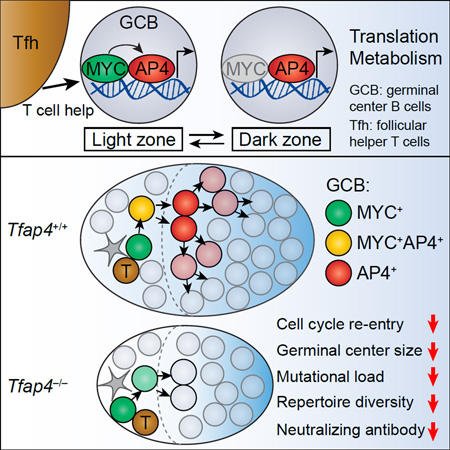
Summary
B cells diversify and affinity-mature their antigen receptor repertoire in germinal centers (GC). GC B cells receive help signals during transient interaction with T cells, yet it remains unknown how these transient T-B interactions in the light zone sustain the subsequent proliferative program of selected B cells that occurs in the anatomically distant dark zone. Here we show that the transcription factor AP4 was required for sustained GC B cell proliferation and subsequent establishment of a diverse and protective antibody repertoire. AP4 was induced by c-MYC during the T-B interactions, maintained by T cell-derived interleukin-21 (IL-21) and promoted repeated rounds of divisions of selected GC B cells. B cell-specific deletion of AP4 resulted in reduced GC sizes and reduced somatic hypermutation coupled with a failure to control chronic viral infection. These results indicate that AP4 integrates T cell-mediated selection and sustained expansion of GC B cells for humoral immunity.
Graphical Abstract
Introduction
Upon infection or vaccination, antigen (Ag)-specific lymphocytes that are present at low frequencies under steady-state conditions undergo rapid clonal expansion to increase the magnitude of adaptive immune responses. While expansion of Ag-specific B cells ensures that sufficient quantities of antibodies are made, it also serves as a template for somatic hypermutation (SHM), affinity maturation, and the subsequent generation of protective humoral memory for long-term immunity (Basso and Dalla-Favera, 2015; Victora and Nussenzweig, 2012). These proliferating Ag-specific B cells form a specialized compartment in peripheral lymphoid organs, the germinal centers (GCs), in which B cells cyclically migrate between the light zone (LZ) and the dark zone (DZ) for selection and subsequent clonal expansion, respectively (Allen et al., 2007; Basso and Dalla-Favera, 2015; Corcoran and Tarlinton, 2016; Victora and Nussenzweig, 2012; Victora et al., 2010). In the LZ, signals from follicular helper T (Tfh) cells facilitate the selection of clones that express B cell receptors (BCRs) with higher affinity for their cognate Ag relative to neighboring clones (Allen et al., 2007; MacLennan, 1994; Rajewsky, 1996; Shih et al., 2002; Victora and Nussenzweig, 2012). The important cues for clonal selection in the LZ include ligation of CD40 on B cells by CD40L on Tfh cells, as well as cytokines, especially interleukin-21 (IL-21) (Basso and Dalla-Favera, 2015; Castigli et al., 1994; Kawabe et al., 1994; Linterman et al., 2010; Renshaw et al., 1994; Victora and Nussenzweig, 2012; Xu et al., 1994; Zotos et al., 2010). GC B cells that receive these signals migrate to the DZ, where they rapidly divide multiple times and accumulate somatic mutations in their immunoglobulin (Ig) genes (Gitlin et al., 2014). GC B cells can then re-enter the LZ for additional rounds of selection followed by clonal expansion for further affinity maturation (Victora et al., 2010). This process allows for the emergence of B cell clones expressing high affinity antibodies that carry multiple Ig mutations (Kocks and Rajewsky, 1988).
The magnitude of expansion of selected B cell clones is programmed by T cell help that B cells receive during their transient interaction with Tfh cells in the LZ. Increased amounts of the cognate peptide Ags presented by B cells to Tfh cells in the context of MHC class II molecules induce elevated production of cytokines, IL-4 and IL-21 by Tfh cells (Shulman et al., 2014), and facilitate rapid expansion of selected B cells in the DZ and affinity maturation (Gitlin et al., 2015; Gitlin et al., 2014). Thus, the transient T-B interaction in the LZ induces gene expression programs that allow selected B cells to sustain their proliferation in the DZ and establish a diverse BCR repertoire. The transcription factor c-MYC regulates proliferation of both pre-GC B cells and GC B cells, while mutations or translocations of the Myc gene are causally linked to GC-derived B cell lymphomas (Basso and Dalla-Favera, 2015; Calado et al., 2012; Dominguez-Sola et al., 2012). Although T cell help controls cell cycle progression of selected B cells by inducing c-MYC, expression of this proto-oncogene is detected only transiently in LZ B cells prior to their proliferation in the DZ (Calado et al., 2012; Dominguez-Sola et al., 2012; Gitlin et al., 2015; Victora et al., 2010). Thus, the identity of nuclear factors that are induced during the T-B interaction in the LZ and continue to be expressed in the DZ to potentiate proliferation of selected B cells remains unknown.
In this study, we dissected the genetic program that is activated in selected B cells during their transient interaction with Tfh cells in the LZ and supports sustained expansion of B cells in the DZ. Our data demonstrated that the transcription factor AP4 was essential for amplification of GC responses. AP4 was induced by c-MYC in B cells that received T cell help through the CD40-CD40L interaction and maintained by IL-21 as they migrated toward the DZ. Although AP4 was dispensable for selection of B cells and their cell cycle progression in the LZ, it was essential for DZ B cells to re-enter additional division cycles after completing the one initiated in the LZ. This AP4-dependent proliferation of DZ B cells amplified the magnitude of GC responses and supported antibody diversification for protection against chronic viral infection. These results demonstrate that AP4 connects selection and enhanced expansion of Ag-specific B cells that occur in distinct locations in the GC.
Results
The c-MYC inducible factor AP4 is expressed in both LZ and DZ GC B cells
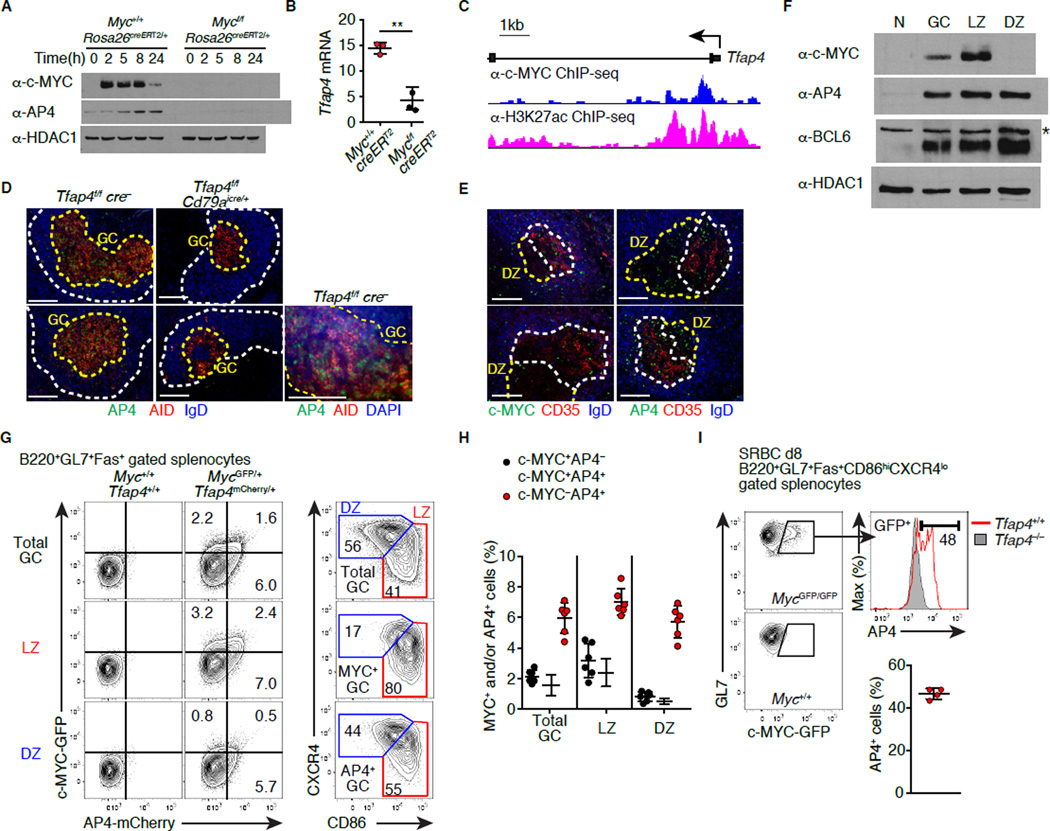
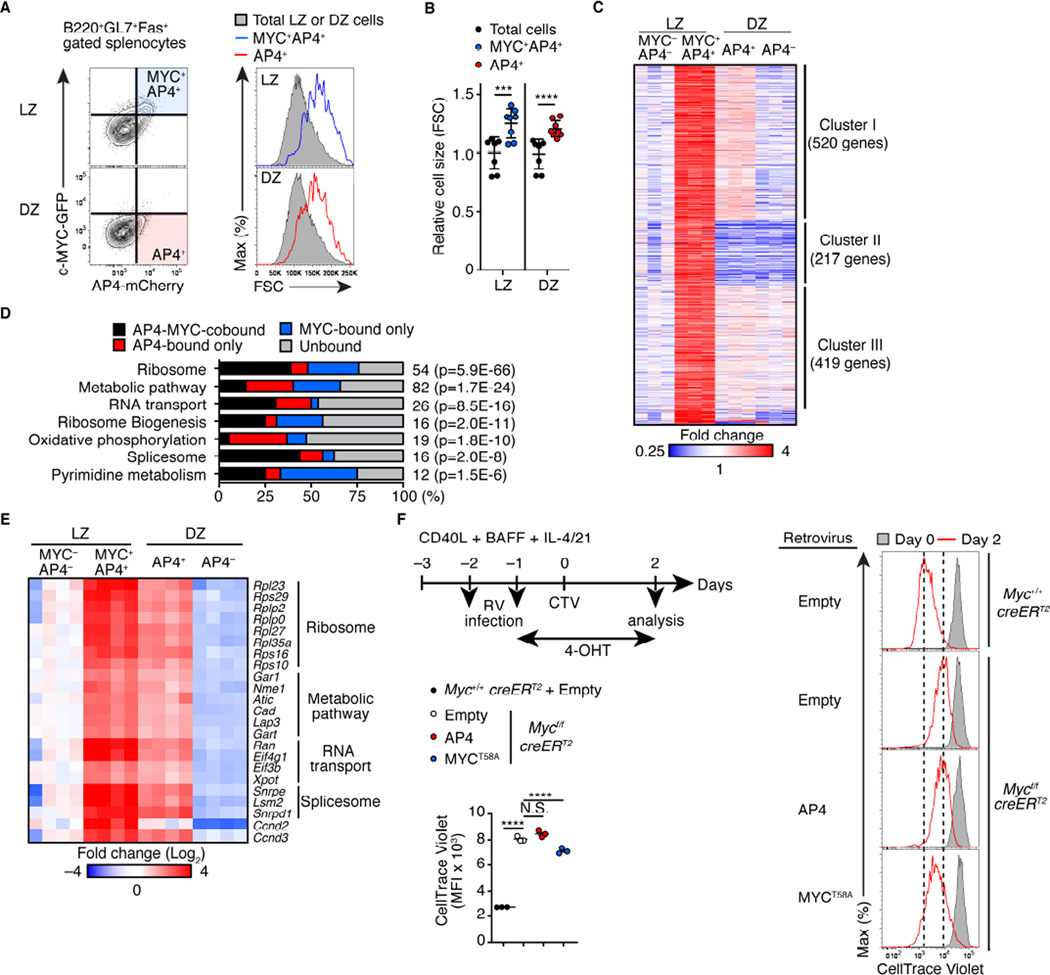
A number of transcription factors are upregulated in GC B cells during clonal selection. The expression of one such factor, AP4 (encoded by Tfap4), is elevated in GC B cells expressing c-MYC compared to those lacking c-MYC based on previously reported gene expression data (Dominguez-Sola et al., 2012). Indeed, AP4 was induced in CD40-stimulated B cells in vitro, but this induction required c-MYC (Figures 1A and 1B). c-MYC bound to an intronic region enriched for the H3K27ac enhancer mark in the Tfap4 locus, suggesting that c-MYC directly regulates Tfap4 expression (Figure 1C). Therefore, we hypothesized that AP4 lies downstream of c-MYC and controls GC B cell proliferation. AP4 protein was detected in splenic GCs, but not in follicular B cells, following immunization with sheep red blood cells (SRBCs) (Figure 1D, left), whereas this staining was absent following B cell-specific deletion of Tfap4 (Figure 1D, middle). The majority of AP4+ cells were also stained for activation induced cytosine deaminase (AID) (Figure 1D, right), which is expressed in both the LZ and DZ cells in mouse GCs (Victora et al., 2012). Within GC, the LZ is marked by the presence of CD23- and CD35-expressing follicular dendritic cells (FDCs) (Calado et al., 2012; Dominguez-Sola et al., 2012; Hardie et al., 1993). As reported previously (Calado et al., 2012; Dominguez-Sola et al., 2012; Victora et al., 2010), c-MYC-expressing cells were detected exclusively in the LZ (Figure 1E, left). In contrast, AP4-expressing cells were distributed in both the LZ and DZ (Figure 1E, right). This result was further confirmed by immunoblotting showing AP4 expression in both CD86hiCXCR4lo LZ and CD86loCXCR4hi DZ GC B populations (Figure 1F). AP4 was also expressed in both LZ and DZ cells derived from human tonsillar GCs, while c-MYC expression was detected only in the LZ (Figure S1A). We next measured AP4 and c-MYC expression at the single cell level using mice harboring genetically targeted alleles that report c-MYC and AP4 protein quantities with GFP and mCherry fluorescence, respectively (Figures 1G, S1B and S1C) (Huang, 2008). In the latter strain, mCherry was fused to the C-terminus of AP4, allowing for detection of AP4 by fluorescence with similar kinetics to endogenous protein (Figures S1D–S1E). In GC B cells, AP4 protein was specifically detected in mCherry+, but not in mCherry− cells (Figure S1F). AP4 was expressed in both LZ and DZ with ~50% of c-MYC+ LZ cells being AP4+ (Figures 1G, 1H and S1G). This was further confirmed by intracellular staining for AP4 in GC B cells (Figure 1I). These results suggest that c-MYC induces AP4 in the LZ, and that its expression persists as GC B cells migrate towards the DZ.
Figure 1. GC B cells express the c-MYC-inducible factor AP4.
(A and B) Expression of AP4 protein and cell number-normalized Tfap4 mRNA (2 hours) in B cells that were harvested from Tamoxifen-treated Mycf/fRosa26creERT2/+ and co-cultured with CD40L-expressing feeders. Data are representative of three experiments.
(C) Binding of c-MYC to the Tfap4 locus in CD40L-activated B cells. Histograms of sequence tags pulled down with anti-c-MYC or anti-H3K27ac antibody are shown.
(D) Staining for AP4, AID and IgD of spleen sections eight days after SRBC immunization. B cell follicles (Fo) and GCs are marked by white and yellow dashed lines, respectively. Data are representative of two experiments.
(E) Staining for AP4 and c-MYC of spleen sections eight days after SRBC immunization. LZ and DZ of GCs defined by the presence of CD35+ FDCs are shown with white and yellow dashed lines, respectively. Data are representative of two experiments.
(F) AP4, c-MYC, and BCL6 protein amounts in naive (N), total B220+GL7+Fas+ GC, CD86hiCXCR4lo LZ and CD86loCXCR4hi DZ B cells eight days after SRBC immunization. Data are representative of three experiments. *: non-specific band.
(G and H) Expression of GFP and mCherry in GC B cells of MycGFP/+Tfap4mCherry/+ mice eight days after SRBC immunization (G). Distribution of AP4-mCherry+ or c-MYC-GFP+ cells in the LZ and DZ defined by CD86 and CXCR4 expression is shown in the right panels (G). Frequencies of AP4- or c-MYC-expressing cells in total GC, LZ and DZ GC B cells from two independent experiments are shown in (H).
(I) Intracellular staining for AP4 in c-MYC-GFP+ LZ GC B cells eight days after SRBC immunization. Staining for AP4 in Tfap4−/− LZ cells is shown as control. Data are pooled from two experiments.
Data in (B, H and I) are shown by means ± SD. Unpaired Student’s t test. n = 3 per group (A and B); n = 4 (D and E); n = 3 (F); n = 6 for (G and H); n = 4 for MycGFPTfap4+/+ and n = 2 for Tfap4– /– mice (I). See also Figure S1. Scale bar, 100 µm.
AP4 is required for normal GC formation in a B cell-intrinsic manner
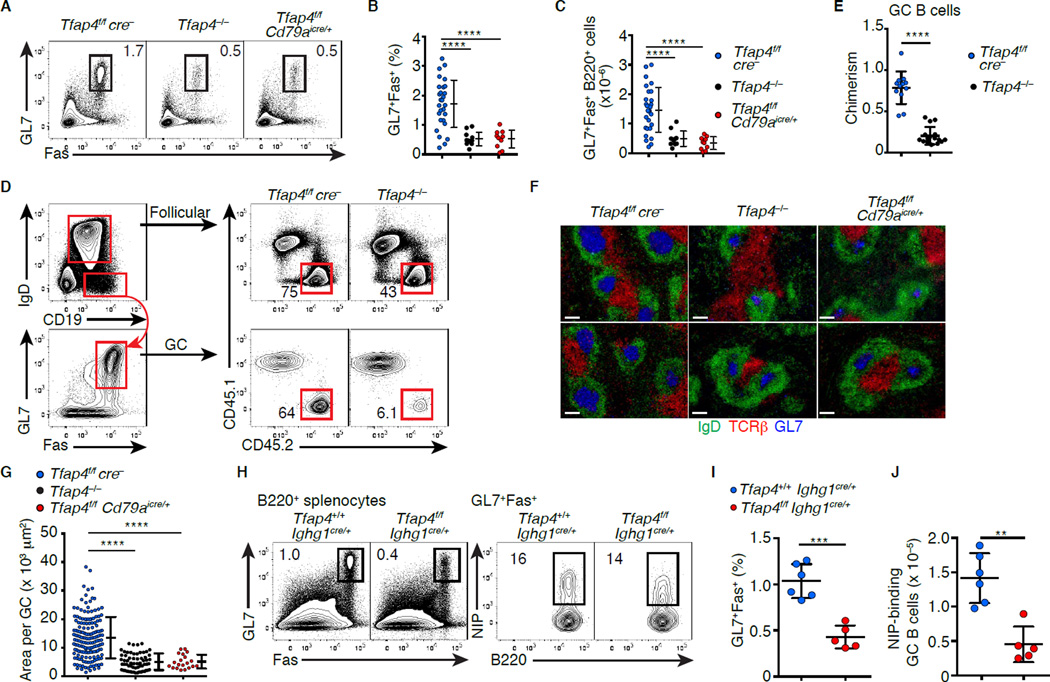
To test whether AP4 is required for GC responses, we immunized Tfap4−/− mice with the T-dependent Ag 4-Hydroxy-3-nitrophenylacetyl-chicken gamma globulin (NP-CGG). Although AP4 was dispensable for normal B cell development in the bone marrow and spleen (Figures S2A–S2F), the frequencies and numbers of B220+GL7+Fas+ GC B cells were reduced in Tfap4−/− mice following immunization (Figures 2A-2C). This defect was also observed in mixed bone marrow chimeras reconstituted with a mixture of WT and Tfap4−/− bone marrow cells (Figures 2D and 2E), indicating that diminished GC response was a cell-intrinsic defect. Consistently, a comparable reduction of GC B cell numbers was seen in Tfap4f/f Cd79aicre/+ mice lacking AP4 specifically in B cells (Hobeika et al., 2006) (Figures 2A-2C). In the absence of AP4, individual splenic GC sizes were smaller (Figures 2F and 2G). GC B cell frequencies in Peyer’s patches were similarly reduced in the absence of AP4 (Figure S3A and S3B). Following immunization with a T-dependent Ag, c-MYC was expressed in activated B cells in a biphasic manner in pre-GC and GC B cells, and essential for both initiation and maintenance of GCs (Calado et al., 2012; Dominguez-Sola et al., 2012). AP4 was also expressed in pre-GC B cells (Figure S3C), and B cell expansion was reduced at the pre-GC stage (Figures S3D and S3E). However, numbers of total and Ag-binding GC B cells were significantly reduced when AP4 was deleted using an Ighg1cre allele in ongoing GCs following immunization (Casola et al., 2006) (Figures 2H-2J), indicating that AP4 was cell-intrinsically required for the maintenance of GCs. This conclusion was further validated by a similar result observed when Tfap4 was deleted in GC B cells by Tamoxifen-inducible cre with the majority of Tfh cells retaining intact Tfap4 (Figures S3F–S3I). In contrast, AP4 was dispensable for a T-independent response to NP-Ficoll (Figures S3J–S3L). These results indicate that AP4 cell-autonomously regulates both GC initiation and maintenance.
Figure 2. AP4 is required for normal GC formation in a B cell-intrinsic manner.
(A–C) Expression of GL7 and Fas in B220+ splenocytes ten days after NP-CGG immunization (A). Statistical analysis of frequencies (B) and absolute numbers (C) of GL7+Fas+ cells in B220+ splenocytes from four independent experiments is shown.
(D and E) Flow cytometric analysis of CD45.2 donor cell contribution to the follicular (IgD+CD19+) and GC (IgD−CD19+GL7+Fas+) B cell compartments in mixed bone marrow chimeras 12 days after NP-CGG immunization (D). Statistical analysis of CD45.2 donor cell contribution to GC normalized to that of follicular B cells from five independent experiments is shown in (E).
(F) Staining for GL7, IgD and TCRβ of spleen sections from mice with indicated genotypes ten days after NP-CGG immunization. Representative data from four independent experiments are shown. Scale bar: 100 µm.
(G) Statistical analysis of the size of each GC in (F). Data are pooled from four independent experiments.
(H–J) Expression of GL7 and Fas in B220+ splenocytes and NIP-allophycocyanin (APC)-binding by B220+GL7+Fas+ cells from Tfap4f/fIghg1cre/+ and control Tfap4+/+Ighg1cre/+ mice ten days after NP-CGG immunization. Statistical analysis of percentages of GL7+Fas+ cells in B220+ cells (I) and the numbers of NIP-binding cells in the spleen (J) from two independent experiments are shown.
Data in (B, C, E, G and I) are shown by means ± SD. One-way ANOVA for (B, C and G). Unpaired Student’s t test for (E, I and J). n = 10 for Tfap4−/− and Tfap4f/fCd79aicre/+ , and n = 28 for Tfap4f/fcre− (A-C, F and G); n = 12 for Tfap4f/fcre− and n = 16 for Tfap4−/− (D and E); n = 5 for Tfap4f/fIghg1cre/+ and n = 6 for Tfap4+/+Ighg1cre/+ mice (H–J). See also Figure S3
AP4 deficiency results in reduced proliferation of DZ B cells
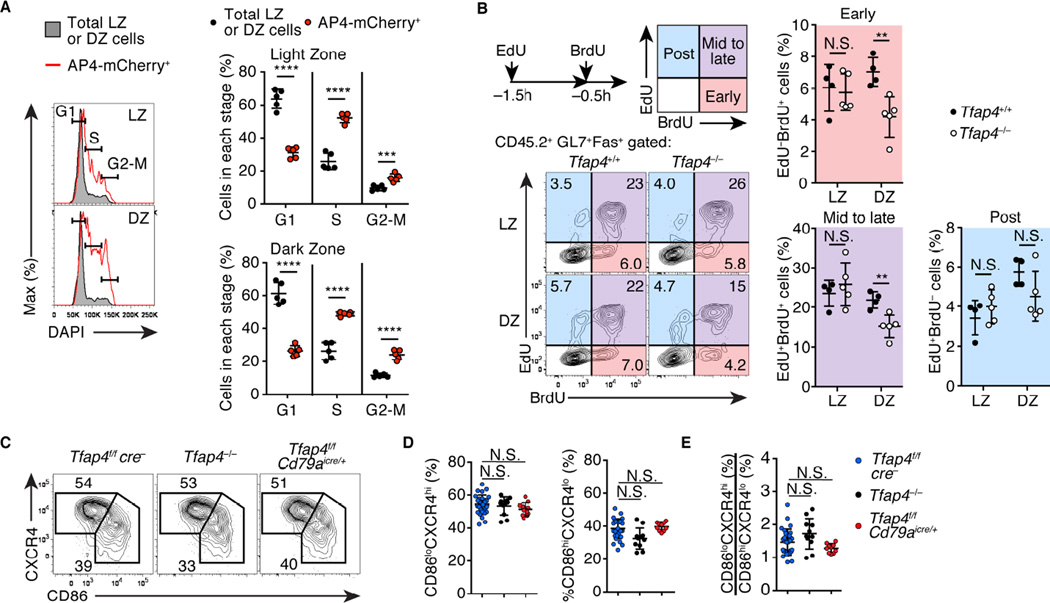
GC B cells induce c-MYC upon selection in the LZ (Calado et al., 2012). The c-MYC induction was unimpaired in Tfap4−/− mice compared to control WT mice (Figure S4A), indicating that AP4 is dispensable for selection of LZ GC B cells. AP4 expression marked actively cycling cells both in the LZ and DZ B cell populations (Figure 3A). A considerable fraction of AP4+ LZ cells were in the S-phase with a reciprocal reduction of cells in the G1-phase, consistent with previous reports that c-MYC+ cells are actively cycling (Calado et al., 2012; Dominguez-Sola et al., 2012). AP4+ DZ cells, which no longer expressed c-MYC, were also enriched for cells in S and G2/M phases of cell cycle, suggesting that continued expression of AP4 supports proliferation of selected GC B cells in the DZ. To assess whether AP4 was required for selected GC B cells to re-enter additional division cycles in the DZ after completing a cycle initiated in the LZ, we examined sequential incorporation of EdU and BrdU (Gitlin et al., 2014) by Tfap4−/− and control Tfap4+/+ GC B cells in mixed bone marrow chimeras (Figure 3B). In the LZ, similar proportions of Tfap4−/− and Tfap4+/+ cells were distributed in distinct cell cycle phases. In contrast, frequencies of DZ cells in the early and mid to late S-phases were significantly reduced in the absence of AP4. These results indicate that AP4 is dispensable for cell cycle progression initiated in the LZ and an initial division in the DZ, but is necessary to promote subsequent divisions. As EdU and BrdU incorporation by CD45.1 WT GC B cells was comparable between Tfap4−/− and control mixed chimeras (Figure S4B), the requirement for AP4 in the cell cycle re-entry was cell-intrinsic. AP4 thus amplifies proliferation of DZ B cells by promoting their re-entry to additional division cycles. Paradoxically, however, LZ to DZ cell number ratios were unaltered in the absence of AP4 (Figures 3C-3E) presumably because impaired expansion of DZ cells may reduce DZ to LZ trafficking of GC B cells, affecting cellularity in both zones.
Figure 3. AP4 enhances GC B cell proliferation through the regulation of cell cycle re-entry in the DZ.
(A) Cell cycle analysis by DNA content measurement of AP4-mCherry+ LZ and DZ cells compared to total LZ and DZ cells from Tfap4mCherry reporter mice eight days after SRBC immunization. Data are pooled from two independent experiments.
(B) Cell cycle analysis by sequential EdU and BrdU labeling of CD45.2 Tfap4−/− and control Tfap4f/fcre− donor LZ and DZ GC B cells in mixed bone marrow chimeras eight days after NP-CGG immunization. Early-, mid to late-, and post-S phases of the cell cycle were defined as previously described (Gitlin et al., 2014). Representative plots and statistical analysis from two independent experiments are shown.
(C–E) Expression of CD86 and CXCR4 in B220+GL7+Fas+ GC B cells from mice with indicated genotypes ten days after NP-CGG immunization. Representative plots (C) and statistical analysis of frequencies of LZ (CD86hiCXCR4lo) and DZ (CD86loCXCR4hi) cells in the GC B cell compartment (D) and DZ to LZ ratios (E) from four independent experiments are shown. Data in (A, B, D and E) are shown by mean ± SD. Unpaired Student’s t test for (A and B) Oneway ANOVA for (D and E). n = 5 (A); n = 4–5 per group (B); n = 10 for Tfap4−/− and Tfap4f/fCd79aicre/+ , and n = 28 for Tfap4f/fcre− (D and E). See also Figure S4.
AP4 sustains GC B cell activation in the DZ
We next defined AP4-dependent gene regulatory programs for GC B cell expansion by profiling gene expression in c-MYC+AP4+ LZ cells and AP4+ DZ cells along with c-MYC−AP4− LZ cells (pre-selection LZ cells) and AP4− DZ cells (post-mitotic DZ cells) (Figure 4A). Since AP4 was expressed in a small fraction of GC B cells, this method would better define AP4-dependent gene expression than directly comparing AP4-sufficient and -deficient GC B cells. A similar approach was used to determine c-MYC target genes (Dominguez-Sola et al., 2012). Consistent with their active cell cycle status (Figure 3A), c-MYC+AP4+ LZ and AP4+ DZ cells were larger in size compared to total LZ and DZ cells, respectively (Figures 4A and 4B). Gene expression profiling by RNAseq identified ~1,200 genes upregulated in c-MYC+AP4+ relative to c-MYC−AP4− LZ cells by more than 1.8-fold, which were clustered into three groups (Figure 4C, and Table S1). Among these genes, those in Cluster I were more highly expressed in AP4+ than AP4− DZ B cells and thus predicted to be relevant to AP4-dependent GC B cell expansion. Genes in Clusters II and III were expressed similarly in AP4+ and AP4− DZ B cells and included Myc and known c-MYC-target genes, such as Batf, Irf4, and Ccnd2 (Dominguez-Sola et al., 2012). Genes in Cluster I were enriched for those in pathways related to protein translation and metabolism (Figure 4D, and Table S1). ChIPseq analysis for AP4 and c-MYC binding using purified GC B cells and activated B cells in vitro, respectively, revealed that ~75% and ~60% of genes in the ribosome and metabolic pathways were directly bound by c-MYC and AP4, respectively, with ~50% and ~20% of those being common direct targets of c-MYC and AP4 (Figures 4E and S4C). These data suggest that c-MYC and AP4 sequentially sustain transcription of genes in ribosome biogenesis and metabolic pathways in selected GC B cells. However, AP4 overexpression was insufficient to fully compensate for c-MYC deficiency in B cells cultured in a condition mimicking GC differentiation (Nojima et al., 2011) (Figure 4F). This result suggests that AP4-dependent proliferation program requires prior c-MYC expression.
Figure 4. AP4-expressing DZ cells maintain activation signatures following c-MYC downregulation.
(A and B) Relative cell sizes (forward scatter) of MYC+AP4+ LZ and DZ B cells from AP4-mCherry and c-MYC-GFP reporter mice eight days after SRBC immunization (A). Statistical analysis from three independent experiments is shown in (B)
(C) RNAseq analysis of MYC−AP4− LZ, MYC+AP4+ LZ, AP4+ DZ and AP4− DZ cells. Expression of genes which were expressed higher by >1.8-fold in MYC+AP4+ LZ cells than MYC−AP4− LZ cells is shown as a heat map following unsupervised clustering analysis that yielded three major clusters.
(D) Pathway analysis of Cluster I genes in (C) combined with ChIPseq analysis showing the frequencies of genes directly bound by AP4, c-MYC, or both in each pathway. The number of genes in each pathway is shown to the right of the bar with P-values for enrichment.
(E) Spike-in qPCR validation of representative AP4-MYC-cobound genes in (D). Expression in four independent samples from two experiments are shown as a heatmap.
(F) Flow cytometric analysis showing CellTrace Violet dilution of Myc-deficient B cells infected with a control retrovirus (RV) or one expressing AP4 or MYCT58A. CD40L-primed Mycf/fRosa26creERT2/+ or Myc+/+Rosa26creERT2/+ splenic B cells were infected with indicated RV, followed by treatment with 4-hydroxytamoxifen (4-OHT), and proliferation was analyzed. Data are representative of two independent experiments.
Data in (B and F) are shown by mean ± SD. Unpaired Student’s t test for (B). One-way ANOVA for (F). n = 6 (A and B); n = 3 (C); n = 3 per group (F). See also Figure S4 and Table S1.
AP4 is required for accumulation of somatic mutations
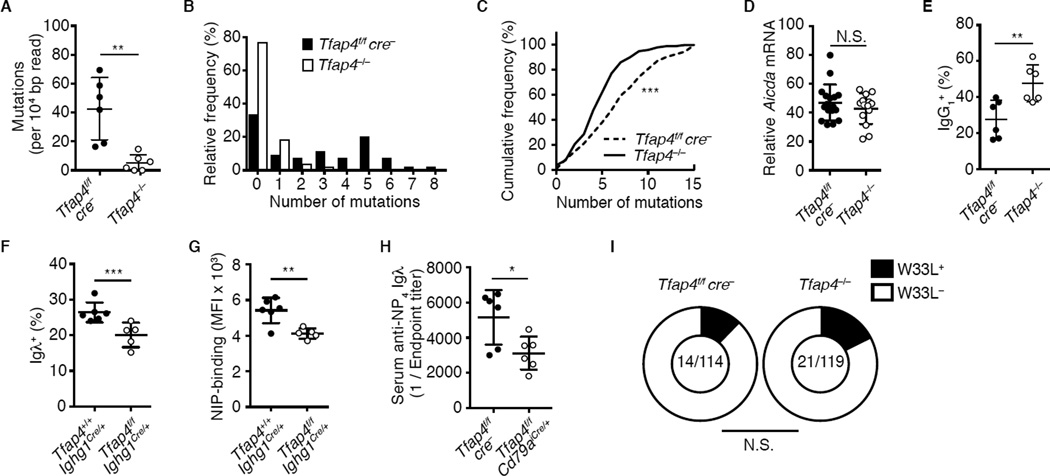
Since expansion of GC B cells is essential for accumulation of somatic mutations and diversification of Ig repertoires, we next determined the frequencies of mutations in the JH4-adjacent intronic region of Igh (Jolly et al., 1997). The overall frequencies of mutations and the frequencies of clones carrying multiple mutations following NP-CGG immunization were substantially decreased in the absence of AP4 (Figures 5A and 5B). The mutation frequency in the VH186.2 BCR, which is over-represented in NP-reactive B cell clones (Allen et al., 1987; Allen et al., 1988), was also reduced (Figure 5C). However, Aicda mRNA expression was unaltered and frequencies of class switched IgG1+ GC B cells were increased in the absence of AP4 (Figures 5D and 5E), suggesting that reduced somatic mutation unlikely resulted from impaired AID activity. The binding avidity of BCR to NP and the surrogate Ag 4-hydroxy-3-iodo-5-nitrophenylacetic acid (NIP) and the frequencies of NP- and NIP-binding clones expressing an Igλ light chain (Allen et al., 1987), as well as NP-specific Igλ antibody titers in serum, were modestly reduced in the absence of AP4 (Figures 5F-5H and S5A–S5K). However, the frequencies of VH186 BCR clones carrying the affinity-enhancing W33L mutation and the ratio of titers between high-affinity anti-NP4 to low-affinity anti-NP15 IgG1 were comparable between Tfap4−/− and control GC B cells (Figure 5I and S5L). These results suggest that AP4 promotes expansion of selected GC B cells, thus diversifying BCR repertoires through somatic hypermutation, although it is dispensable for the selection of high-affinity clones and affinity maturation of anti-NP antibodies.
Figure 5. AP4 is necessary for the accumulation of somatic mutations.
(A) Frequencies of mutations in an JH4 adjacent intronic region in Tfap4−/− and control Tfap4f/fcre− donor GC B cells from mixed bone marrow chimeras 12 days after NP-CGG immunization. Data are pooled from three independent experiments.
(B) Frequencies of clones carrying multiple mutations in (A).
(C) Cumulative frequency distribution of VH186.2 somatic mutations in CD45.2 donor GC B cells in (A). Data are pooled from three independent experiments.
(D and E) Expression of Aicda mRNA and frequencies of IgG1+ in CD45.2 donor-derived GC B cells in (A). Data are pooled from three independent experiments.
(F) Frequencies of Igλ+ cells in NIP-APC-binding GC B cells from Tfap4f/fIghg1cre/+ and control Tfap4+/+Ighg1cre/+ mice ten days after NP-CGG immunization. Data are pooled from two independent experiments.
(G) NIP-APC-binding by GC B cells in (F). Data are pooled from two independent experiments. (H) NP4-specific serum Igλ titers in Tfap4f/fCd79aicre/+ and control Tfap4f/fcre− mice 56 days after NP-CGG immunization. Data are pooled from two independent experiments.
(I) Frequencies of the W33L mutation in VH186.2 BCR+ clones in CD45.2 GC B cells in (A). Numbers of W33L bearing clones and total number of clones sequenced are shown. Data are pooled from four independent experiments.
Data in (A, D, E, F-H) are shown by means ± SD, with unpaired Student’s t test. Kolmogorov-Smirnov test for (C). c2 test with Yates correction for (I). n = 6 for each group (A-C, E-I); n = 17 for Tfap4f/fcre− and n = 14 for Tfap4−/− (D). See also Figure S5.
AP4 is required for control of chronic viral infection
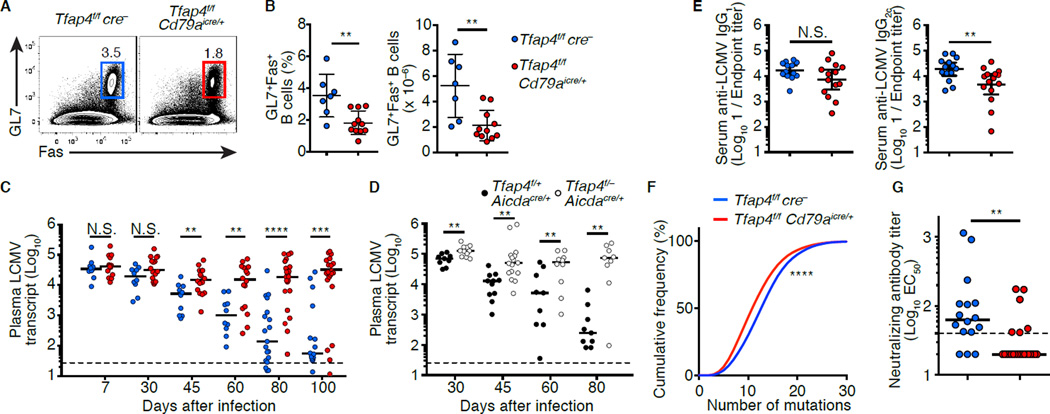
Our results demonstrate that AP4 contributes to diversification of BCR repertoire by promoting expansion of DZ GC B cells. Antibody diversification and accumulation of somatic mutations are critical for the generation of protective antibodies against chronic viral infections in mammals, as exemplified by broadly neutralizing antibodies (bnAbs) against HIV that harbor high frequencies of mutations (Halper-Stromberg et al., 2014; Klein et al., 2013). These bnAbs against HIV emerge years after the initial infection, raising the possibility that maximal diversification of the BCR repertoire by GCs as the virus mutates is essential for their emergence (Mascola and Haynes, 2013). In mice, the generation of high affinity antibodies during GC responses is required for control of chronic LCMV infection (Bergthaler et al., 2009; Harker et al., 2011). LCMV clone 13 (LCMV-c13) infection of Tfap4f/f Cd79aicre/+ mice induced GC formation; however, the numbers and frequencies of GC B cells were significantly reduced compared to control Tfap4f/f cre− mice (Figures 6A and 6B) despite normal numbers of CXCR5+PD-1+ Tfh cells in the mutants (Figure S6A–S6C). Consistent with previous studies (Harker et al., 2011; Wherry et al., 2003), the serum viral titers in control mice started to decline 30 days after infection, and viremia was mostly resolved by day 100 (Figure 6C). In Tfap4f/f Cd79aicre/+ mice, despite comparable viremia 7 days post-infection, viral loads remained high until day 100 (Figure 6C). Similarly defective control of viremia was seen in Tfap4f/– Aicdacre/+ compared to Tfap4f/+ Aicdacre/+ mice (Figure 6D). These results indicate that AP4 expression in B cells is essential for control of chronic LCMV infection. Consistent with the presence of GCs in Tfap4f/f Cd79aicre/+ mice, AP4 was dispensable for the production of class switched anti-LCMV antibodies (Figures 6E, S6D and S6E). However, AP4-deficient GC B cells accumulated reduced numbers of mutations (Figure 6F). Consequently, heat-inactivated immune sera from Tfap4f/f Cd79aicre/+ mice were minimally able to neutralize LCMV in vitro, in sharp contrast to their WT counterparts (Figures 6G and S6F). These results demonstrate that induced and sustained AP4 expression in Ag-specific B cells is required for maximizing the magnitude of GC responses and for generating protective antibodies against viral infections.
Figure 6. AP4 is essential for humoral responses against chronic viral infection.
(A and B) Frequencies and absolute numbers of B220+GL7+Fas+ cells in the spleens of Tfap4f/fCd79aicre/+ and control Tfap4f/fcre− mice 30 days after infection with LCMV-c13. Data are pooled from two independent experiments.
(C and D) Analysis of plasma LCMV loads in Tfap4f/fCd79aicre/+ and control Tfap4f/fcre− mice (C) and Tfap4f/-Aicdacre/+ and control Tfap4f/+Aicdacre/+ mice (D) after infection. Data are pooled from six (C) and three (D) independent experiments.
(E) Titers of LCMV-binding serum IgG in Tfap4f/fCd79aicre/+ and control Tfap4f/fcre− mice 80 days after infection. Data are pooled from six independent experiments.
(F) Cumulative frequency distribution of somatic mutations of the Igh variable regions in GC B cells in (B). Data are pooled from two independent experiments.
(G) Neutralizing serum antibody titers from Tfap4f/fCd79aicre/+ and control Tfap4f/fcre− mice 80 days after LCMV infection. Neutralizing titers were shown as 1/EC50. Data are pooled from six independent experiments.
Data in (B and E) are show by means ± SD. Lines in (C, D and G) represent medians. Unpaired Student’s t test for (B and E). Mann-Whitney’s ranked test for (C, D and G). Kolmogorov-Smirnov test for (F). n = 7 for Tfap4f/fcre− and n = 11 for Tfap4f/fCd79aicre/+ (A and B); for each time point n = 10–19 for Tfap4f/fcre− and n = 16–25 for Tfap4f/fCd79aicre/+ (C); for each time point n = 9–11 per group (D); n = 12 for Tfap4f/fcre− and n = 14 for Tfap4f/fCd79aicre/+ (E); n = 6 per group (F); n = 14 for Tfap4f/fcre− and n = 25 for Tfap4f/fCd79aicre/+ (G). See also Figure S6.
AP4 is sustained by IL-21R signals following induction by c-MYC
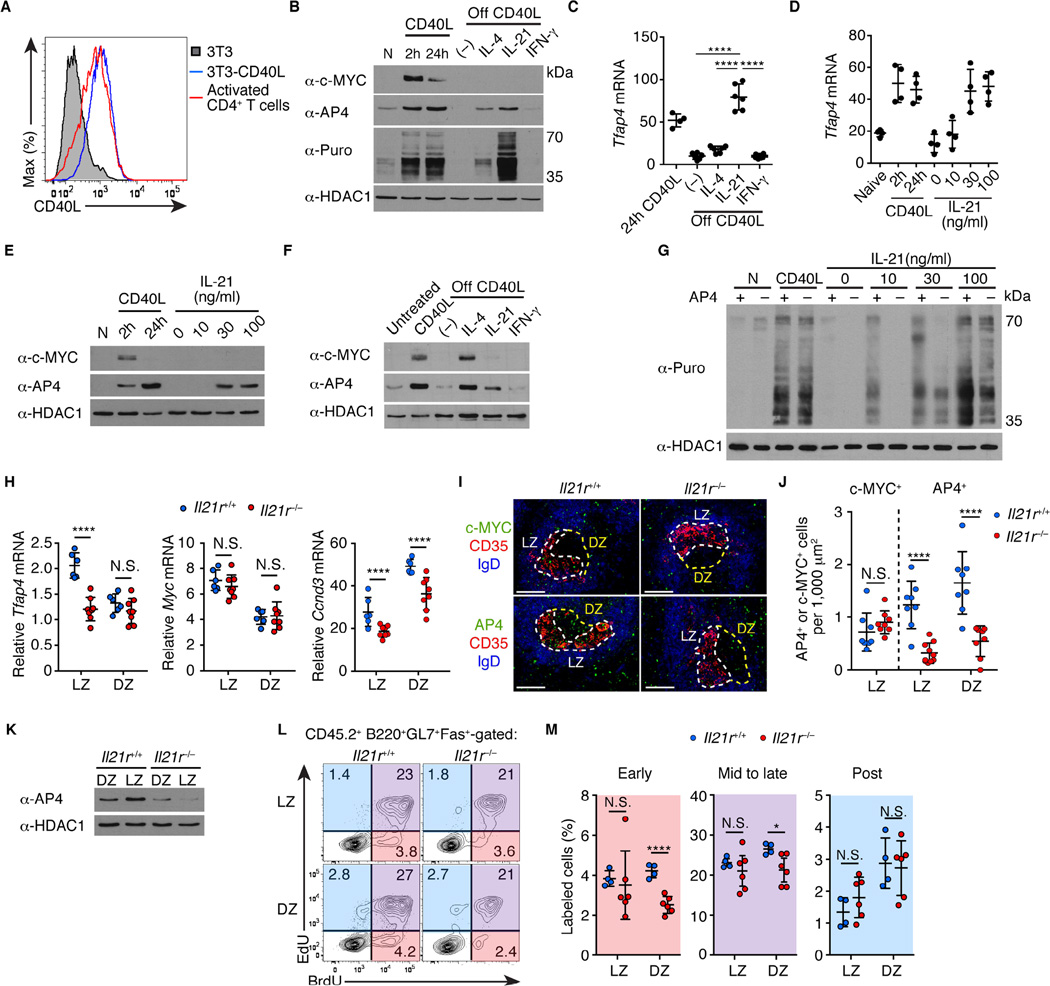
The finding that AP4 is induced by c-MYC in selected GC B cells, but is maintained longer than c-MYC, suggests that help signals derived from Tfh cells may sustain AP4 expression during migration of LZ B cells towards the DZ. To test this, we first induced c-MYC-dependent AP4 expression in B cells by co-culturing them on NIH3T3 feeders engineered to express CD40L at an amount comparable to activated CD4+ T cells (Figure 7A). Under these conditions, both c-MYC and AP4 protein quantities increased rapidly (Figure 7B). During GC reactions, selected B cells contact Tfh cells and receive survival and proliferative signals through a CD40-CD40L ligation, and subsequently lose this contact as they migrate to the DZ (Allen et al., 2004; Castigli et al., 1994; Kawabe et al., 1994; MacLennan, 1994; Renshaw et al., 1994; Victora and Nussenzweig, 2012; Victora et al., 2010; Xu et al., 1994). Indeed, deprivation of CD40 stimulation reduced AP4 expression at both transcript and protein levels (Figures 7B and 7C). Importantly, subsequent stimulation of the CD40-primed cultured B cells with IL-21 sustained AP4 expression despite the absence of c-MYC protein, while Tfh-derived cytokines, IL-4 and IFN-γ, showed only a modest and no effect, respectively (Figure 7B). IL-21 increased the amount of AP4 in a dose-dependent manner (Figures 7D and 7E). IL-21 was also sufficient to sustain AP4 expression in cultured GC B cells ex vivo following stimulation with CD40L (Figure 7F). However, IL-21 was insufficient to increase the AP4 amount without priming of B cells with CD40L (Figure S7A). Furthermore, co-stimulation of B cells with IL-21 and CD40L reduced AP4 mRNA amounts compared to CD40L stimulation alone due to an unknown reason, although AP4 protein amounts were unchanged possibly due to compensation by IL-21-mediated elevation of protein translation (Figure S7B and S7C). Thus, IL-21 can maintain AP4 expression after c-MYC expression is downregulated in B cells in vitro. In addition to upregulation of AP4 mRNA (Figure 7C), global protein translation as measured by a puromycin pulse labeling (Nathans, 1964) was markedly elevated by IL-21 (Figure 7B). Interestingly, IL-21-mediated increase of protein translation was reduced in AP4-deficient B cells (Figure 7G).
Figure 7. IL-21 sustains expression of AP4 after c-MYC downregulation.
(A) Flow cytometric analysis showing expression of CD40L on NIH-3T3 cells infected with a CD40L-expressing retrovirus compared to that in CD4+ T cells polarized in vitro for 2 days.
(B) AP4 and c-MYC protein amounts and puromycin (Puro) incorporation in naive (N), stimulated B cells with CD40L, and those subsequently stimulated with cytokines without CD40L (off CD40L). (–) denotes no cytokines added. Data are representative of three independent experiments.
(C) Cell-number normalized Tfap4 mRNA expression in activated B cells from (B). Data are pooled from two independent experiments.
(D and E) Cell-number normalized Tfap4 mRNA and protein expression in B cells stimulated with CD40L and those subsequently stimulated with IL-21. Data are pooled from three independent experiments.
(F) AP4 and c-MYC protein expression in GC B cells ex vivo that were untreated, CD40L-stimulated, and those subsequently stimulated with cytokines without CD40L signals (off CD40L). (–) denotes no cytokine added. GC B cells were purified eight days after SRBC immunization. Data are representative of two independent experiments.
(G) Puromycin incorporation by Tfap4f/–Cd79aicre/+ and control Tfap4f/+Cd79aicre/+ naive B cells (N), CD40L-stimulated B cells, and those subsequently stimulated with different concentrations of IL-21 without CD40L. Data are representative of two independent experiments.
(H) Tfap4, Myc, Ccnd3 mRNA amounts in Il21r+/+ and Il21r−/− CD86hiCXCR4lo LZ and CD86loCXCR4hi DZ GC B cells from mixed bone marrow chimeras (Figure S7F) ten days after NP-CGG immunization. Data are pooled from three independent experiments.
(I and J) Staining for c-MYC and AP4 expression of spleen sections from mice in (H). LZ and DZ defined as in Figure 1E. Data are representative of three independent experiments. Scale bar, 100 µm. Statistical analysis of c-MYC- and AP4-expressing cell frequencies per 1,000 µm2 of LZ or DZ is shown in (J). Data are pooled from three independent experiments.
(K) AP4 protein amounts in LZ and DZ GC B cells in (H). Data are representative of three independent experiments.
(L and M) Cell cycle analysis by sequential EdU and BrdU labeling of CD45.2 Il21r−/− and control Il21r+/+ donor LZ and DZ GC B cells from mixed bone marrow chimeras in (H). Data are pooled from two independent experiments.
Data in (C, D, H, J and M) are shown by means ± SD. One-way ANOVA for (C), and unpaired Student’s t test for (H, J and M). n = 4–6 (B–E); n = 2 (F); n = 2 per group (G); n = 6 for Il21r+/+ and n = 8 for Il21r−/− (H); n = 4 for Il21r+/+ and n = 6 for Il21r−/− (I–M). See also Figure S7.
Therefore, prior expression of AP4 induced by CD40 signals and c-MYC amplifies the effect of IL-21 on protein translation, including accumulation of its own protein, which may be retained in a subset of DZ cells passively or post-transcriptionally. IL-21 is also necessary for upregulation of Bcl6 and Aicda transcripts (Ettinger et al., 2005; Linterman et al., 2010). However, expression of these genes were not affected in IL-21 stimulated Tfap4−/− B cells in vitro (Figure S7D and S7E), indicating that AP4 plays a targeted role in controlling the IL-21-dependent GC B cell program .
To validate this finding in vivo, we generated bone marrow chimeras by reconstituting mice with a mixture of bone marrow cells from Igh−/− (75%) and Il21r−/− (25%) mice (Figure S7F). In the resulting chimeras, all B cells were exclusively derived from Il21r−/− bone marrow cells, while a large proportion of non-B cells, including Tfh cells, were sufficient for Il21r (Figures S7G–S7J). Consistent with previous reports (Linterman et al., 2010; Zotos et al., 2010), GC responses were blunted in bone marrow chimeras with Il21r−/− B cells following NP-CGG immunization, with a significant reduction in the amounts of Bcl6 and Aicda transcripts compared to control chimera with Il21r+/+ B cells (Figure S7K–S7N). In these chimeras, Il21r−/− LZ B cells expressed significantly reduced Tfap4, but not Myc transcripts compared to Il21r+/+ LZ B cells (Figure 7H). Ccnd3, a direct AP4 target in GC B cells (Figures 4E and S4C), was also downregulated in Il21r−/− GC B cells (Figure 7H). While frequencies of c-MYC+ cells were similar between Il21r−/− and Il21r+/+ GCs, frequencies of AP4+ cells as well as AP4 protein amounts were markedly diminished in both LZ and DZ of Il21r−/− GCs (Figures 7I-7K). Furthermore, early and mid to late S-phase cells in the DZ, but not in the LZ, were reduced in the absence of IL-21R signals, which is similar to the phenotype observed in AP4-deficient GCs (Figures 7L and 7M). Thus, IL-21 amplifies GC response through sustaining AP4 expression.
Discussion
In the present work, we have defined a molecular mechanism by which T cell help signals received in the LZ programs repeated rounds of proliferation of selected GC B cells in the anatomically distant DZ. Studies using a DEC205-targeted Ag-delivery have demonstrated that amounts of cognate Ag that GC B cells capture and present to Tfh cells in the LZ regulate the magnitude of expansion of the selected B cells in the DZ through upregulation of c-MYC target genes (Gitlin et al., 2015; Gitlin et al., 2014). However, expression of c-MYC is restricted to a small subset of LZ GC B cells, and it thus remains unknown how GC B cell proliferation initiated in the LZ is sustained in the DZ after they lose contact with Tfh cells and expression of c-MYC. Our results indicate that AP4 connects the selection in the LZ to sustained proliferation in the DZ. AP4 is induced by CD40 signals in a c-MYC-dependent manner and its expression is maintained after the c-MYC decay. AP4 is maintained by the cytokine IL-21, which is upregulated in Tfh cells during their transient interaction with Ag-presenting GC B cells (Shulman et al., 2014). IL-21 may continue to provide T cell help to selected B cells during the T-B interaction and during their migration towards the DZ to express important genes for GC reactions, including AP4. This effect is mediated at the transcriptional level, and also at the translational level in part in an AP4-dependent manner. Thus, IL-21 may specifically act on selected B cells that have upregulated AP4. However, it may not reach the DZ B cells since IL-21R-dependent upregulation of AP4 at the transcript level was observed only in the LZ B cells.
GCs are essential for the generation of protective antibodies that accumulate multiple somatic mutations in their Ig genes. Recent studies have identified bnAbs against HIV that are characterized by substantially high quantities of somatic hypermutations (Mascola and Haynes, 2013), suggesting that enhanced BCR diversification by sustained GC responses is critical for the generation of these antibodies. Strains of LCMV have been used to study both humoral and cellular immune responses against chronic viral infection in mice. GC responses and expression of AID are essential for the control of chronic LCMV infection (Bergthaler et al., 2009; Harker et al., 2011). Our data indicate that AP4-mediated durable GC response is required for the control of chronic LCMV infection in a B cell-intrinsic manner. This was evidenced by reduced accumulation of somatic mutations and the impaired development of antibodies with neutralizing activities in the absence of AP4. Although these results indicate that AP4 is required for diversification of antibody repertoire and subsequent affinity maturation, AP4 deficiency had no substantial impact on antibody affinity maturation to NP. This can be explained by recent reports that anti-hapten antibody affinity is robustly increased by a few dominant mutations, whereas affinity maturation against complex protein Ag requires sequential accumulation of many mutations during GC reaction (Kuraoka et al., 2016; Tas et al., 2016).
We previously demonstrated that AP4 maximizes acute CD8+ T cell responses by sustaining c-MYC-initiated gene expression program (Chou et al., 2014). Together with our current study, both T and B cells seem to be dependent on restricted c-MYC expression and subsequently utilize its downstream factor AP4 to achieve the maximal magnitude of adaptive immune response. After induction by c-MYC, expression of AP4 is sustained in an IL-2 or IL-21-dependent manner in CD8+ T or GC B cells, respectively, and compensates for the early termination of c-MYC expression (Chou et al., 2014). It is unclear why activated lymphocytes utilize c-MYC only for a brief window and terminate its expression prematurely before the completion of their clonal expansion. Although c-MYC is critical for the establishment of highly proliferative states of lymphocytes upon Ag receptor stimulation, prolonged expression of c-MYC may make the cells prone to apoptosis (Sander et al., 2012) or increase the risk of transformation in the presence of genotoxic stress during GC responses. Aberrant expression of c-MYC has been causally linked to increased incidence of pre-B cell leukemia and B cell lymphoma (Adams et al., 1985). The Myc locus is also efficiently targeted by AID (Kato et al., 2012). Therefore, restriction of c-MYC at both protein and transcript levels may be necessary to prevent transformation during GC responses, although extensive lymphocyte proliferation is critical for protection against virulent pathogens.
Collectively, our results demonstrate that direct T cell help during the T-B interaction and potential remote help mediated by IL-21 sequentially induce transient expression of c-MYC and sustained expression of AP4 to drive sufficient expansion of GC B cells following clonal selection by Tfh cells. Such sustained GC responses are necessary for the generation of mutated antibodies with protective activity against chronic viral infection in the mouse model and may also be important for the generation of the anti-HIV bnAbs in humans. In conjunction with our previous study, the current data suggest that activated B and T cells both utilize a transcription factor cascade in which c-MYC hands off its role to AP4, a relay that is required to maximize clonal expansion to drive effective antiviral immunity.
Experimental Procedures
Mice
C57BL/6N and B6-CD45.1 mice were purchased from Charles River Laboratory. Rosa26creERT2 (Ventura et al., 2007), Ighg1cre (Casola et al., 2006), Aicdacre (Robbiani et al., 2008), Igh−/− (µMT) (Kitamura et al., 1991), and Il21r−/− mice (Frohlich et al., 2007) were from JAX. Tfap4f (Chou et al., 2014), Tfap4− (Egawa and Littman, 2011), Mycf (de Alboran et al., 2001), Cd79aicre (Hobeika et al., 2006) and MycGFP (Huang, 2008) alleles have been described. A Tfap4mCherry allele was generated by inserting a sequence encoding the mCherry fluorescence protein to the carboxyl terminus of AP4 (Figures S1B and S1C). NotI linearized plasmid DNA was electroporated into JM8.N4 ES cells (KOMP Repository), and G418/Gancyclovir double-resistant clones were screened for homologous recombination by Southern blotting. Correctly targeted clones were injected into B6(Cg)-Tyrc-2J blastocysts, and chimeric males were crossed to EIIa-cre females (Lakso et al., 1996) (Jackson Laboratory) to obtain germ-line transmission and to delete the selection cassette. To generate bone marrow chimeras, B6-CD45.1 mice were lethally irradiated (10.5 Gy) and reconstituted with donor bone marrow cells for at least 8 weeks before experiments. All mice were maintained in the C57BL/6 background, were housed in a specific pathogen-free facility at Washington University in St. Louis, and were analyzed at 6–14 weeks of age unless stated otherwise. Both sexes were included without randomization or blinding. All experiments were performed according to a protocol approved by Washington University’s Animal Studies Committee.
LCMV Infection
Mice were infected with 2 × 106 plaque-forming units (PFU) of LCMV-c13 by intravenous injection. For the quantification of serum viral load, RNA was extracted from 10 µl of serum from LCMV-infected mice using Trizol (Life Technologies). 1 µl of ERCC RNA Spike-In Control Mixes, at a concentration of 1:10,000, was added to the lysate before RNA extraction and reverse transcription. The amounts of the LCMV NP transcript relative to that of ‘spiked-in’ RNA were determined by real-time quantitative RT-PCR as previously described (Chou et al., 2014; McCausland and Crotty, 2008). In vitro neutralization assay was performed according to a published protocol with some modifications (Seiler et al., 1998). Vero cells were plated at 5 × 104 cells per well in a flat-bottom 96-well plate one day before infection. Heat-inactivated (one hour at 55 °C) sera from LCMV-infected and control mice were serially diluted with media and incubated with an equal volume of viral supernatant containing 300 PFU of LCMV clone 13 at 37 °C for 90 min. Infectivity of the mixtures was determined by intracellular staining for LCMV NP of Vero cells that were incubated with the mixture for 48 hours (Korns Johnson and Homann, 2012). For each dilution of serum samples, the percentage of LCMV NP+ Vero cells treated with LCMV-immune serum was normalized to that treated with control serum. A neutralization curve and an EC50 value were calculated using the non-linear fit equation “log (inhibitor) vs. normalized response -- variable slope” in Prism 6 software (Graphpad).
Immunization protocols and treatments
Mice were immunized with either 8–10 × 108 SRBC (Lampire), 100 µg of NP-CGG (Biosearch) precipitated in alum, or 100 µg of NP-Ficoll (Biosearch) in PBS by intraperitoneal injection. NP-and NIP-APC were prepared by conjugating allophycocyanin (Sigma-Aldrich) with NP or NIP ester (Biosearch) as previously described (McHeyzer-Williams and McHeyzer-Williams, 2004).
In vitro B cell culture
CD40L-expressing feeders were generated by infecting NIH-3T3 cells with a retrovirus expressing CD40L and sort-purified based on surface CD40L expression. The feeders were seeded in RPMI-1640 supplemented with 10% FBS at a density of 4 × 106 cells per 6-, or 12-well plate, or 100mm dish one day before adding splenic B cells purified using anti-CD19-microbeads (Miltenyi). B cells were removed from feeders by gentle shaking and pipetting. For liquid culture of B cells, CD40L-activated B cells were incubated in medium containing IL-4 (eBioscience), IL-21 (Peprotech), or IFN-γ (Peprotech) for additional 16–18 hours. To measure protein translation, B cells were treated with 10 µg/ml of puromycin (Sigma) for 10 min at 37°C before lysis. CD40L- and BAFF-expressing feeders (Nojima et al., 2011) were used for culturing of GC B cells ex vivo.
Statistical analysis
P values were calculated with an unpaired two-tailed Student’s t-test for two-group comparisons and by one-way ANOVA for multi-group comparisons with the Tukey post-hoc test using Prism 6 software. For the comparison of cumulative frequencies between two datasets and the frequency of W33L+ clones between two groups, the Kolmogorov-Smirnov test and χ2 test with Yates correction were used, respectively. P values of <0.05 were considered significant. N.S., not significant; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Supplementary Material
Acknowledgments
We thank B.P. Sleckman for c-MYC-GFP mice, F.W. Alt for MycF mice, E. Oltz for Cd79aicre/+mice, J.M. White for ES cell microinjection, S. Hsiung, Y. Wang, R. Wong and S. Faragasan for technical support, C-S. Hsieh, K.M. Murphy and E. Oltz for discussion. Supported by the Lucille P. Markey Pathway Program (C.C.), the National Institutes of Health (P30 AR048335 to the Rheumatic Diseases Core Center; R01 AI097244 and R56 AI114593 to T.E.; R01 AI099108 to D.B.; UL1 TR000448 to the Washington University Institute of Clinical and Translational Sciences; P30 CA91842 to the Siteman Cancer Center), and the Edward Mallinckrodt Jr. Foundation (T.E.). D.B. is a New York Stem Cell Foundation-Robertson Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Number
ChIPseq and RNAseq data are available at the NCBI Gene Expression Omnibus under an accession number GSE80669.
Author Contributions
C.C. and T.E. designed experiments. C.C., D.J.V., E.T., and M.H. performed experiments. M. Ce. and M.Co provided critical reagents, C.C., D.B. and T.E. analyzed data. T.E. wrote manuscript with editorial comments from the coauthors.
The authors declare no competing financial conflict of interest related to this study.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Allen CD, Ansel KM, Low C, Lesley R, Tamamura H, Fujii N, Cyster JG. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat Immunol. 2004;5:943–952. doi: 10.1038/ni1100. [DOI] [PubMed] [Google Scholar]
- Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen D, Cumano A, Dildrop R, Kocks C, Rajewsky K, Rajewsky N, Roes J, Sablitzky F, Siekevitz M. Timing, genetic requirements and functional consequences of somatic hypermutation during B-cell development. Immunol Rev. 1987;96:5–22. doi: 10.1111/j.1600-065x.1987.tb00506.x. [DOI] [PubMed] [Google Scholar]
- Allen D, Simon T, Sablitzky F, Rajewsky K, Cumano A. Antibody engineering for the analysis of affinity maturation of an anti-hapten response. The EMBO journal. 1988;7:1995–2001. doi: 10.1002/j.1460-2075.1988.tb03038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Dalla-Favera R. Germinal centres and B cell lymphomagenesis. Nat Rev Immunol. 2015;15:172–184. doi: 10.1038/nri3814. [DOI] [PubMed] [Google Scholar]
- Bergthaler A, Flatz L, Verschoor A, Hegazy AN, Holdener M, Fink K, Eschli B, Merkler D, Sommerstein R, Horvath E, et al. Impaired antibody response causes persistence of prototypic T cell-contained virus. PLoS Biol. 2009;7:e1000080. doi: 10.1371/journal.pbio.1000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado DP, Sasaki Y, Godinho SA, Pellerin A, Kochert K, Sleckman BP, de Alboran IM, Janz M, Rodig S, Rajewsky K. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol. 2012;13:1092–1100. doi: 10.1038/ni.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, Hao Z, Waisman A, Egert A, Ghitza D, Rajewsky K. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci U S A. 2006;103:7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castigli E, Alt FW, Davidson L, Bottaro A, Mizoguchi E, Bhan AK, Geha RS. CD40-deficient mice generated by recombination-activating gene-2-deficient blastocyst complementation. Proc Natl Acad Sci U S A. 1994;91:12135–12139. doi: 10.1073/pnas.91.25.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C, Pinto AK, Curtis JD, Persaud SP, Cella M, Lin CC, Edelson BT, Allen PM, Colonna M, Pearce EL, et al. c-Myc-induced transcription factor AP4 is required for host protection mediated by CD8+ T cells. Nat Immunol. 2014;15:884–893. doi: 10.1038/ni.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran LM, Tarlinton DM. Regulation of germinal center responses, memory B cells and plasma cell formation-an update. Current opinion in immunology. 2016;39:59–67. doi: 10.1016/j.coi.2015.12.008. [DOI] [PubMed] [Google Scholar]
- de Alboran IM, O’Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Victora GD, Ying CY, Phan RT, Saito M, Nussenzweig MC, Dalla-Favera R. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol. 2012;13:1083–1091. doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T, Littman DR. Transcription factor AP4 modulates reversible and epigenetic silencing of the Cd4 gene. Proc Natl Acad Sci U S A. 2011;108:14873–14878. doi: 10.1073/pnas.1112293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger R, Sims GP, Fairhurst AM, Robbins R, da Silva YS, Spolski R, Leonard WJ, Lipsky PE. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. J Immunol. 2005;175:7867–7879. doi: 10.4049/jimmunol.175.12.7867. [DOI] [PubMed] [Google Scholar]
- Frohlich A, Marsland BJ, Sonderegger I, Kurrer M, Hodge MR, Harris NL, Kopf M. IL-21 receptor signaling is integral to the development of Th2 effector responses in vivo. Blood. 2007;109:2023–2031. doi: 10.1182/blood-2006-05-021600. [DOI] [PubMed] [Google Scholar]
- Gitlin AD, Mayer CT, Oliveira TY, Shulman Z, Jones MJ, Koren A, Nussenzweig MC. HUMORAL IMMUNITY. T cell help controls the speed of the cell cycle in germinal center B cells. Science. 2015;349:643–646. doi: 10.1126/science.aac4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509:637–640. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halper-Stromberg A, Lu CL, Klein F, Horwitz JA, Bournazos S, Nogueira L, Eisenreich TR, Liu C, Gazumyan A, Schaefer U, et al. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell. 2014;158:989–999. doi: 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DL, Johnson GD, Khan M, MacLennan IC. Quantitative analysis of molecules which distinguish functional compartments within germinal centers. Eur J Immunol. 1993;23:997–1004. doi: 10.1002/eji.1830230502. [DOI] [PubMed] [Google Scholar]
- Harker JA, Lewis GM, Mack L, Zuniga EI. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 2011;334:825–829. doi: 10.1126/science.1208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, Reth M. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci U S A. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CYBAL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- Jolly CJ, Klix N, Neuberger MS. Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic acids research. 1997;25:1913–1919. doi: 10.1093/nar/25.10.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato L, Begum NA, Burroughs AM, Doi T, Kawai J, Daub CO, Kawaguchi T, Matsuda F, Hayashizaki Y, Honjo T. Nonimmunoglobulin target loci of activation-induced cytidine deaminase (AID) share unique features with immunoglobulin genes. Proc Natl Acad Sci U S A. 2012;109:2479–2484. doi: 10.1073/pnas.1120791109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe T, Naka T, Yoshida K, Tanaka T, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T, Kikutani H. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1:167–178. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Rajewsky K. Stepwise intraclonal maturation of antibody affinity through somatic hypermutation. Proc Natl Acad Sci U S A. 1988;85:8206–8210. doi: 10.1073/pnas.85.21.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korns Johnson D, Homann D. Accelerated and improved quantification of lymphocytic choriomeningitis virus (LCMV) titers by flow cytometry. PloS one. 2012;7:e37337. doi: 10.1371/journal.pone.0037337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka M, Schmidt AG, Nojima T, Feng F, Watanabe A, Kitamura D, Harrison SC, Kepler TB, Kelsoe G. Complex Antigens Drive Permissive Clonal Selection in Germinal Centers. Immunity. 2016;44:542–552. doi: 10.1016/j.immuni.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman MA, Beaton L, Yu D, Ramiscal RR, Srivastava M, Hogan JJ, Verma NK, Smyth MJ, Rigby RJ, Vinuesa CG. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J Exp Med. 2010;207:353–363. doi: 10.1084/jem.20091738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan IC. Germinal centers. Annual review of immunology. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCausland MM, Crotty S. Quantitative PCR technique for detecting lymphocytic choriomeningitis virus in vivo. Journal of virological methods. 2008;147:167–176. doi: 10.1016/j.jviromet.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHeyzer-Williams LJ, McHeyzer-Williams MG. Analysis of antigen-specific B-cell memory directly ex vivo. Methods Mol Biol. 2004;271:173–188. doi: 10.1385/1-59259-796-3:173. [DOI] [PubMed] [Google Scholar]
- Nathans D. Puromycin Inhibition of Protein Synthesis: Incorporation of Puromycin into Peptide Chains. Proc Natl Acad Sci U S A. 1964;51:585–592. doi: 10.1073/pnas.51.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Haniuda K, Moutai T, Matsudaira M, Mizokawa S, Shiratori I, Azuma T, Kitamura D. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun. 2011;2:465. doi: 10.1038/ncomms1475. [DOI] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Renshaw BR, Fanslow WC, 3rd, Armitage RJ, Campbell KA, Liggitt D, Wright B, Davison BL, Maliszewski CR. Humoral immune responses in CD40 ligand-deficient mice. J Exp Med. 1994;180:1889–1900. doi: 10.1084/jem.180.5.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, Nussenzweig MC. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander S, Calado DP, Srinivasan L, Kochert K, Zhang B, Rosolowski M, Rodig SJ, Holzmann K, Stilgenbauer S, Siebert R, et al. Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer cell. 2012;22:167–179. doi: 10.1016/j.ccr.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler P, Kalinke U, Rulicke T, Bucher EM, Bose C, Zinkernagel RM, Hengartner H. Enhanced virus clearance by early inducible lymphocytic choriomeningitis virus-neutralizing antibodies in immunoglobulin-transgenic mice. J Virol. 1998;72:2253–2258. doi: 10.1128/jvi.72.3.2253-2258.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih TA, Meffre E, Roederer M, Nussenzweig MC. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat Immunol. 2002;3:570–575. doi: 10.1038/ni803. [DOI] [PubMed] [Google Scholar]
- Shulman Z, Gitlin AD, Weinstein JS, Lainez B, Esplugues E, Flavell RA, Craft JE, Nussenzweig MC. Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science. 2014;345:1058–1062. doi: 10.1126/science.1257861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tas JM, Mesin L, Pasqual G, Targ S, Jacobsen JT, Mano YM, Chen CS, Weill JC, Reynaud CA, Browne EP, et al. Visualizing antibody affinity maturation in germinal centers. Science. 2016;351:1048–1054. doi: 10.1126/science.aad3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla-Favera R, Nussenzweig MC. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 2012;120:2240–2248. doi: 10.1182/blood-2012-03-415380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annual review of immunology. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Foy TM, Laman JD, Elliott EA, Dunn JJ, Waldschmidt TJ, Elsemore J, Noelle RJ, Flavell RA. Mice deficient for the CD40 ligand. Immunity. 1994;1:423–431. doi: 10.1016/1074-7613(94)90073-6. [DOI] [PubMed] [Google Scholar]
- Zotos D, Coquet JM, Zhang Y, Light A, D’Costa K, Kallies A, Corcoran LM, Godfrey DI, Toellner KM, Smyth MJ, et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J Exp Med. 2010;207:365–378. doi: 10.1084/jem.20091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.