytomatosis viral oncogene homolog | NM_001003246 |
| PFDN5 | Prefoldin subunit 5 | NM_001251949 |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α | XM_545208.4 |
| PTEN | Phosphatase and tensin homolog | NM_001003192 |
| TP53 | Tumor protein p53 | NM_001003210 |
| ACTB | β-actin | XM_536888 |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | NM_001003142 |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | NM_001003357 |
4.2.4. Housekeeping Genes
Three different housekeeping genes (HKG) were used: ACTB (β-actin), GAPDH (glyceraldehyde 3-phosphate dehydrogenase), and HPRT1 (hypoxanthine phosphoribosyltransferase 1) (Table 4).
4.2.5. Multiplexed Branched-DNA (b-DNA) Assay
Gene expression was analyzed using the QuantiGene 2.0 Plex Assay (Affymetrix, Santa Clara, CA, USA). Probe sets for the target genes were custom designed based on their accession numbers (Table 4) from Affymetrix.
Briefly, each bead set is coated with a reagent specific to a certain target in order to detect a special analyte in the analyzed material. Multiple readings are made on each bead set resulting in an individual fluorescent signal for each assay. The multiplex branched-DNA assay permits rapid and accurate analysis of up to 100 targets within a single specimen.
In preliminary experiments, an evaluation of the technology on the biological material was performed in order to identify the ideal sample input for the specimens. These analyses revealed that 250 and 200 ng of RNA were sufficient amounts to perform the assay for FF and FFPE specimens, respectively. Moreover, considering that the amount of duplicate measurements of certain samples intra-assay were consistent (represented by a low coefficient of variation), a single measurement was considered sufficient. The assay was carried out according to the manufacturer’s protocol.
4.3. Histologic Classification
All samples were evaluated and diagnosed using hematoxylin and eosin staining. Samples were separated into the following four groups: (1) healthy mammary tissue (non-neoplastic tissue); (2) lobular hyperplasia (non-neoplastic tissue); (3) benign tumor; and (4) malignant tumor.
4.4. Data and Statistical Analysis
Specimens with expression values below the limit of detection (LOD—background plus 3 times standard deviation of the background) were excluded (according to recommendations by the assay provider). Samples were normalized against the average of the three HKG. Data were not normally distributed and therefore the Mann-Whitney U test was performed using the software SAS Enterprise Guide 7.1 (SAS Institute Inc., Cary, NC, USA). Results were classified as significant (* p < 0.05), very significant (** p < 0.01), and extremely significant (*** p < 0.001).
5. Conclusions
In conclusion, based on the outcomes of the present study, the suppressor genes TP53, FOXO3, PTEN and PFDN5 are considered as potential markers for predicting canine mammary tumors. The results herein are in line with the literature, revealing low expression of these genes in malignant tumors, suggesting they play a role in cellular growth. Further investigations are needed to prove if these genes alone play a role in general tumor progression or if they, in combination with other genes, confer an important predisposition to the disease with further development of mammary tumors. Moreover, the clinical follow-up data of the patients would assist in the interpretation of the gene expression results herein described. A comparison of the expression between already existing methods for gene expression measurements and multiplex branched-DNA assay would enable the establishment of a cutoff for each target gene in the different histologic groups.
The multiplex technology is certainly a method which enables the analysis of several samples which present small amounts of starting material of FF and FFPE tissue. Moreover, several target genes are concomitantly analyzed, thereby reducing hands-on time and costs.
Acknowledgments
The authors would like to thank CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior)-Brazil for providing funding to carry out this research. The authors would also like to thank Frances Sherwood-Brock for proofreading the manuscript and Martin Beyerbach for his support with statistical analysis.
Author Contributions
Ingo Nolte and Hugo Murua Escobar performed the primary study design, manuscript editing and final approval; Marion Hewicker-Trautwein checked and diagnosed all samples utilized herein; Saskia Willenbrock participated in the study design and provided technical assistance; Bertram Brenig provided tumor samples and participated in the manuscript editing; Florenza Lüder Ripoli, Annika Mohr, and Susanne Conradine Hammer performed the experiments; Florenza Lüder Ripoli and Annika Mohr analyzed the data; Florenza Lüder Ripoli wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bronden L.B., Nielsen S.S., Toft N., Kristensen A.T. Data from the danish veterinary cancer registry on the occurrence and distribution of neoplasms in dogs in denmark. Vet. Rec. 2010;166:586–590. doi: 10.1136/vr.b4808. [DOI] [PubMed] [Google Scholar]
- 2.Bostock D.E. Canine and feline mammary neoplasms. Br. Vet. J. 1986;142:506–515. doi: 10.1016/0007-1935(86)90107-7. [DOI] [PubMed] [Google Scholar]
- 3.Dobson J.M. Breed-predispositions to cancer in pedigree dogs. ISRN Vet. Sci. 2013 doi: 10.1155/2013/941275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Von Bomhard D. Epidemiologie. In: Nolte I., Nolte M., editors. Praxis der Onkologie bei Hund und Katze, 1. Auflage. Enke Verlag; Stuttgart, Deutschland: 2001. pp. 104–108. [Google Scholar]
- 5.Betz D., Schoenrock D., Mischke R., Baumgartner W., Nolte I. Postoperative treatment outcome in canine mammary tumors: Multivariate analysis of the prognostic value of pre- and postoperatively available information. Tierärztliche Praxis Ausgabe K Kleintiere Heimtiere. 2012;40:235–242. [PubMed] [Google Scholar]
- 6.Karayannopoulou M., Kaldrymidou E., Constantinidis T.C., Dessiris A. Histological grading and prognosis in dogs with mammary carcinomas: Application of a human grading method. J. Comp. Pathol. 2005;133:246–252. doi: 10.1016/j.jcpa.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Duraiyan J., Govindarajan R., Kaliyappan K., Palanisamy M. Applications of immunohistochemistry. J. Pharm. Bioallied Sci. 2012;4:S307–309. doi: 10.4103/0975-7406.100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tavasoly A., Golshahi H., Rezaie A., Farhadi M. Classification and grading of canine malignant mammary tumors. Vet. Res. Forum: Int. Q. J. 2013;4:25–30. [PMC free article] [PubMed] [Google Scholar]
- 9.Cassali G.D., Lavalle G.E., Ferreira E., Estrela-Lima A., de Nardi A.B., Bertagnolli A.C., Alessi A.C., Daleck C.R., Salgado B.S., Fernandes C.G., et al. Consensus for the diagnosis, prognosis and treatment of canine mammary tumors. Braz. J. Vet. Pathol. 2011;4:153–180. [Google Scholar]
- 10.Seidal T., Balaton A.J., Battifora H. Interpretation and quantification of immunostains. Am. J. Sur. Pathol. 2001;25:1204–1207. doi: 10.1097/00000478-200109000-00013. [DOI] [PubMed] [Google Scholar]
- 11.Flagella M., Bui S., Zheng Z., Nguyen C.T., Zhang A., Pastor L., Ma Y., Yang W., Crawford K.L., McMaster G.K., et al. A multiplex branched DNA assay for parallel quantitative gene expression profiling. Anal. Biochem. 2006;352:50–60. doi: 10.1016/j.ab.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Knudsen B.S., Allen A.N., McLerran D.F., Vessella R.L., Karademos J., Davies J.E., Maqsodi B., McMaster G.K., Kristal A.R. Evaluation of the branched-chain DNA assay for measurement of rna in formalin-fixed tissues. J. Mol. Diagn. 2008;10:169–176. doi: 10.2353/jmoldx.2008.070127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pabinger S., Rödigerb S., Kriegnera A., Vierlingera K., Weinhäusel A. A survey of tools for the analysis of quantitative PCR (qPCR) data. Elsevier. 2014;1:23–33. doi: 10.1016/j.bdq.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Queiroga F.L., Raposo T., Carvalho M.I., Prada J., Pires I. Canine mammary tumours as a model to study human breast cancer: Most recent findings. In Vivo. 2011;25:455–465. [PubMed] [Google Scholar]
- 15.Hennecke S., Beck J., Bornemann-Kolatzki K., Neumann S., Murua Escobar H., Nolte I., Hammer S.C., Hewicker-Trautwein M., Junginger J., Kaup F.J., et al. Prevalence of the prefoldin subunit 5 gene deletion in canine mammary tumors. PLoS ONE. 2015;10:1589. doi: 10.1371/journal.pone.0131280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivera P., Melin M., Biagi T., Fall T., Haggstrom J., Lindblad-Toh K., von Euler H. Mammary tumor development in dogs is associated with BRCA1 and BRCA2. Cancer Res. 2009;69:8770–8774. doi: 10.1158/0008-5472.CAN-09-1725. [DOI] [PubMed] [Google Scholar]
- 17.Klopfleisch R., Gruber A.D. Increased expression of BRCA2 and RAD51 in lymph node metastases of canine mammary adenocarcinomas. Vet. Pathol. 2009;46:416–422. doi: 10.1354/vp.08-VP-0212-K-FL. [DOI] [PubMed] [Google Scholar]
- 18.Nieto A., Perez-Alenza M.D., del Castillo N., Tabanera E., Castano M., Pena L. BRCA1 expression in canine mammary dysplasias and tumours: Relationship with prognostic variables. J. Comp. Pathol. 2003;128:260–268. doi: 10.1053/jcpa.2002.0631. [DOI] [PubMed] [Google Scholar]
- 19.Ochiai K., Morimatsu M., Tomizawa N., Syuto B. Cloning and sequencing full length of canine BRCA2 and RAD51 cDNA. J. Vet. Med. Sci. 2001;63:1103–1108. doi: 10.1292/jvms.63.1103. [DOI] [PubMed] [Google Scholar]
- 20.Enginler S.O., Akis I., Toydemir T.S., Oztabak K., Haktanir D., Gunduz M.C., Kirsan I., Firat I. Genetic variations of BRCA1 and BRCA2 genes in dogs with mammary tumours. Vet. Res. Commun. 2014;38:21–27. doi: 10.1007/s11259-013-9577-7. [DOI] [PubMed] [Google Scholar]
- 21.Muto T., Wakui S., Takahashi H., Maekawa S., Masaoka T., Ushigome S., Furusato M. P53 gene mutations occurring in spontaneous benign and malignant mammary tumors of the dog. Vet. Pathol. 2000;37:248–253. doi: 10.1354/vp.37-3-248. [DOI] [PubMed] [Google Scholar]
- 22.Chu L.L., Rutteman G.R., Kong J.M., Ghahremani M., Schmeing M., Misdorp W., van Garderen E., Pelletier J. Genomic organization of the canine p53 gene and its mutational status in canine mammary neoplasia. Breast Cancer Res. Treat. 1998;50:11–25. doi: 10.1023/A:1006010526813. [DOI] [PubMed] [Google Scholar]
- 23.Kanae Y., Endoh D., Yokota H., Taniyama H., Hayashi M. Expression of the pten tumor suppressor gene in malignant mammary gland tumors of dogs. Am. J. Vet. Res. 2006;67:127–133. doi: 10.2460/ajvr.67.1.127. [DOI] [PubMed] [Google Scholar]
- 24.Qiu C., Lin D., Wang J., Wang L. Expression and significance of pten in canine mammary gland tumours. Res. Vet. Sci. 2008;85:383–388. doi: 10.1016/j.rvsc.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Ressel L., Millanta F., Caleri E., Innocenti V.M., Poli A. Reduced pten protein expression and its prognostic implications in canine and feline mammary tumors. Vet. Pathol. 2009;46:860–868. doi: 10.1354/vp.08-VP-0273-P-FL. [DOI] [PubMed] [Google Scholar]
- 26.Beck J., Hennecke S., Bornemann-Kolatzki K., Urnovitz H.B., Neumann S., Strobel P., Kaup F.J., Brenig B., Schutz E. Genome aberrations in canine mammary carcinomas and their detection in cell-free plasma DNA. PLoS ONE. 2013;8:1589. doi: 10.1371/journal.pone.0075485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borge K.S., Nord S., van Loo P., Lingjaerde O.C., Gunnes G., Alnaes G.I., Solvang H.K., Luders T., Kristensen V.N., Borresen-Dale A.L., et al. Canine mammary tumours are affected by frequent copy number aberrations, including amplification of myc and loss of pten. PLoS ONE. 2015;10:1589. doi: 10.1371/journal.pone.0126371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu W.L., Huang H.M., Liao J.W., Wong M.L., Chang S.C. Increased survival in dogs with malignant mammary tumours overexpressing HER-2 protein and detection of a silent single nucleotide polymorphism in the canine HER-2 gene. Vet. J. 2009;180:116–123. doi: 10.1016/j.tvjl.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Kano R., Yano T., Nagamatsu K., Maruyama H., Kamata H., Hasegawa A. Effectiveness of small interfering RNA (siRNA) against the Mcl-1 gene in a canine mammary gland tumor cell line. Res. Vet. Sci. 2009;87:64–66. doi: 10.1016/j.rvsc.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Ripoli F.L., Mohr A., Hammer S.C., Willenbrock S., Hewicker-Trautwein M., Hennecke S., Escobar H.M., Nolte I. A comparison of fresh frozen vs. formalin-fixed, paraffin-embedded specimens of canine mammary tumors via branched-DNA assay. Int. J. Mol. Sci. 2016 doi: 10.3390/ijms17050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim M.S., Kim T., Kong S.Y., Kwon S., Bae C.Y., Choi J., Kim C.H., Lee E.S., Park J.K. Breast cancer diagnosis using a microfluidic multiplexed immunohistochemistry platform. PLoS ONE. 2010;5:1589. doi: 10.1371/journal.pone.0010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivera P., von Euler H. Molecular biological aspects on canine and human mammary tumors. Vet. Pathol. 2011;48:132–146. doi: 10.1177/0300985810387939. [DOI] [PubMed] [Google Scholar]
- 33.Mavaddat N., Antoniou A.C., Easton D.F., Garcia-Closas M. Genetic susceptibility to breast cancer. Mol. Oncol. 2010;4:174–191. doi: 10.1016/j.molonc.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gadzicki D., Evans D.G., Harris H., Julian-Reynier C., Nippert I., Schmidtke J., Tibben A., van Asperen C.J., Schlegelberger B. Genetic testing for familial/hereditary breast cancer—comparison of guidelines and recommendations from the UK, France, the Netherlands and Germany. J. Commun. Genet. 2011;2:53–69. doi: 10.1007/s12687-011-0042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Accili D., Arden K.C. Foxos at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/S0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 36.Gasco M., Shami S., Crook T. The p53 pathway in breast cancer. Breast Cancer Res. 2002;4:70–76. doi: 10.1186/bcr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S.I., Puc J., Miliaresis C., Rodgers L., McCombie R., et al. Pten, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 38.Fujioka Y., Taira T., Maeda Y., Tanaka S., Nishihara H., Iguchi-Ariga S.M., Nagashima K., Ariga H. Mm-1, a c-Myc-binding protein, is a candidate for a tumor suppressor in leukemia/lymphoma and tongue cancer. J. Biol. Chem. 2001;276:45137–45144. doi: 10.1074/jbc.M106127200. [DOI] [PubMed] [Google Scholar]
- 39.Jiang Y., Zou L., Lu W.Q., Zhang Y., Shen A.G. Foxo3a expression is a prognostic marker in breast cancer. PLoS ONE. 2013;8:1589. doi: 10.1371/journal.pone.0070746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Leeuwen I.S., Hellmen E., Cornelisse C.J., van den Burgh B., Rutteman G.R. P53 mutations in mammary tumor cell lines and corresponding tumor tissues in the dog. Anticancer Res. 1996;16:3737–3744. [PubMed] [Google Scholar]
- 41.Yang P., Du C.W., Kwan M., Liang S.X., Zhang G.J. The impact of p53 in predicting clinical outcome of breast cancer patients with visceral metastasis. Sci. Rep. 2013 doi: 10.1038/srep02246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu C., Wang J. A physical mechanism and global quantification of breast cancer. PLoS ONE. 2016;11:1589. doi: 10.1371/journal.pone.0157422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bose S., Crane A., Hibshoosh H., Mansukhani M., Sandweis L., Parsons R. Reduced expression of PTEN correlates with breast cancer progression. Hum. Pathol. 2002;33:405–409. doi: 10.1053/hupa.2002.124721. [DOI] [PubMed] [Google Scholar]
- 44.Chen J., Lindblom P., Lindblom A. A study of the PTEN/MMAC1 gene in 136 breast cancer families. Hum. Genet. 1998;102:124–125. [PubMed] [Google Scholar]
- 45.Li X., Wang Q., Fu L., Liu M., Yu X. Expression of PTEN, p53 and EGFR in the molecular subtypes of breast carcinoma and the correlation among them. Med. Sci. 2015;40:973–978. doi: 10.11817/j.issn.1672-7347.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 46.Sivaraman V.S., Wang H., Nuovo G.J., Malbon C.C. Hyperexpression of mitogen-activated protein kinase in human breast cancer. J. Clin. Investig. 1997;99:1478–1483. doi: 10.1172/JCI119309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun S., Zhang W., Cui Z., Chen Q., Xie P., Zhou C., Liu B., Peng X., Zhang Y. High mobility group box-1 and its clinical value in breast cancer. Onco Targets Ther. 2015;8:413–419. doi: 10.2147/OTT.S73366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fusco A., Fedele M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 49.Levine D.A., Bogomolniy F., Yee C.J., Lash A., Barakat R.R., Borgen P.I., Boyd J. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin. Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 50.O’Driscoll L., Cronin D., Kennedy S.M., Purcell R., Linehan R., Glynn S., Larkin A., Scanlon K., McDermott E.W., Hill A.D., et al. Expression and prognostic relevance of Mcl-1 in breast cancer. Anticancer Res. 2004;24:473–482. [PubMed] [Google Scholar]
- 51.Ross J.S., Fletcher J.A., Linette G.P., Stec J., Clark E., Ayers M., Symmans W.F., Pusztai L., Bloom K.J. The HER-2/NEU gene and protein in breast cancer 2003: Biomarker and target of therapy. Oncologist. 2003;8:307–325. doi: 10.1634/theoncologist.8-4-307. [DOI] [PubMed] [Google Scholar]
- 52.Tsuda H. Gene and chromosomal alterations in sporadic breast cancer: Correlation with histopathological features and implications for genesis and progression. Breast Cancer. 2009;16:186–201. doi: 10.1007/s12282-009-0124-x. [DOI] [PubMed] [Google Scholar]
- 53.Sana M., Irshad S. A review on breast cancer biomarkers BRCA1 and BRCA2. Res. Cancer Tumor. 2012;1:1–4. [Google Scholar]