Abstract
A number of common driver mutations have been identified in melanoma, but other genetic or epigenetic aberrations are also likely to play a role in the pathogenesis of melanoma and present potential therapeutic targets. Translocations of the anaplastic lymphoma kinase (ALK), for example, have been reported in spitzoid melanocytic neoplasms leading to kinase-fusion proteins that result in immunohistochemically detectable ALK expression. In this study, we sought to determine whether ALK was also expressed in non-spitzoid primary and metastatic cutaneous melanomas. ALK immunohistochemistry (IHC) was performed on 603 melanomas (303 primary and 300 metastatic tumors) from 600 patients. ALK IHC expression was identified in 7 primary and 9 metastatic tumors. In 5 of 7 primary tumors and in 6 of 9 metastatic lesions, the majority of tumor cells were immunoreactive for ALK. In the other two primary and three metastatic lesions, positive staining was identified in less than half of the tumor cells. ALK-positivity was found in the presence or absence of BRAF or NRAS mutations. In contrast to prior observations with ALK-positive Spitz tumors, none of the ALK-positive melanomas harbored a translocation. Instead, the ALK-positive melanomas predominantly expressed the recently described ALK isoform, ALKATI, which lacks the extracellular and transmembrane domains of wild-type ALK, consists primarily of the intracellular tyrosine kinase domain, and originates from an alternative transcriptional initiation (ATI) site within the ALK gene. The findings are clinically relevant as patients with metastatic melanoma who have ALK expression may potentially benefit from treatment with ALK kinase inhibitors.
Keywords: ALK, alternative transcriptional initiation, anaplastic lymphoma kinase, diagnosis, immunohistochemistry, melanoma, pathology
INTRODUCTION
A number of common driver mutations have been identified in cutaneous melanoma. A recent analysis by the Cancer Genome Atlas Network broadly classified melanomas into four subtypes with regard to their mutation profile: mutant BRAF, mutant RAS, mutant NF1, and triple wild-type (WT) tumors1. The latter group encompasses melanomas with molecular aberrations that are infrequent in cutaneous melanoma, such as mutations of KIT2, 3, GNA114, and GNAQ5, or genomic rearrangements of BRAF6, 7 and RAF18. In addition to these aberrations on which this classification is based, the pathogenesis of melanomas also depends on several additional genetic and epigenetic changes, including DNA copy number changes9-12, translocations13-16, DNA methylation17, and alternative transcriptional initiation18.
The study of spitzoid melanocytic neoplasms, for example, has revealed a number of genomic aberrations11, 19. The most common type of aberration in this subclass of melanocytic tumors are genomic rearrangements involving various receptor tyrosine kinases, such as ALK, ROS1, NTRK1, RET, or MET, and the serine-threonine kinases BRAF13, 20. The rearrangements link the kinase domain of these signaling proteins to a wide range of fusion partners, leading to highly expressed and kinase-active proteins. The resulting chimeric proteins stimulate multiple oncogenic signaling pathways and promote tumor initiation and progression13.
Recently, a new ALK isoform was detected in cutaneous melanoma as well as in other cancer types21. This ALK transcript is expressed from a de novo alternative transcription initiation (ATI) site in ALK intron 19, and was accordingly named ALKATI. Melanomas with ALKATI expression also showed positive staining by ALK immunohistochemistry (IHC)18. This observation prompted us to examine the frequency of ALK expression in a large set of primary and metastatic melanomas and to determine whether ALK immunoreactivity in melanoma is associated with ALK translocations or the truncated ALKATI isoform. Since ALK aberrations in cancer represent a therapeutic opportunity for patients22-27, such information would be helpful to determine the what extent pharmacological ALK inhibitors may become a treatment option for patients with metastatic melanoma. If ALK expression correlated with therapeutic response to ALK kinase inhibitors such as crizotinib or ceritinib, it would enhance the role of pathologists in contributing to personalized treatment plans.
MATERIALS AND METHODS
Patients
A total number of 603 melanomas, consisting of 303 primary cutaneous and 300 metastatic tumors, from 600 melanoma patients, were examined utilizing formalin-fixed and paraffin-embedded tissues.
Immunohistochemical Analysis
Five micron thick tissue sections were cut from tissue blocks. 534 lesions were evaluated by tissue microarray (TMA); the remainder on conventional full tumor sections. Each tumor sample found to be ALK positive on a TMA was re-analyzed on a whole tumor section.
An automated IHC system (Ventana BenchMark XT, Ventana Medical Systems, Inc., Tucson, AZ) was used with a standard avidin-biotin technique. The commercially available rabbit monoclonal antibody (ALK D5F3 XP Rabbit mAb #3633, Cell Signaling Technology) was used to evaluate ALK expression. The staining result was recorded as the percentage of immunoreactive tumor cells per total number of tumor cells, using a whole tumor section. 0 indicated no staining at all. Any staining of 1 – 24% of tumor cells was scored as 1+. If 25-49% were positive, it was designated as 2+. If 50-74% of cells were positive, it was recorded as 3+. If 75% or more of the tumor cells were positive, this result was scored as 4+.
RNA extraction and NanoString
To accurately distinguish and quantify the expression of wild-type ALK, translocated ALK, and ALKATI in formalin-fixed, paraffin-embedded tumor samples, we used a NanoString nCounter assay with probes for ALK exons 1–19, intron 19, and exons 20–29. For this assay, RNA was extracted from 5-µm-thick formalin-fixed, paraffin-embedded tissue sections using the High Pure miRNA Isolation Kit (Roche) according to the manufacturers instructions. After isolation, the RNA was frozen at −80C, quantified, and used for the NanoString nCounter assay.
Details of the NanoString nCounter Analysis System (NanoString Technologies) were reported previously18, 28. In brief, two sequence-specific probes were constructed for ALK exons 1-19, intron 19, and exons 20-29, respectively. Four control genes (RPS13, RPL27, RPS20 and ACTB) were used for normalization. The probes were complementary to a 100 bp region of the target mRNA. 100 ng of total RNA from each sample was hybridized, the raw data were normalized to the standard curve generated via the nCounter system, and the average value of the two probes in each target region (exons 1-19, intron 19, exons 20-29) were printed in bar charts using GraphPad Prism software 6.0.
Mutation Status
The BRAF or NRAS mutation status of the ALK-positive metastatic melanomas was obtained from the patients’ medical records. Sequence data were available in all but one case. The methods for mutation analysis varied depending on which laboratory the tissue was sent to, and included Sanger sequencing and Sequenom mass spectrometry. In one case without sequencing data, the BRAFV600E mutation was determined by immunohistochemistry using the anti-BRAF V600E (VE1) antibody (Ventana).
RESULTS
Immunohistochemical Results
Metastatic melanoma
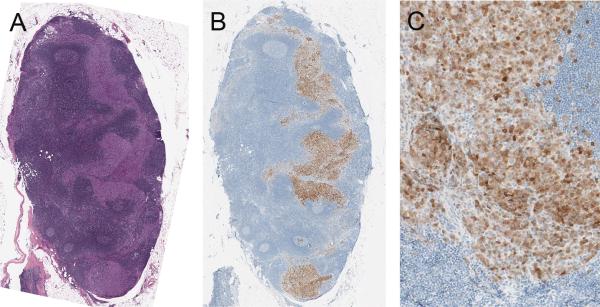
We examined 300 tumor samples of metastatic melanoma for immunoreactivity for ALK. Nine (3%) were positive. Positive labeling in the majority of tumor cells was found in 6 of 300 (2%) cases (Table 1, Fig. 1). In three cases less than half of the tumor cells were immunoreactive for ALK (Table 1, Fig. 2). The pattern of staining was predominantly cytoplasmic, but nuclear labeling was also observed (Fig. 2). The staining intensity was variable not only from lesion to lesion, but could also vary within a tumor (Fig. 1), with subpopulations of cells displaying strong labeling while other cells stained only weakly. In tumors in which only a minority of tumor cells was ALK-positive, the positive cells tended to be scattered randomly throughout the tumor as isolated cells or small clusters (Fig. 2). The overall staining intensity of those cases tended to be weak.
Table 1.
Clinical and pathologic features of ALK-positive metastatic melanoma
| Case | Age/Gender | Site of Metastasis | Site of primary | Thickness of primary (mm) | IHC | Mutations |
|---|---|---|---|---|---|---|
| 1 | 63M | LN | Back | 2.7 | 4+ | BRAF-WT |
| 2 | 56M | LN | Back | 0.95 | 4+ | BRAF-WT |
| 3 | 56M | LN | Unknown | NA | 1+ | NRASQ61K |
| 4 | 67F | Soft tissue | Scalp | 2.5 | 1+ | BRAFV600K |
| 5 | 52F | Brain | Back | 2.2 | 1+ |
BRAFV600E
BRAFK601E |
| 6 | 38M | LN | Back | 3.2 | 4+ | BRAF-WT |
| 7 | 61M | LN | Back | 7.8 | 4+ | BRAF-WT |
| 8 | 65F | LN | Toe | 2.7 | 4+ | BRAF-WT |
| 9 | 51M | LN | Calf | 0.65 | 3+ | Negative for BRAFV600E* |
Abbreviations:
F = female; M = male; LN = lymph node; IHC = immunohistochemistry; mm = millimeter IHC Proportion: 1 + = < 25% of tumor cells are positive; 2+ = 25-49% of tumor cells are positive; 3+ = 50-74% of tumor cells are positive; 4+ = ≥ 75% of tumor cells are positive WT = wild type (no BRAF mutations detected by Sanger sequencing or Sequenom analysis);
immunohistochemical analysis using the monoclonal antibody VE1
Figure 1.
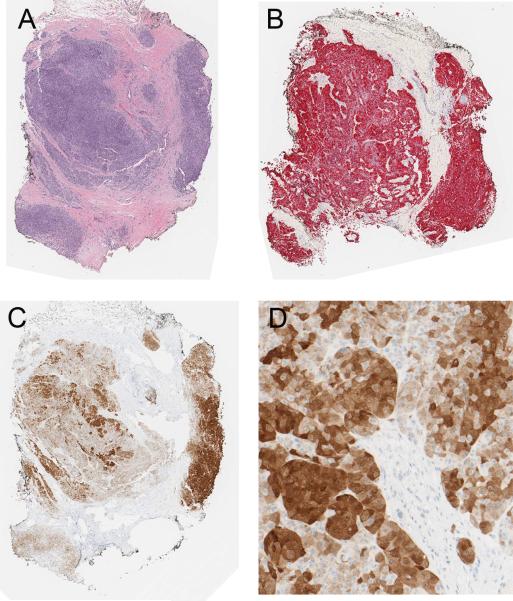
Metastatic melanoma expressing ALK in most tumor cells (case 9 of Table 1). A, Malignant amelanotic epithelioid tumor cells (H&E stain). B, The tumor cells are homogenously positive for melan-A. C, Most of the tumor cells are positive for ALK by immunohistochemistry. D, There is a range of staining intensity within the tumor from weak to strong.
Figure 2.
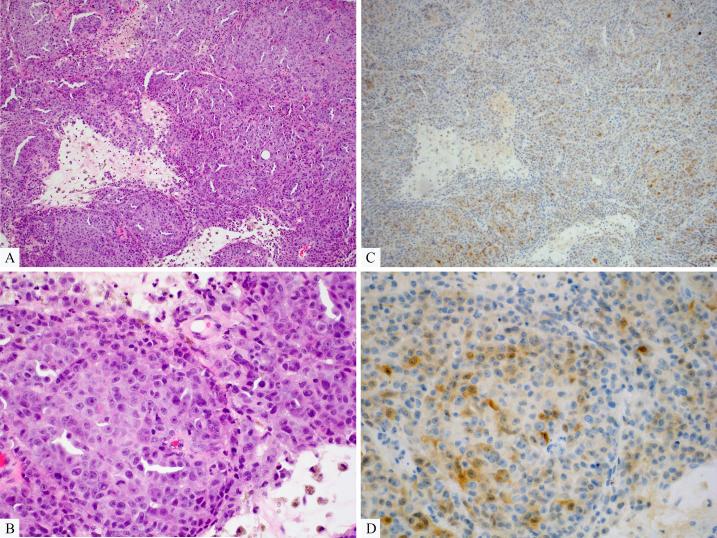
Metastatic melanoma expressing ALK in a minor subpopulation of tumor cells (case 5 of Table 1). A, Predominantly amelanotic melanoma with foci of necrosis (H&E stain). B, The tumor is composed of large atypical epithelioid cells (H&E stain). C, ALK Immunohistochemistry reveals a minor population of weakly immunopositive cells scattered throughout the tumor. D, Both cytoplasmic and rare nuclear labeling are seen.
In hematoxylin and eosin-stained sections, metastatic ALK-positive melanomas were predominantly composed of amelanotic epithelioid melanocytes (Figs. 1 and 2). None of the tumors was a spindle cell melanoma. Mitoses and foci of necrosis were common.
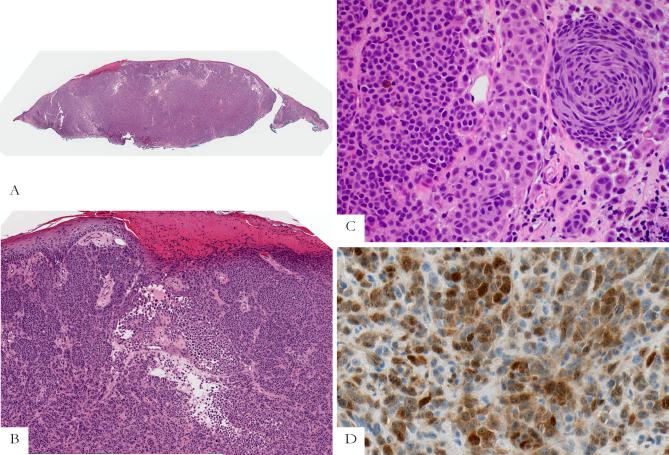
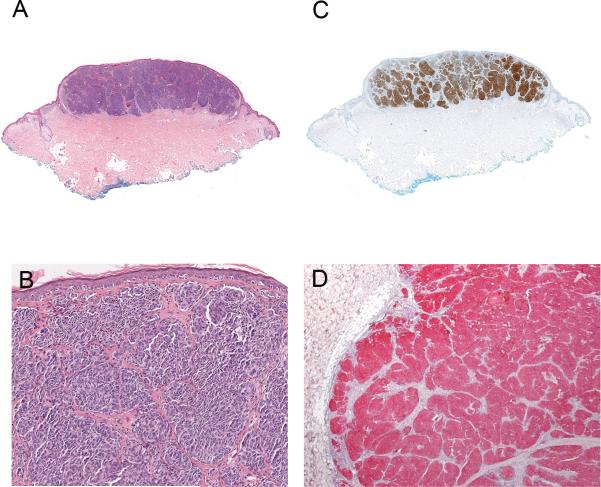
Six of the patients with metastatic ALK-positive melanomas were men and three were women. The clinical and pathologic findings are summarized in Table 1. The patients’ ages at the time of diagnosis of metastatic melanoma ranged from 38 – 67 years (mean = 56.5 years; median = 56 years). One patient had metastatic melanoma in the lymph node with no known primary tumor. All other patients had known primary cutaneous tumors, predominantly from the skin of the back. The thickness of the primary tumors ranged from 0.65 to 7.8 mm (mean = 2.8 mm; median = 2.6 mm). Four of the primary melanomas were nodular, two were of superficial spreading type, one was subungual, and one was unclassified. In three cases of ALK-positive metastatic melanoma, histologic sections of the corresponding primary tumors were available and could be tested for ALK expression. All were immunoreactive for ALK. The proportion of immunoreactive cells in the metastases was similar to the primary melanomas (Figs. 3 – 5).
Figure 3.
Primary nodular melanoma (case 4 of Table 2; corresponding metastasis shown in Fig. 4). A, Silhouette of the primary nodular melanoma. B, The tumor is ulcerated. Nests and sheets of predominantly amelanotic epithelioid melanoma cells are present in the dermis. C, Most tumor cells have epithelioid cell features, but there is also a cluster of fusiform melanocytes. D, Most tumor cells are positive for ALK. Both nuclear and cytoplasmic labeling are seen.
Figure 5.
Primary nodular melanoma (case 1 of Table 2) with metastasis. A, Silhouette of the primary nodular melanoma. B, The tumor cells are amelanotic and epithelioid in appearance forming large nests and sheet-like aggregates. C, Most tumor cells are strongly immunopositive for ALK. D, The tumor cells of the corresponding metastasis are strongly and homogenously positive for ALK.
Table 1 also documents the BRAF and NRAS mutation status of the ALK-positive metastatic melanomas. ALK-positivity was found in melanomas with and without associated BRAF or NRAS mutations. Melanomas with high expression of ALK (expressed by the majority of tumor cells) tended to be wild type for BRAF and NRAS, while the tumors with documented BRAF or NRAS mutation expressed ALK only in a minority of melanocytes (Table 1).
Primary cutaneous melanoma
We examined 303 primary cutaneous melanomas for ALK expression by IHC. Seven tumors (2.3%) were immunopositive for ALK. Six patients were male and one was female. The patients’ ages ranged from 38 – 72 years of age (mean = 56 years; median = 63 years). Four tumors were located on the back, two on the lower extremity, and one on the face. Tumor thickness ranged from 0.95 to 10 mm (mean = 5 mm; median = 4.3 mm). The tumor mitotic rates ranged from 1 to 24 mitoses per mm2 (mean = 7; median = 4). Five of the tumors were nodular melanomas, one was a subungual melanoma; and one was unclassified. Five melanomas were amelanotic and two tumors were focally pigmented. Two were ulcerated. The tumors were predominantly composed of epithelioid melanocytes. In some tumors a minor population of fusiform melanocytes was also present (Fig. 3). The features of the primary ALK-positive melanomas are summarized in Table 2.
Table 2.
Clinical and pathologic features of primary ALK-positive melanomas
| Case | Age/Gender | Site | Thickness (mm) | Ulcer | TMR (#/mm2) | Type | IHC | ALKATI |
|---|---|---|---|---|---|---|---|---|
| 1 | 63M | Back | 2.7 | Absent | 2 | Nodular | 4+ | Present |
| 2 | 46M | Back | 0.95 | Absent | 3 | Unclassified | 4+ | Present |
| 3 | 72M | Back | 10 | Absent | 4 | Nodular | 4+ | Present |
| 4 | 38M | Back | 3.2 | Present | 24 | Nodular | 4+ | Present |
| 5 | 65M | Toe | 5.2 | Present | 1 | Subungual | 2+ | Not done* |
| 6 | 70F | Face | 4.3 | Absent | 8 | Nodular | 3+ | Present |
| 7 | 39M | Leg | 9 | Absent | 7 | Nodular | 2+ | Present |
Abbreviations:
F = female; M = male; LN = lymph node; TMR = tumor mitotic rate;
IHC = immunohistochemistry; # = number; mm - millimeter
Test could not be performed (inadequate RNA due to prior decalcification of tissue)
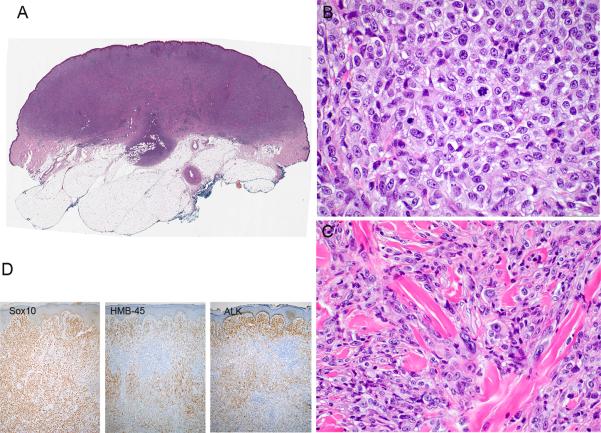
Similar to metastatic tumor tissue, the proportion of primary tumor cells, which were immunoreactive for ALK, was variable. In five melanomas, most tumor cells were positive for ALK. In four tumors, more than 75% of the tumor cells were positive (Table 2, Figs. 3 - 5), with a similar proportion of IHC-positive cells between primary tumor and corresponding metastasis. In two primary melanomas less than half of the tumor cells was immunoreactive for ALK (Table 2, Fig. 6). Their corresponding sentinel lymph nodes were negative. Both patients had so far remained free of disease (Oct 2015).
Figure 6.
Primary nodular melanoma with only a subpopulation of ALK-positive tumor cells (case 7 of Table 2). A, Silhouette of an amelanotic primary nodular melanoma. B, The tumor is composed of large epithelioid melanocytes, some in mitosis, arranged in sheet-like aggregates. C, In other areas of the tumor collagen bundles separate tumor cells with pleomorphic nuclei, some displaying prominent nucleoli. D, Immunohistochemical analysis documents that all tumor cells are positive for Sox10, but only a subpopulation is positive for HMB-45 and/or ALK.
NanoString Analysis
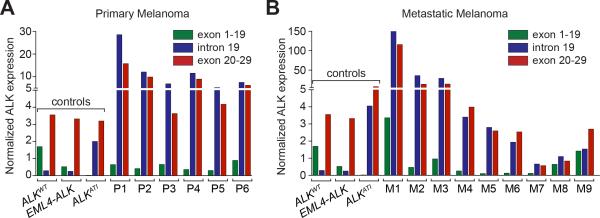
A NanoString nCounter assay29 with probes in ALK exons 1-19, intron 19, and exons 20-29, was used to confirm the presence of ALKATI, the recently described novel ALK transcript18. Of the seven IHC ALK-positive primary melanomas, adequate RNA was obtained from six; one subungual tumor yielded inadequate RNA quality for further analysis due to prior decalcification of the tissue. All six remaining primary tumors analysed by the NanoString nCounter assay contained the ALKATI isoform (Fig. 7A). Levels of intact wild-type ALK were very low. All 9 ALK-positive metastases contained the ALKATI novel transcript, but three of them (M1, M8 and M9) showed co-expression of ALKATI and wild-type ALK (Fig. 7B).
Figure 7.
Quantitative mRNA profiling using Nanostring nCounter to distinguish and quantify the expression of wild-type ALK, translocated ALK, and ALKATI in clinical specimens. Tumors expressing wild-type ALK (ALKWT) show expression of ALK exons 1–19 and exons 20–29, but not intron 19. Tumors with ALK translocations show only expression of the ALK exons 20–29. ALKATI-expressing tumors show expression of the alternative transcriptional start site in ALK intron 19, as well as expression of the exons 20–29, containing the transforming kinase domain. A, ALKATI expression of primary melanoma samples. B, ALKATI expression of primary melanoma samples.
DISCUSSION
While early-stage primary cutaneous melanoma is surgically curable, the prognosis is less favorable for patients with regional metastasis, and survival has historically been poor for patients with distant metastatic disease30. However, the recent development of selective kinase inhibitors31-36 has provided new therapeutic opportunities for advanced stage metastatic melanoma patients with activating mutations of BRAF and NRAS. Recent trials using FDA approved kinase inhibitors have confirmed that targeting activating mutations in melanoma can have dramatic anti-tumor responses, although they are rarely durable37, 38.
While mutations of BRAF, NRAS and NF1 are the most common alterations in melanoma1, a number of less common genomic aberrations have been identified, which might also provide new therapeutic opportunities20, 39. The identification of new targets continues to increase therapeutic options for cancer patients40. We have recently found a new isoform of the ALK receptor-tyrosine kinase in melanoma and other cancer types. This novel isoform is expressed as the result of alternative transcriptional initiation, and we recently reported one patient harboring this isoform who had a temporary partial response to crizotinib18.
In this study, we sought to determine the frequency of ALK expression in melanoma. After we determined ALK-positivity using IHC, we used a NanoString nCounter to assess whether immunoreactivity for ALK reflects expression of wild-type ALK, ALK translocations (as in Spitz nevi/tumors)13 or the presence of the recently identified novel ALKATI isoform18.
The frequency of ALK-positivity in metastatic cutaneous melanoma was 3%, if any, even focal staining was included. The frequency of ALK expression was 2% in the case of positive staining of the majority of tumor cells. Among primary melanomas, the frequency of positive labeling was 2.3% for any staining and 1.7% for staining of the majority of tumor cells. The frequency of the IHC ALK expression in this series of 603 melanomas is considerably lower than the initially reported frequency of ~11% (38 of 334 melanomas)18, which was assessed by gene expression profiling using the RNA-seq dataset of the TCGA project1. This discrepancy is likely related to methodology. First, RNA-seq assesses the presence of the ALK mRNA transcript and might be more sensitive than ALK IHC, which detects the expression of the ALK protein. If that is the case, it will be of interest to determine whether low expression of ALKATI detectable by RNA-seq, but not by immunohistochemistry is clinically relevant. This is best addressed in the context of a prospective clinical trial comparing anti-ALK targeted therapy responses to various levels of ALKATI expression by different methods. Second, in several melanomas with ALK expression not all of the tumors cells expressed ALK. As most of our cases were initially screened by TMAs containing small core biopsies from randomly selected tumor areas, it is likely that some ALK-expressing tumor cell populations were missed by sampling bias (the cores on the TMA did not contain any ALK-expressing tumor cells, but other areas of the tumor may have expressed ALK). Thus, our screening method may have led to under-recognition of tumors with ALKATI expression, in which only a subpopulation of tumor cells was immunoreactive for ALK. On the other hand, it is also possible that the process for selecting tumors to the TCGA project led to enrichment of melanomas expressing ALKATI. Considering these methodological limitations, the frequency of ALKATI-expressing melanomas can be estimated to range somewhere in between 2 and 10% of cutaneous melanomas.
Since positive staining for ALK was observed in a similar proportion of tumor cells and at a similar level of staining intensity in the few cases of matching primary and metastatic tumors, ALK-positivity and the presence of the truncated ALKATI isoform likely reflect an oncogenic mechanism of melanoma in primary tumors, which persists in metastases thereof. Given the small number of ALK-positive primary and metastatic melanomas available to date it is difficult to assess to what extent ALK-positivity and ALKATI expression may also develop during tumor progression or treatment-related clonal tumor selection.
The small number of ALKATI-expressing melanomas makes it also difficult to ascertain whether there is a distinct morphologic phenotype that corresponds to the IHC and molecular findings. Preliminary observations suggest that most ALK-positive melanomas are primary nodular melanomas predominantly composed of amelanotic epithelioid or mixed spindle and epithelioid cells.
With regard to an association of ALK expression and mutation status, we found ALK immunoreactivity in metastatic tumors with or without BRAF or NRAS mutations. This confirms prior observations in the TCGA dataset that ALKATI can be expressed independently of different mutations, including BRAF, NRAS and KIT. This suggests that efforts to identify patients with ALK-positive melanomas should include cutaneous melanomas with and without known mutations. The frequency of associated BRAF mutations in our series may be slightly underestimated due to the use of IHC in one case.
It is of interest that none of the ALK-positive primary or metastatic melanomas harbored an ALK translocation. Such translocations are typical of ALK-positive spitzoid neoplasms13, 14, 41. While some of our patients with ALK-positive metastatic melanoma have died of metastatic disease, to date we have not yet learned of a patient with a primary Spitz tumor containing an ALK translocation, who has died of metastatic melanoma (KJB, unpublished observations, Oct 2015). While more long-term follow-up of patients with ALK-positive spitzoid neoplasms is needed, our observations suggest a fundamental difference in the biology between tumors designated as “atypical Spitz tumor” or “spitzoid melanoma” and non-spitzoid ALK-positive melanoma. Age of presentation is another difference. Most spitzoid neoplasms affect children, adolescents and young adults, while most patients with ALK-positive non-spitzoid melanomas in this series were middle-aged or older.
It is also noteworthy that in spitzoid tumors with ALK-translocations usually all melanocytes of a tumor are strongly positive for ALK by IHC13, 41. In contrast, in ALKATI-expressing melanomas not all tumor cells are ALK immunoreactive and the staining intensity is often variable. The most plausible explanation for this phenomenon is that genomic rearrangements, such as ALK translocation, are stable changes of the DNA sequence and put the ALK kinase domain under the control of a strong and constitutively expressed promoter. However, in ALKATI-expressing tumors no recurrent genetic aberrations are found at and around the ALK locus, indicating that the transcriptional activation is independent of genetic aberrations at the ALK locus. In addition, integrated genomic analysis of DNA-, RNA-, and ChIP-seq data revealed that both ALK alleles are actively transcribed suggesting that the transcriptional activation of ALKATI depends on trans-acting elements, such as transcription factors or chromatin modifiers21. Thus, ALKATI-expression is probably determined by the concerted expression of several transcription factors and transcriptional repressors as well as the epigenetic landscape and the accessibility of transcription factor binding sites. ALKATI-expression may therefore vary dependent on the stage of the cell cycle or the tumor microenvironment.
In conclusion, we have documented that protein expression of ALK can be detected in a small subset of melanomas using IHC, with variable immunoreactivity ranging from focal weak to strong and diffuse labeling. All ALK-positive melanomas in this series contained the ALKATI isoform, and not an ALK translocation. Given the demonstrated efficacy of ALK kinase inhibitors in other cancers types, our findings suggest a potential therapeutic opportunity for patients with ALK-positive metastatic melanomas. Since IHC confirmation of ALK expression is important for identifying tumors that can be targeted by ALK-inhibitors27, pathologists play an important role in the process of identifying treatment options for patients with metastatic melanoma. Our findings should encourage pathologists to collaborate with oncologists to offer ALK testing for patients, in which there may be no other therapeutic opportunity.
Figure 4.
ALK-positive melanoma metastasis in lymph node (corresponding to the primary tumor shown in Fig. 3). A, H&E stained section of lymph node with amelanotic metastatic melanoma. B, The tumor cells are immunoreactive for ALK. C, There is a range from weak to strong staining intensity, both nuclear and cytoplasmic.
Acknowledgements
The authors wish to thank Danielle Leng and Allyne Manzo for the help with the figures.
This work was supported by grants from the Harry J. Lloyd Trust - Translational Research Grant to K.J.B and T.W., and the Charles H. Revson Senior Fellowship to T.W. Research reported in this publication was supported in part by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748.
Footnotes
Disclosures:
The authors have disclosed that they have no significant relationships with, or financial interest in, any commercial companies pertaining to this article.
REFERENCES
- 1.Cancer Genome Atlas N: Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvajal RD, Antonescu CR, Wolchok JD, Chapman PB, Roman RA, Teitcher J, Panageas KS, Busam KJ, Chmielowski B, Lutzky J, Pavlick AC, Fusco A, Cane L, Takebe N, Vemula S, Bouvier N, Bastian BC, Schwartz GK. KIT as a therapeutic target in metastatic melanoma. Jama. 2011;305:2327–34. doi: 10.1001/jama.2011.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 4.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, Sozen MM, Baimukanova G, Roy R, Heguy A, Dolgalev I, Khanin R, Busam K, Speicher MR, O'Brien J, Bastian BC. Mutations in GNA11 in uveal melanoma. The New England journal of medicine. 2010;363:2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, Simpson EM, Barsh GS, Bastian BC. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botton T, Yeh I, Bastian BC. Melanoma BRAF fusions--letter. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:6631. doi: 10.1158/1078-0432.CCR-14-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botton T, Yeh I, Nelson T, Vemula SS, Sparatta A, Garrido MC, Allegra M, Rocchi S, Bahadoran P, McCalmont TH, LeBoit PE, Burton EA, Bollag G, Ballotti R, Bastian BC. Recurrent BRAF kinase fusions in melanocytic tumors offer an opportunity for targeted therapy. Pigment cell & melanoma research. 2013;26:845–51. doi: 10.1111/pcmr.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, Han B, Cao Q, Cao X, Suleman K, Kumar-Sinha C, Dhanasekaran SM, Chen YB, Esgueva R, Banerjee S, LaFargue CJ, Siddiqui J, Demichelis F, Moeller P, Bismar TA, Kuefer R, Fullen DR, Johnson TM, Greenson JK, Giordano TJ, Tan P, Tomlins SA, Varambally S, Rubin MA, Maher CA, Chinnaiyan AM. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nature medicine. 2010;16:793–8. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson MA, Zhao F, Khare S, D'Andrea K, Wubbenhorst B, Roszik J, Woodman SE, Rimm DL, Kirkwood JM, Kluger HM, Schuchter LM, Lee SJ, Flaherty KT, Nathanson KL. Copy number changes are associated with response to treatment with carboplatin, paclitaxel, and sorafenib in melanoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015 doi: 10.1158/1078-0432.CCR-15-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bastian BC, LeBoit PE, Pinkel D. Genomic approaches to skin cancer diagnosis. Archives of dermatology. 2001;137:1507–11. doi: 10.1001/archderm.137.11.1507. [DOI] [PubMed] [Google Scholar]
- 11.Bastian BC, Wesselmann U, Pinkel D, Leboit PE. Molecular cytogenetic analysis of Spitz nevi shows clear differences to melanoma. The Journal of investigative dermatology. 1999;113:1065–9. doi: 10.1046/j.1523-1747.1999.00787.x. [DOI] [PubMed] [Google Scholar]
- 12.Bauer J, Bastian B. Genomic analysis of melanocytic neoplasia. Advances in dermatology. 2005;21:81–99. doi: 10.1016/j.yadr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Wiesner T, He J, Yelensky R, Esteve-Puig R, Botton T, Yeh I, Lipson D, Otto G, Brennan K, Murali R, Garrido M, Miller VA, Ross JS, Berger MF, Sparatta A, Palmedo G, Cerroni L, Busam KJ, Kutzner H, Cronin MT, Stephens PJ, Bastian BC. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nature communications. 2014;5:3116. doi: 10.1038/ncomms4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh I, de la Fouchardiere A, Pissaloux D, Mully TW, Garrido MC, Vemula SS, Busam KJ, LeBoit PE, McCalmont TH, Bastian BC. Clinical, histopathologic, and genomic features of Spitz tumors with ALK fusions. The American journal of surgical pathology. 2015;39:581–91. doi: 10.1097/PAS.0000000000000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hantschke M, Mentzel T, Rutten A, Palmedo G, Calonje E, Lazar AJ, Kutzner H. Cutaneous clear cell sarcoma: a clinicopathologic, immunohistochemical, and molecular analysis of 12 cases emphasizing its distinction from dermal melanoma. The American journal of surgical pathology. 2010;34:216–22. doi: 10.1097/PAS.0b013e3181c7d8b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiuru M, Hameed M, Busam KJ. Compound clear cell sarcoma misdiagnosed as a Spitz nevus. Journal of cutaneous pathology. 2013;40:950–4. doi: 10.1111/cup.12197. [DOI] [PubMed] [Google Scholar]
- 17.Neumann LC, Weinhausel A, Thomas S, Horsthemke B, Lohmann DR, Zeschnigk M. EFS shows biallelic methylation in uveal melanoma with poor prognosis as well as tissue-specific methylation. BMC cancer. 2011;11:380. doi: 10.1186/1471-2407-11-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiesner T, Lee W, Obenauf AC, Ran L, Murali R, Zhang QF, Wong EW, Hu W, Scott SN, Shah RH, Landa I, Button J, Lailler N, Sboner A, Gao D, Murphy DA, Cao Z, Shukla S, Hollmann TJ, Wang L, Borsu L, Merghoub T, Schwartz GK, Postow MA, Ariyan CE, Fagin JA, Zheng D, Ladanyi M, Busam KJ, Berger MF, Chen Y, Chi P. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature. 2015;526:453–7. doi: 10.1038/nature15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bastian BC, LeBoit PE, Pinkel D. Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. The American journal of pathology. 2000;157:967–72. doi: 10.1016/S0002-9440(10)64609-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh I, Botton T, Talevich E, Shain AH, Sparatta AJ, de la Fouchardiere A, Mully TW, North JP, Garrido MC, Gagnon A, Vemula SS, McCalmont TH, LeBoit PE, Bastian BC. Activating MET kinase rearrangements in melanoma and Spitz tumours. Nature communications. 2015;6:7174. doi: 10.1038/ncomms8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiesner T, Lee W, Obenauf AC, Ran L, Murali R, Zhang QF, Wong EW, Hu W, Scott SN, Shah RH, Landa I, Button J, Lailler N, Sboner A, Gao D, Murphy DA, Cao Z, Shukla S, Hollmann TJ, Wang L, Borsu L, Merghoub T, Schwartz GK, Postow MA, Ariyan CE, Fagin JA, Zheng D, Ladanyi M, Busam KJ, Berger MF, Chen Y, Chi P. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature. 2015 doi: 10.1038/nature15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boolell V, Alamgeer M, Watkins DN, Ganju V. The Evolution of Therapies in Non-Small Cell Lung Cancer. Cancers. 2015;7:1815–46. doi: 10.3390/cancers7030864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carpenter EL, Mosse YP. Targeting ALK in neuroblastoma--preclinical and clinical advancements. Nature reviews Clinical oncology. 2012;9:391–9. doi: 10.1038/nrclinonc.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw AT. Combining inhibitors of ALK and ROS1 with other agents for the treatment of non-small cell lung cancer. Clinical advances in hematology & oncology : H&O. 2015;13:282–4. [PubMed] [Google Scholar]
- 25.Normanno N, Rachiglio AM, Roma C, Fenizia F, Esposito C, Pasquale R, La Porta ML, Iannaccone A, Micheli F, Santangelo M, Bergantino F, Costantini S, De Luca A. Molecular diagnostics and personalized medicine in oncology: challenges and opportunities. Journal of cellular biochemistry. 2013;114:514–24. doi: 10.1002/jcb.24401. [DOI] [PubMed] [Google Scholar]
- 26.Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, Wu YL, Thomas M, O'Byrne KJ, Moro-Sibilot D, Camidge DR, Mok T, Hirsh V, Riely GJ, Iyer S, Tassell V, Polli A, Wilner KD, Janne PA. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. The New England journal of medicine. 2013;368:2385–94. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 27.Mosse YP, Wood A, Maris JM. Inhibition of ALK signaling for cancer therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:5609–14. doi: 10.1158/1078-0432.CCR-08-2762. [DOI] [PubMed] [Google Scholar]
- 28.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature biotechnology. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 29.Reis PP, Waldron L, Goswami RS, Xu W, Xuan Y, Perez-Ordonez B, Gullane P, Irish J, Jurisica I, Kamel-Reid S. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC biotechnology. 2011;11:46. doi: 10.1186/1472-6750-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coit DG, Andtbacka R, Anker CJ, Bichakjian CK, Carson WE, 3rd, Daud A, Dimaio D, Fleming MD, Guild V, Halpern AC, Hodi FS, Jr., Kelley MC, Khushalani NI, Kudchadkar RR, Lange JR, Lind A, Martini MC, Olszanski AJ, Pruitt SK, Ross MI, Swetter SM, Tanabe KK, Thompson JA, Trisal V, Urist MM, McMillian N, Ho M, National Comprehensive Cancer N Melanoma, version 2.2013: featured updates to the NCCN guidelines. Journal of the National Comprehensive Cancer Network : JNCCN. 2013;11:395–407. doi: 10.6004/jnccn.2013.0055. [DOI] [PubMed] [Google Scholar]
- 31.Salama AK, Kim KB. Trametinib (GSK1120212) in the treatment of melanoma. Expert opinion on pharmacotherapy. 2013;14:619–27. doi: 10.1517/14656566.2013.770475. [DOI] [PubMed] [Google Scholar]
- 32.Kainthla R, Kim KB, Falchook GS. Dabrafenib for treatment of BRAF-mutant melanoma. Pharmacogenomics and personalized medicine. 2014;7:21–9. doi: 10.2147/PGPM.S37220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fisher R, Larkin J. Vemurafenib: a new treatment for BRAF-V600 mutated advanced melanoma. Cancer management and research. 2012;4:243–52. doi: 10.2147/CMAR.S25284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O'Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA, Group B-S. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ribas A, Flaherty KT. BRAF targeted therapy changes the treatment paradigm in melanoma. Nature reviews Clinical oncology. 2011;8:426–33. doi: 10.1038/nrclinonc.2011.69. [DOI] [PubMed] [Google Scholar]
- 36.Flaherty L, Hamid O, Linette G, Schuchter L, Hallmeyer S, Gonzalez R, Cowey CL, Pavlick A, Kudrik F, Curti B, Lawson D, Chapman PB, Margolin K, Ribas A, McDermott D, Flaherty K, Cranmer L, Hodi FS, Day BM, Linke R, Hainsworth J. A single-arm, open-label, expanded access study of vemurafenib in patients with metastatic melanoma in the United States. Cancer journal. 2014;20:18–24. doi: 10.1097/PPO.0000000000000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, Dummer R, McMahon M, Stuart DD. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–5. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puzanov I, Amaravadi RK, McArthur GA, Flaherty KT, Chapman PB, Sosman JA, Ribas A, Shackleton M, Hwu P, Chmielowski B, Nolop KB, Lin PS, Kim KB. Long-term outcome in BRAF(V600E) melanoma patients treated with vemurafenib: Patterns of disease progression and clinical management of limited progression. European journal of cancer. 2015;51:1435–43. doi: 10.1016/j.ejca.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shain AH, Garrido M, Botton T, Talevich E, Yeh I, Sanborn JZ, Chung J, Wang NJ, Kakavand H, Mann GJ, Thompson JF, Wiesner T, Roy R, Olshen AB, Gagnon A, Gray JW, Huh N, Hur JS, Busam KJ, Scolyer RA, Cho RJ, Murali R, Bastian BC. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nature genetics. 2015 doi: 10.1038/ng.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dietel M, Johrens K, Laffert MV, Hummel M, Blaker H, Pfitzner BM, Lehmann A, Denkert C, Darb-Esfahani S, Lenze D, Heppner FL, Koch A, Sers C, Klauschen F, Anagnostopoulos I. A 2015 update on predictive molecular pathology and its role in targeted cancer therapy: a review focussing on clinical relevance. Cancer gene therapy. 2015 doi: 10.1038/cgt.2015.39. [DOI] [PubMed] [Google Scholar]
- 41.Busam KJ, Kutzner H, Cerroni L, Wiesner T. Clinical and pathologic findings of Spitz nevi and atypical Spitz tumors with ALK fusions. The American journal of surgical pathology. 2014;38:925–33. doi: 10.1097/PAS.0000000000000187. [DOI] [PMC free article] [PubMed] [Google Scholar]