Abstract
Generating sequence data of a defined community composed of organisms with complete reference genomes is indispensable for the benchmarking of new genome sequence analysis methods, including assembly and binning tools. Moreover the validation of new sequencing library protocols and platforms to assess critical components such as sequencing errors and biases relies on such datasets. We here report the next generation metagenomic sequence data of a defined mock community (Mock Bacteria ARchaea Community; MBARC-26), composed of 23 bacterial and 3 archaeal strains with finished genomes. These strains span 10 phyla and 14 classes, a range of GC contents, genome sizes, repeat content and encompass a diverse abundance profile. Short read Illumina and long-read PacBio SMRT sequences of this mock community are described. These data represent a valuable resource for the scientific community, enabling extensive benchmarking and comparative evaluation of bioinformatics tools without the need to simulate data. As such, these data can aid in improving our current sequence data analysis toolkit and spur interest in the development of new tools.
Subject terms: Next-generation sequencing, Microbial communities, DNA sequencing, Metagenomics
Background & Summary
By definition, benchmark studies aim to provide standards that can be used to evaluate the performance of a process. The field of nucleic acid sequencing and sequence data processing has witnessed immense developments towards optimizing the balance of sequencing cost, precision and overall applicability to real-world questions. This progress has routinely relied on experimental setups of defined nature to critically rate novel approaches. In recent years, mock communities have been assisting in a variety of laboratory and computational test experiments, which resulted in quantitative and qualitative evaluation of corresponding studied methods. For example, mock communities were generated for the comparison of DNA extraction methods1–3, for the development of a dual-index sequencing and curation pipeline for Illumina MiSeq generated amplicon sequence data4–8, and to evaluate the Ion Torrent sequencing platform for gene-targeted studies9,10. Similarly, Pabinger et al.11–13 used a mock community to benchmark MEMOSys, a web-based platform for metabolic models. The jumpstart consortium human microbiome project (HMP) data generation working group established a standardized protocol for ensuring high throughput consistency of 16S rRNA gene amplification and sequencing protocols by implementing a synthetic mock community of 21 known organisms, before finalizing their HMP 16S 454 protocol14–16. The HMP DNA and sequence data resources have not only enabled comprehensive characterization of the human microbiota, e.g.17–19, but also the use and development of a variety of advanced analysis tools. For example, chimera screening tools UCHIME and Chimera Slayer1,3, the OTU construction pipeline UPARSE4,6–8, and fine-tuned workflows for amplicon gene studies9 used HMP data generated from mock communities.
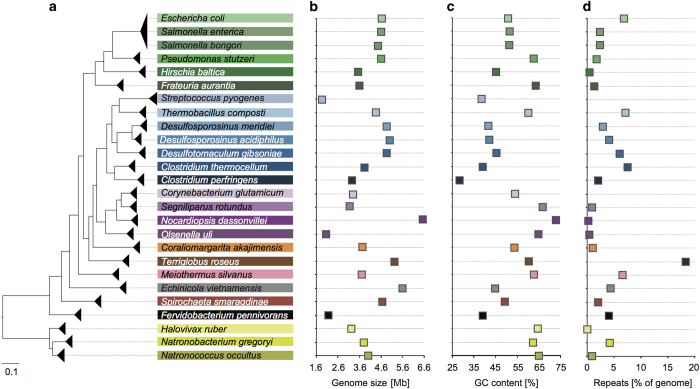
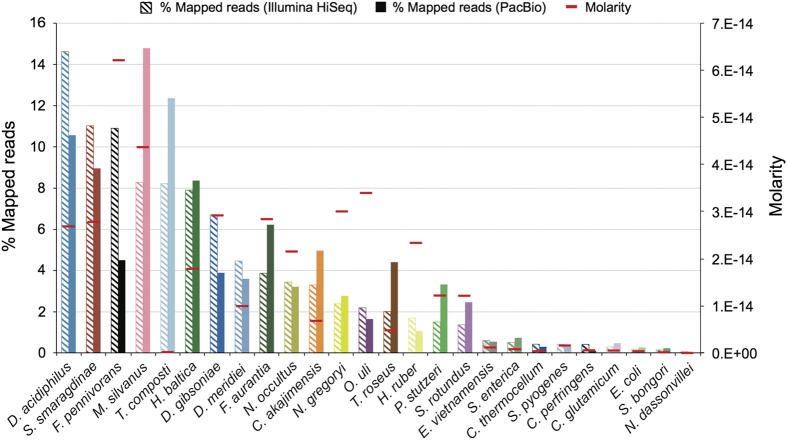
In contrast to the HMP mock, the synthetic community described here, MBARC-26 (Mock Bacteria ARchaea Community), is composed of organisms isolated from heterogeneous soil and aquatic environments as well as derived from human, bovine and frog (Table 1). MBARC-26 consists of 23 bacterial and 3 archaeal strains, belonging to the phyla Acidobacteria, Actinobacteria, Bacteroidetes, Deinococcus-Thermus, Firmicutes, (Alpha- and Gamma-)Proteobacteria, Spirochaetes, Thermotogae, Verrucomicrobia and Euryarchaeota. Genome sizes span 1.8–6.5 Mbp, GC contents vary between 28.4–72.7%, and repeat content ranges from 0–18.3% (Fig. 1, Table 1). All genomes are available as finished genome sequences in GenBank (Table 1). MBARC-26 DNA was shotgun sequenced on Illumina HiSeq 2000 and PacBio RSII sequencing platforms (Table 2). We provide detailed descriptions of organism characteristics (Table 1), sample processing, including DNA extraction and quantification, sequencing library creation, and sequencing procedures (Table 2). Data statistics encompass sequencing throughput characteristics (Table 2), community structure according to read mapping to reference genomes and according to molarity (Fig. 2, Supplementary Table 1, Supplementary Fig. 1), quantitative comparison between Illumina and PacBio datasets (Fig. 3a, Table 1, Supplementary Figs 2 and 3), % genome coverage and fold coverage by sequencing platform (Fig. 3b), and GC content analysis (Supplementary Fig. 3). Due to inherent sequencing technology differences11,13, these two datasets are characterized by platform-, run mode-, and chemistry-specific read length, data throughput, GC and amplification bias, and error rate. We point out that our quantitative results are directly correlated to the respective sample preparation and sequencing methods used, as these have been shown to critically affect community representation14,20.
Table 1. Genome statistics of each mock community member.
| Organism | Isolation source | GenBank Accession ID | Genome size [bp] | GC [%] | % repeats | # of scaffolds | # of 16S copies |
|---|---|---|---|---|---|---|---|
| Genome size includes chromosomes and plasmids. All genomes are available as finished sequences. Phylum associations for each strain are abbreviated as follows: AD—Acidobacteria, AT—Actinobacteria, B—Bacteroidetes, D—Deinococcus-Thermus, E—Euryarchaeota, F—Firmicutes, P—Proteobacteria, S—Spirochaetes, T—Thermotogae, V—Verrucomicrobia. Isolation sources were obtained from literature on respective strains, where available. GC content is based on genome size. Genomes without NCBI repeat region annotation are denoted with an *. | |||||||
| Terriglobus roseus DSM 18391 (AD) | Soil | NC_018014 | 5227858 | 60.3 | 18.3 | 1 | 2 |
| Corynebacterium glutamicum ATCC 13032 (AT) | Sewage | NC_003450 | 3309401 | 53.8 | NA* | 1 | 6 |
| Nocardiopsis dassonvillei DSM 43111 (AT) | Soil | NC_014211 | 6543312 | 72.7 | 0.2 | 2 | 5 |
| Olsenella uli DSM 7084 (AT) | Human gingival crevice | NC_014363 | 2051896 | 64.7 | 0.46 | 1 | 1 |
| Segniliparus rotundus DSM 44985 (AT) | Human sputum | NC_014168 | 3157527 | 66.8 | 0.92 | 1 | 1 |
| Echinicola vietnamensis DSM 17526 (B) | Seawater collected in a mussel farm | NC_019904 | 5608040 | 44.8 | 4.34 | 1 | 4 |
| Meiothermus Silvanus DSM 9946 (D) | Hot spring (50 °C) | NC_014212 | 3721669 | 62.7 | 6.54 | 3 | 2 |
| Clostridium perfringens ATCC 13124 (F) | Bovine | NC_008261 | 3256683 | 28.4 | 2.02 | 1 | 20 |
| Clostridium thermocellum ATCC 27405 (F) | Various | NC_009012 | 3843301 | 39 | 7.51 | 1 | 4 |
| Desulfosporosinus acidiphilus SJ4 DSM 22704 (F) | Pond sediment | NC_018068 | 4991181 | 42.1 | 4.08 | 3 | 9 |
| Desulfosporosinus meridiei DSM 13257 (F) | Aquifer groundwater | NC_018515 | 4873567 | 41.8 | 2.89 | 1 | 11 |
| Desulfotomaculum gibsoniae DSM 7213 (F) | Freshwater mud | NC_021184 | 4855529 | 45.5 | 5.99 | 1 | 8 |
| Streptococcus pyogenes M1 GAS SF370 (F) | Infected wound | NC_002737 | 1852441 | 38.5 | NA* | 1 | 6 |
| Thermobacillus composti KWC4, DSM 18247 (F) | Composting reactor | NC_019897 | 4355525 | 60.1 | 7.14 | 2 | 5 |
| Escherichia coli K-12, MG1655 (P) | Human stool | NC_000913 | 4639675 | 50.8 | 6.7 | 1 | 7 |
| Frateuria aurantia DSM 6220 (P) | Lilium auratium | NC_017033 | 3603458 | 63.4 | 1.32 | 1 | 4 |
| Hirschia baltica ATCC 49814 (P) | Brackish water | NC_012982 | 3540114 | 45.2 | 0.45 | 2 | 2 |
| Pseudomonas stutzeri RCH2 (P) | Cr-contaminated aquifer | NC_019936 | 4600489 | 62.5 | 1.83 | 4 | 4 |
| Salmonella bongori NCTC 12419 (P) | African frog | NC_015761 | 4460105 | 51.3 | 2.36 | 1 | 7 |
| Salmonella enterica subsp. arizonae serovar RSK2980 (P) | Animal tissue | NC_010067 | 4600800 | 51.4 | 2.42 | 1 | 7 |
| Spirochaeta smaragdinae DSM 11293 (S) | Oil field | NC_014364 | 4653970 | 49 | 2.01 | 1 | 2 |
| Fervidobacterium pennivorans DSM 9078 (T) | Hot mud of spa | NC_017095 | 2166381 | 39 | 4.04 | 1 | 2 |
| Coraliomargarita akajimensis DSM 45221 (V) | Seawater | NC_014008 | 3750771 | 53.6 | 1.07 | 1 | 2 |
| Halovivax ruber XH-70 (E) | Saline lake | CP003050.1 | 3223876 | 64.3 | NA* | 1 | 2 |
| Natronobacterium gregoryi SP2 (E) | Solar saltworks | NC_019792.1 | 3788356 | 62.2 | 4.22 | 1 | 3 |
| Natronococcus occultus DSM 3396 (E) | Lake | NC_019974.1 | 4314118 | 64.7 | 0.91 | 3 | 4 |
Figure 1. Characteristics of MBARC-26 community.
Community members display diversity in phylogenetic distribution and relatedness (a), genome size (b), GC content (c), and repeat content normalized by genome size (d). Shades of the same color in (a) denote the same phylum association: Green—Proteobacteria, blue—Actinobacteria, purple—Firmicutes, yellow—Euryarchaeota.
Table 2. Sequence Statistics by sequencing platform.
| Platform | Illumina | PacBio |
|---|---|---|
| Model | HiSeq-HO 2000 | RS II |
| Library chemistry | TruSeq paired-end cluster kit v3 | SMRTbell template preparation kit |
| Sequencing chemistry | TruSEq SBS sequencing kit 200 cycles v3 | P4C2 |
| Run mode | 2x150 | 1x120 min |
| # of raw reads | 355,875,608 | 300,584 |
| # of filtered reads | 347,963,988 | 53,654 |
| Average insert size [bp] | 219±43 | 1,041±576 |
| Average quality score (filtered reads) | Read 1: 33.47, Read 2: 32.04 | 0.976 |
Figure 2. MBARC-26 community composition and relative abundance distribution, as based on Illumina and PacBio read mapping and mean DNA molarity.
Mock community members are grouped and arranged in order of % mapped sequences (Illumina). The observed discrepancy between molarity and % mapped PacBio and Illumina sequences in T. composti is likely due to contamination as T. composti was previously found to occur as laboratory contaminant in various shotgun metagenome datasets (unpublished data). The smaller discrepancies are expected due to DNA quantification spreads and platform biases. Colors denote phylum association as defined in Fig. 1.
Figure 3. Quantitative comparison of MBARC-26 Illumina and PacBio shotgun sequence datasets.
(a) Community representation according to % mapped sequences for each mock community member in the PacBio (x-axis) and Illumina (y-axis) shotgun sequence datasets. (b) Percent chromosome coverage and fold coverage of each mock community genome by sequencing platform using unassembled sequences. Colors denote phylum association as defined in Fig. 1.
To date, several studies already utilized MBARC-26 and took advantage of its purposefully selected characteristics. Availability of complete reference genomes and relative abundance spread of individual constituents enabled determining lower limits of various metagenome library preparation protocols14. MBARC-26 was also used to develop a new full-length 16S rRNA gene amplicon sequencing protocol called PhyloTags17 and allowed for quantitative comparison of amplicon to shotgun sequence data and bias evaluation associated with GC content. Using the MBARC-26 Illumina metagenome dataset and corresponding single-cell sequence data Bremges et al. developed MeCorS, a metagenome-enabled single-cell read correction tool21. To further encourage the use of this mock community, we report the release of molarity and shotgun sequence datasets of MBARC-26.
Perpetual community efforts to develop improved DNA sequence analysis software with various applications for shotgun sequence data requires standardized and well-characterized data for benchmark experiments. MBARC-26 was validated according to the specific sample processing tools using a variety of commonly used quality control methods, is accompanied by data statistics, and meant to enable method development and evaluation while enabling reproducibility of research findings.
Methods
These methods are expanded from descriptions in our previous work17.
Cultivation and DNA extraction
DNA from Escherichia coli, Salmonella bongori, Salmonella enterica, Clostridium perfringens, Clostridium thermocellum and Streptococcus pyogenes was purchased from the American Type- Culture Collection (ATCC, Manassas, VA, USA). DNA from Fervidobacterium pennivorans, Thermobacillus composti and Corynebacterium glutamicum was extracted using phenol–chloroform extraction, as described in (ref. 22). DNA from Desulfosporosinus acidiphilus, Desulfosporosinus meridiei, Desulfotomaculum gibsoniae, Echinicola vietnamensis, Frateuria aurantia, Natronococcus occultus, Olsenella uli and Terriglobus roseus was isolated using the Jetflex Genomic DNA Purification Kit (Genomed GmbH, Loehne, Germany). DNA from Hirschia baltica was extracted using the Blood and Cell Culture DNA Maxi Kit (Qiagen, Valencia, CA, USA). DNA from Meiothermus silvanus, Nocardiopsis dassonvillei and Segniliparus rotundus was extracted using the Qiagen Genomic 500 DNA Kit (Qiagen, Hilden, Germany). DNA from Pseudomonas stutzeri was isolated using the Wizard Genomic DNA Purification Kit (Promega Corp., Madison, WI, USA). DNA from Coraliomargarita akajimensis, Halovivax ruber, Natronobacterium gregoryi and Spirochaeta smaragdinae was extracted using the Masterpure Gram-Positive DNA Purification Kit (Epicentre, Madison, WI, USA). All DNA extracts were quantified using the PicoGreen assay and the Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA, USA) (Supplementary Fig. 1). Each sample was quantified in quadruplicate. Samples were pooled at varying ratios to generate the mock community (Fig. 2, Supplementary Table 1).
Library creation and sequencing
For Illumina library creation, 100 ng of genomic DNA of MBARC-26, brought up to a total of 100 μl using TE, was sheared to 300 bp using the Covaris LE200 (Covaris, Inc., Woburn, MA, USA) and size-selected using AMPure XP beads (Beckman Coulter, Brea, CA, USA): 60 μl of beads were added to 100 μl of sample. The sample was then incubated at room temperature (RT) for 5 min. Beads were pelleted using a magnetic particle concentrator (MPC) (Thermo Fisher Scientific, South San Francisco, CA, USA) until liquid was clear. The supernatant was removed and transferred to a new tube. 30 μl of AMPure XP beads were then added for the second bead size selection. The mixture was pulse vortexed, quickly spun and incubated at RT for 5 min. Beads were pelleted using a magnetic particle concentrator (MPC) (Thermo Fisher Scientific, South San Francisco, CA, USA) until liquid was clear. The supernatant was then discarded without disturbing the beads and 200 μl of freshly prepared 75% ethanol (EtOH) was added, followed by a 30 s incubation to wash the beads. EtOH was discarded before the wash step with EtOH was repeated for a total of two washes. Afterwards, the sample was placed on a thermocycler (Eppendorf, Hamburg, Germany) with the lid open and incubated at 37 °C until the beads were dry and residual EtOH had evaporated. The beads were re-suspended in 53 μl of EB buffer (Qiagen, Redwood City, CA, USA), vortexed, quickly spun and incubated at RT for 1 min. Beads were pelleted using an MPC until liquid was clear (Thermo Fisher Scientific, South San Francisco, CA, USA). 50 μl of supernatant was then transferred to a new tube. The DNA fragment size was assessed using the Agilent Bioanalyzer 2100 High Sensitivity Kit (Agilent Technologies, Palo Alto, CA, USA) before proceeding to end repair.
The fragments were treated with the Kapa Library Preparation Kit ORIGIN (Kapa Biosystems, Wilmington, MA, USA) for the following steps: For end-repair 26 μl MilliQ water, 9 μl 10X End Repair Buffer, and 5 μl End Repair Enzyme were combined in a 1.5 ml tube. The cocktail was vortexed and quickly spun, then stored on ice. 40 μl of End Repair cocktail was added to the 50 μl DNA sample. The mixture was vortexed and quickly spun, before incubation at 30 °C for 30 min in a thermocycler (Eppendorf, Hamburg, Germany). After incubation, 126 μl of AMPure XP beads (Beckman Coulter, Brea, CA, USA) were added to 90 μl of End Repair sample, pulse vortexed, quickly spun, and incubated at RT for 5 min. Beads were pelleted using a MPC until liquid was clear. The supernatant was then discarded without disturbing the beads. The beads were washed twice with 200 μl of freshly prepared 75% EtOH with an incubation time of 30 s. After washing, the sample was incubated at 37 °C in a thermocycler with the lid open until residual EtOH had evaporated. For DNA elution, 17.5 μl of EB buffer was added. The sample was vortexed, quickly spun, and incubated at RT for 1 min, before beads were pelleted on a MPC. 15 μl of supernatant was then transferred to a new tube.
For A-tailing, 9 μl of MilliQ water, 3 μl of 10X A-Tailing Buffer and 3 μl of A-Tailing Enzyme were combined in this order in a 1.5 ml tube. The cocktail was vortexed and quickly spun. 15 μl of the A-Tailing cocktail was added to the 15 μl sample. The mixture was vortexed and quickly spun. The samples were then incubated in a thermocycler at 30 °C for 30 min, followed by 5 min at 70 °C.
Adaptor ligation was immediately performed thereafter: 9 μl of 5X Ligation Buffer and 5 μl of ligase were combined in a 1.5 ml tube, vortexed and spun. The mixture was pulse vortexed and quickly spun. 14 μl of adaptor ligation cocktail were added to the 30 μl sample, before 1 μl of 18 μM adaptor was added to the ligation mixture for a final concentration of 400 nM. The mixture was incubated in a thermocycler at 20 °C for 15 min.
After adaptor ligation, 5 μl of EB Buffer was added to 45 μl of adaptor-ligated sample. The sample was size-selected and washed twice with 45 μl of AMPure XP beads as described previously. After the first clean-up step, the sample was eluted with 52 μl of EB Buffer and 45 μl of supernatant was transferred to a clean tube. After the second clean-up step, the sample was eluted with 25 μl of EB Buffer. 23 μl of supernatant was transferred to a clean tube. The sample was quality-controlled and quantified using an Agilent Bioanalyzer 2100 High Sensitivity Kit.
The prepared Illumina library was further quantified by using the Kapa Biosystems next-generation sequencing library qPCR kit according to the manufacturer’s guidelines (Kapa Biosystems, Wilmington, MA, USA). The amplification products were run on a Roche LightCycler 480 real-time PCR instrument for quantification (Roche Holding AG, Basel, Switzerland). The quantified library was then prepared for sequencing on the Illumina HiSeq sequencing platform (Illumina, Inc., San Diego, CA, USA). First, the TruSeq paired-end cluster kit, v3, and Illumina’s cBot instrument were used to generate a clustered flowcell for sequencing (Illumina, Inc., San Diego, CA, USA). Sequencing of the flowcell was performed on the Illumina HiSeq2000 sequencer using a TruSeq SBS sequencing kit 200 cycles, v3, following a 2x150 indexed run recipe (Illumina, Inc., San Diego, CA, USA) (Table 2). This resulted in 355,875,608 raw reads.
For PacBio library creation, 5 μg of gDNA was sheared using a Covaris LE220 to generate 2 kb fragments (Covaris, Inc., Woburn, MA, USA). The sheared DNA fragments were then prepared according to the SMRTbell template preparation kit guidelines (Pacific Biosciences, Menlo Park, CA, USA). Briefly, DNA fragments were treated with DNA damage repair mix, end-repaired, and 5’ phosphorylated. PacBio hairpin adapters were then ligated to the fragments to create SMRTbell template for sequencing. The SMRTbell templates were purified using exonuclease treatments and size-selected using AMPure PB beads (Pacific Biosciences, Menlo Park, CA, USA) (Table 2).
Sequencing primers were annealed and v. P4 sequencing polymerase was bound to the SMRTbell templates. The prepared SMRTbell template libraries were then sequenced on a Pacific Biosciences RSII sequencer using v. C2 chemistry and 1x120 min sequencing movie run times (Pacific Biosciences, Menlo Park, CA, USA). This resulted in 300,584 raw reads (Table 2).
Sequence QC
Illumina shotgun reads were filtered using BBDuk (filterk=27, trimk=27; http://jgi.doe.gov/data-and-tools/bb-tools/) to remove Illumina adapters, known Illumina artifacts, phiX, and to quality-trim both ends to Q12. Resulting reads containing more than one ‘N’, or with quality scores (before trimming) averaging less than 8 over the read, or length under 40 bp after trimming, were discarded. Remaining reads were mapped to a masked version of human HG19, dog, cat, and mouse with BBMap (http://jgi.doe.gov/data-and-tools/bb-tools/), discarding all hits exceeding 93% identity. This resulted in 347,963,988 filtered reads with average insert size of 219±43 bp.
Quality filtering and error correction of PacBio sequences was performed using the RS_ReadsOfInsert protocol v. 2.3.0 in SMRT Portal (minimum subread length: 50 bp; minimum read quality: 75%). This resulted in 53,654 quality-filtered subreads with average read length of 1,041±576 bp.
Mapping, repeat regions, and phylogenetic tree construction
High quality Illumina and PacBio sequences were mapped to their bacterial and archaeal reference genomes using BBMap with parameters bbmap.sh, ambig=toss (Illumina) and mapPacBio.sh, ambig=toss (PacBio), respectively. Numbers of mapped sequences were normalized to the respective whole genome and chromosome lengths of reference organisms (Supplementary Table 1). Unmapped sequences amounted to 2,105 (3.92%) and 3,777 (7.04%) PacBio sequences, when mapped against genome and chromosome references, respectively. In the Illumina dataset, 8,981,844 (2.58%) and 18,088,260 (5.20%) Illumina sequences remained unmapped, when mapped against genome and chromosome reference, respectively. Repeat regions reported here were retrieved from NCBI GenBank23 on May 16, 2016. They include tandem, inverted, flanking, terminal, direct and dispersed repeat types. For phylogenetic tree construction, full-length 16S rRNA gene sequences were aligned using the SINA aligner24 including 10 neighbors at 95% minimum identity for classification against the SILVA, RDP, greengenes, LTP, and EMBL databases25. The alignment was masked using the SILVA-compatible 1,349 Lane mask26. Tree construction was performed using FastTree27.
Data Records
Filtered shotgun sequences generated on the Illumina and PacBio platforms are publically available through NCBI (Data Citation 1 and Data Citation 2).
Technical Validation
To assess the quality of genomic DNA received, we used the PicoGreen assay and the Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA, USA). Each sample was quantified in quadruplicate. Samples were pooled at varying ratios to generate the mock community (Supplementary Fig. 1).
Both shotgun sequence datasets were screened for adapters, artifacts, according to quality scores (Illumina: Q12; PacBio: 75%), number of ‘N’, read length (Illumina: min 40 bp, PacBio: min 50 bp), and contaminant sequences related to human, dog, cat, and mouse.
Additional information
How to cite: Singer, E. et al. Next generation sequencing data of a defined microbial mock community. Sci. Data 3:160081 doi: 10.1038/sdata.2016.81 (2016).
Supplementary Material
Acknowledgments
This work was conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, and was supported under Contract No. DE-AC02-05CH11231.
Footnotes
The authors declare no competing financial interests.
Data Citations
- 2016. NCBI Sequence Read Archive. SRX1836716
- 2016. NCBI Sequence Read Archive. SRX1836715
References
- Edgar R. C., Haas B. J., Clemente J. C., Quince C. & Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D. et al. Comparison of DNA Extraction Methods for Microbial Community Profiling with an Application to Pediatric Bronchoalveolar Lavage Samples. PLoS ONE 7, e34605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J. et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Research 21, 494–504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. S., Baker B. J., Thomas B. C., Singer S. W. & Banfield J. F. EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biology 12, R44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K. & Schloss P. D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. et al. A large-scale benchmark study of existing algorithms for taxonomy-independent microbial community analysis. Briefings in Bioinformatics 13, 107–121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Meth. 10, 996–998 (2013). [DOI] [PubMed] [Google Scholar]
- Langille M. G. I. et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences Nature Publishing Group 31, 814–821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. C., Morrison H. G., Benjamino J., Grim S. L. & Graf J. Analysis, Optimization and Verification of Illumina-Generated 16S rRNA Gene Amplicon Surveys. PLoS ONE 9, e94249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. et al. Evaluation of the Ion Torrent Personal Genome Machine for Gene-Targeted Studies Using Amplicons of the Nitrogenase Gene nifH . Appl. Environ. Microbiol. 81, 4536–4545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Carneiro M. O. & Schatz M. C. The advantages of SMRT sequencing. Genome Biology 14, 405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabinger S. & Trajanoski Z. MEMOSys: Platform for Genome-Scale Metabolic Models. Encyclopedia of Metagenomics 361–366 (2015). [Google Scholar]
- Minoche A. E., Dohm J. C. & Himmelbauer H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biology 12, R112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers R. M. et al. Impact of library preparation protocols and template quantity on the metagenomic reconstruction of a mock microbial community. BMC Genomics 16, 1–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, T. H. M. P. A framework for human microbiome research. Nature 486, 215–221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumpstart Consortium Human Microbiome Project Data Generation Working Group. Evaluation of 16S rDNA-Based Community Profiling for Human Microbiome Research. PLoS ONE 7, e39315 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer E. et al. High-resolution phylogenetic microbial community profiling. ISME J 10, 2020–2032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J. et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. PNAS 107, 7503–7508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, T. H. M. P. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. B. et al. Library preparation methodology can influence genomic and functional predictions in human microbiome research. PNAS 112, 14024–14029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremges A. et al. MeCorS: Metagenome-enabled error correction of single cell sequencing reads. Bioinformatics 2199–2201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore D. D. & Dowhan D. Preparation and Analysis of DNA. Current Protocols in Molecular Biology (1995). [Google Scholar]
- Benson D. A. et al. GenBank. Nucleic Acids Research 41, D36–D42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E., Peplies J. & Glockner F. O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research 41, D590–D596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J. in Nucleic acid techniques in bacterial systematics (eds. Stackebrandt E. & Goodfellow M. ) 115–176 Wiley. [Google Scholar]
- Price M. N., Dehal P. S. & Arkin A. P. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2016. NCBI Sequence Read Archive. SRX1836716
- 2016. NCBI Sequence Read Archive. SRX1836715