ABSTRACT
Bacillus subtilis possesses different enzymes for the utilization of plant cell wall polysaccharides. This includes a gene cluster containing galactan degradation genes (ganA and ganB), two transporter component genes (ganQ and ganP), and the sugar-binding lipoprotein-encoding gene ganS (previously known as cycB). These genes form an operon that is regulated by GanR. The degradation of galactan by B. subtilis begins with the activity of extracellular GanB. GanB is an endo-β-1,4-galactanase and is a member of glycoside hydrolase (GH) family 53. This enzyme was active on high-molecular-weight arabinose-free galactan and mainly produced galactotetraose as well as galactotriose and galactobiose. These galacto-oligosaccharides may enter the cell via the GanQP transmembrane proteins of the galactan ABC transporter. The specificity of the galactan ABC transporter depends on the sugar-binding lipoprotein, GanS. Purified GanS was shown to bind galactotetraose and galactotriose using thermal shift assay. The energy for this transport is provided by MsmX, an ATP-binding protein. The transported galacto-oligosaccharides are further degraded by GanA. GanA is a β-galactosidase that belongs to GH family 42. The GanA enzyme was able to hydrolyze short-chain β-1,4-galacto-oligosaccharides as well as synthetic β-galactopyranosides into galactose. Thermal shift assay as well as electrophoretic mobility shift assay demonstrated that galactobiose is the inducer of the galactan operon regulated by GanR. DNase I footprinting revealed that the GanR protein binds to an operator overlapping the −35 box of the σA-type promoter of Pgan, which is located upstream of ganS.
IMPORTANCE Bacillus subtilis is a Gram-positive soil bacterium that utilizes different types of carbohydrates, such as pectin, as carbon sources. So far, most of the pectin degradation systems and enzymes have been thoroughly studied in B. subtilis. Nevertheless, the B. subtilis utilization system of galactan, which is found as the side chain of the rhamnogalacturonan type I complex in pectin, has remained partially studied. Here, we investigated the galactan utilization system consisting of the ganSPQAB operon and its regulator ganR. This study improves our knowledge of the carbohydrate degradation systems of B. subtilis, especially the pectin degradation systems. Moreover, the galactan-degrading enzymes may be exploited for the production of galacto-oligosaccharides, which are used as prebiotic substances in the food industry.
INTRODUCTION
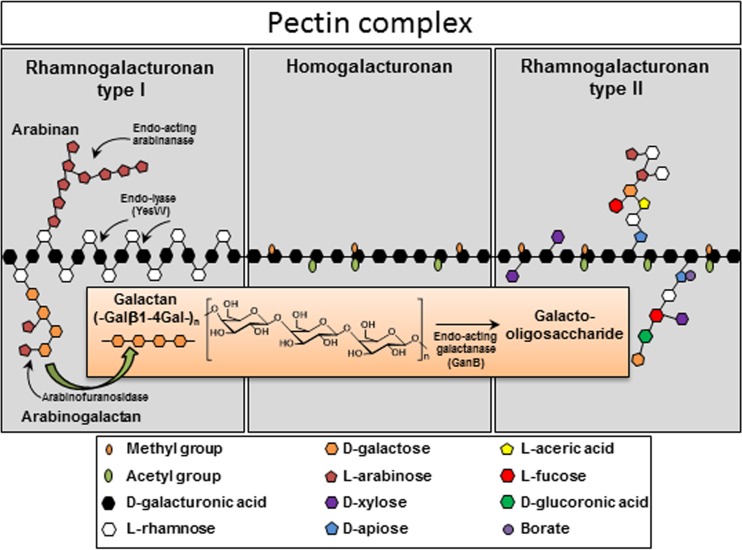
Soil microorganisms are usually able to degrade a variety of polysaccharides and carbohydrates found in soil, including the remains of plants and especially the plant cell wall. The primary cell wall of plants is composed of different polysaccharide components, including cellulose, hemicellulose, and pectin, while the secondary cell walls are rigidified by lignin (1, 2). Among them, pectin is a family of galacturonic acid-rich polysaccharides that consists of the following three major pectic polysaccharide types: a linear component of polygalacturonan without branches called homogalacturonan (HG) and branched chains of rhamnogalacturonan type I (RG-I) and rhamnogalacturonan type II (RG-II) (Fig. 1). HG forms a linear backbone chain of α-1,4-linked d-galacturonic acid monomers, which are methyl esterified. Other pectic polysaccharides have the same HG backbone with d-xylose side residues in xylogalacturonan or with highly branched polysaccharides, including xylose, glucuronic acid, rhamnose, arabinose, apiose, galacturonic acid, fucose, and galactose, in RG-II. On the other hand, RG-I consists of a backbone chain of α-1,4-d-galacturonic acid-α-1,2-l-rhamnose with branches of arabinogalactan, galactan, and arabinan (3, 4) (Fig. 1).
FIG 1.
Structure of the pectin complex and degradation of the arabinogalactan side chains of rhamnogalacturonan type I.
Bacillus subtilis, as a common soil saprophytic bacterium, is able to degrade pectin and metabolize it through a RG-I degradation pathway (5). Principally, RG-I is degraded by the action of three types of enzymes, RG-I hydrolases, RG-I lyases, and RG-I-specific acetylesterases (for a review, see reference 6). To break down the backbone chain of the pectin polymer, B. subtilis encodes two pectate lyases, Pel (7) and PelC (8). Also, a putative pectate lyase (ywoF) and a pectin lyase (pelB) exist in the genome of B. subtilis (9, 10). At least two gene clusters, namely, yesOPQRSTUVWXYZ and ytePRSTU, are involved in the degradation of RG-I (5). Based on similar models proposed by Itoh et al. (11) and Silva et al. (6), the RG-I backbone is primarily deacetylated by the RG acetylesterases YesT and YesY (12) and then attacked by extracellular RG endolyase YesW, cleaving the α-(1→4) bonds of the RG-I backbone to produce unsaturated galacturonate oligomers with different lengths. These unsaturated products are further degraded by the intracellular exolyase YesX to convert these oligomers to unsaturated disaccharides (13). The final step is the release of a single unsaturated galacturonate (5-dehydro-4-deoxy-d-galacturonate) by YesR and YteR (11). The RG-I side chains, including galactan, arabinan, and arabinogalactan, are dissociated from the backbone by two extracellular arabinan endo-1,5-α-l-arabinosidases, namely, AbnB (14) and AbnA (7, 15), releasing the α-l-arabinofuranoside residues as well as galactans from the arabinogalactan branched chains of RG-I. These enzymes provide oligoarabinans, which can be taken up via the AraNPQ transporter. Afterward, the intracellular oligoarabinan can be further degraded into arabinofuranose by α-l-arabinofuranosidases AbfA and Xsa and finally delivered to the pentose phosphate pathway by the araABDLMNPQ-abfA cluster (14, 16).
In addition to arabinan and arabinogalactan, galactan chains are also released during the degradation of pectin. In Bacillus subtilis, an extracellular enzyme, named galactanase GalA, was found to be able to degrade the galactan portion of arabinogalactan (17). Studying the glycoside hydrolases (GHs) of B. subtilis revealed an arrangement of a GH family 42 β-galactosidase gene, lacA, neighboring a GH family 53 enzyme galA gene. Cloning the two genes enabled Escherichia coli to grow with galactan as a carbon source (18). The previously mentioned GalA was later annotated as GanB (arabinogalactan endo-β-1,4-galactosidase) and belongs to the pentacistronic operon cycB-ganPQAB, which is regulated by GanR (also known as LacR) located upstream of this operon (19, 20). CycB (renamed GanS) is a sugar-binding lipoprotein that is reported to bind cyclodextrin (21) and forms the ABC transporter together with GanP and GanQ (22). Finally, the ganA gene (also known as lacA) encodes a β-galactosidase that is able to degrade substrates, such as ortho-nitrophenyl-β-galactopyranoside (oNP-β-Gal) or 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (18, 19). Prior to this study, the importance of the cycB-ganPQAB operon for the growth of B. subtilis with galactan was shown (23). In this study, we characterized the function of GanB and GanA and their substrate specificity and thereby expanded our knowledge of galactan degradation by B. subtilis. Beside this, the uptake of the products and the specific inducer of the galactan utilization system were investigated.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All strains used in this study are listed in Table S1 in the supplemental material. Escherichia coli JM109 was used for plasmid propagation and gene expression throughout this study. E. coli strains containing the desired plasmids were selected on LB plates supplemented with ampicillin (100 μg/ml) or spectinomycin (100 μg/ml) depending on the plasmid selection marker. To overexpress the desired genes in E. coli, LB medium with ampicillin (100 μg/ml) was inoculated with an overnight culture of E. coli carrying the target plasmid with a starting optical density at 600 nm (OD600) of 0.05. After 2 h of incubation at 37°C with shaking at 200 rpm, 0.2% l-rhamnose was added to the bacterial culture at an OD600 of about 0.3, and the cells were harvested by centrifugation after 4 h of incubation at 30°C. The cell pellet was kept at −20°C for further studies.
Bacillus subtilis KM0 (strain 168 with a functional TrpC) was used as the wild-type (wt) strain during this study. B. subtilis knockout strains (BKE) were provided by Bacillus Genetic Stock Center (BGSC; Columbus, OH). B. subtilis transformants were selected on LB plates containing spectinomycin (100 μg/ml) or erythromycin (5 μg/ml).
The tryptophan auxotroph strains were cultivated on Spizizen minimal plates (24) containing 50 μg/ml tryptophan. To investigate the regulation of the galactan utilization genes in vivo, B. subtilis strains were inoculated to 40 ml LB medium with a starting OD600 of 0.05. After 2 h of incubation at 37°C, 0.2% lupin galactan or 0.2% glucose (alone or in combination) was added to the bacterial culture, and the GanA β-galactosidase and GanB galactanase activities of each sample were measured after 6 h of cultivation at 37°C. All experiments were carried out in triplicate, and the mean values were used for further analysis.
Modification of B. subtilis knockout mutants.
The B. subtilis knockout strains (BKE strains) provided by Bacillus Genetic Stock Center (BGSC) contained an erythromycin resistance gene flanked by loxP sites. In order to remove this antibiotic marker gene, the BKE strains were transformed with pJOE6732.1 expressing Cre recombinase under the control of a constitutive Pxyl promoter originating from Bacillus megaterium QM B1551. After resolving the loxP sites, the antibiotic-sensitive strains were screened based on erythromycin or spectinomycin sensitivity. Plasmid pJOE6732.1 is derived from pAMβ1, which contains the minimal repDE genes, the replication origin without copF, the stabilizing genes resβ and topβ, and partition and toxin-antitoxin genes (25). Due to its high instability, pJOE6732.1 can easily be cured. To repair the trpC2 mutation in the desired strains, plasmid pKAM041, containing the functional trpC gene, and its flanking regions were used.
Measurement of the GanB galactanase activity.
Different substrates were used to measure the β-galactanase activity. As a natural substrate, arabinogalactan from larch wood (Sigma-Aldrich, Munich, Germany) or galactan from lupin, which was treated with arabinofuranosidase (Megazyme, Wicklow, Ireland), was used. The enzymatic reaction was carried out by adding appropriate amounts of enzyme (crude extracts or purified His6-GanB) to 1% galactan in a total volume of 40 μl potassium phosphate buffer (0.1 M, pH 6.5). After incubation at 37°C, samples were withdrawn, and the reaction was stopped by heating for 10 min at 95°C. The products of the enzymatic reactions were further qualitatively analyzed by thin-layer chromatography or by high-performance anion-exchange chromatography (HPAEC). In order to calculate the GanB activity in units, the reduced sugars released from the lupin galactan degradation were determined by the 3,5-dinitrosalicylic acid (DNS) assay (26). Briefly, appropriate amounts of His6-GanB (0.5 μg) in 40 μl of potassium phosphate buffer (0.1 M, pH 6.5) were mixed with 40 μl of 2% lupin galactan. The reaction mixture was incubated for 15 min at 37°C, and the reaction was stopped with 300 μl dinitrosalicylic acid reagent. After heating at 95°C for 20 min, 1 ml H2O was added and kept for 20 min on ice. The absorption of the developed orange color was measured at 540 nm. One unit was defined as the release of 1 μmol of reducing sugar (here galactose) per minute determined by the DNS assay.
A standard GanB activity assay was performed with azurine-cross-linked-potato (AZCL) galactan (product 38127; Sigma-Aldrich, Munich, Germany) as the substrate in a colorimetric assay. The enzyme reaction was carried out with 0.2% AZCL galactan in a total volume of 250 μl containing variable amounts of enzyme. The reaction mixture was incubated for 15 min at 37°C and stopped with 250 μl of 0.4 M sodium borate. After centrifugation for 5 min at 13,000 × g, 100 μl of the blue colored supernatant was diluted with 900 μl H2O, and its absorbance was measured at 595 nm. As the blank, the reaction mixture ingredients without enzyme were used.
In order to test the degradation of galactotetraose, galactotriose, and galactobiose by GanB, appropriate amounts of purified GanB were incubated with these substrates, with the final concentration of 5 mM in a total volume of 10 μl. The product composition of the reaction mixture was analyzed by high-pressure liquid chromatography (HPLC).
Measurement of β-galactosidase activity.
β-Galactosidase activity was determined by measuring the rate of p-nitrophenyl-β-d-galactopyranoside (pNP-β-Gal) hydrolysis as described earlier (27). The β-galactosidase activity toward lactose was calculated from the amount of released glucose according to Placier et al. (27). One unit of the enzyme activity was defined as the release of 1 μmol of pNP or glucose per minute, respectively. The substrates, galactotetraose, galactotriose, and galactobiose, were used with the final concentration of 5 mM in a total volume of 10 μl. The product composition was analyzed by HPLC as described before. Finally, all β-galactosidase assays were carried out at 37°C using 0.1 M potassium phosphate buffer (pH 6.5).
Analysis of galacto-oligosaccharides by chromatographic methods.
Qualitative and quantitative chromatographic methods were used for the analysis of galacto-oligosaccharides generated after the degradation of galactan. In the qualitative analysis, the separation of galacto-oligosaccharides was performed using thin-layer chromatography (TLC) on Silica Gel 60 F254 plates (Merck, Darmstadt, Germany). The mobile phase was a mixture of acetone–H2O (87:13, vol/vol). Carbohydrate spots were visualized by soaking the TLC plates with a solution containing 10.5 g ammonium sulfate, 0.5 g cerium sulfate, and 15 ml concentrated H2SO4 in 245 ml H2O. The carbohydrate blue spots were developed after backing the plates at 80°C. Quantitative analysis of individual galacto-oligosaccharide fractions was performed using high-performance anion-exchange chromatography (HPAEC) with pulsed amperometric detection (ESA Coulochem III; ESA Inc., Chelmsford, MA). The samples were separated on a CarboPac PA1 column (Dionex, Idstein, Germany) using the mobile phase of 0.22 M NaOH and 0.02 M sodium acetate with a flow rate of 0.75 ml/min as described previously (28). Individual carbohydrates, such as galactose, lactose, and β-1,4-galactobiose (β-d-Gal-[1→4]-β-d-Gal) (product G9662; Sigma, Munich, Germany), were used as standards during HPLC analysis.
Isolation of galacto-oligosaccharides.
Galacto-oligosaccharides used as a substrate for enzymatic reactions or in mass spectrometry analysis were isolated after enzymatic digestion of lupin galactan by the purified GanB enzyme in a preparative approach. One percent lupin galactan was treated with 12 μg purified His6-GanB in 0.1 M potassium phosphate buffer (pH 6.5) in a total volume of 800 μl. The reaction was stopped after 30 min of incubation at 37°C by heating for 10 min at 95°C. After centrifugation for 10 min at 13,000 × g, 400 μl of supernatant was loaded onto a Bio-Gel P-2 column (250 ml; Bio-Rad, Munich, Germany) and separated with double-distilled water (ddH2O) as the mobile phase. The 1-ml fractions were collected after a flowthrough of 70 ml and analyzed by HPLC. Fractions 15 to 21 were pooled as P1, fractions 26 to 31 were pooled as P2, and fraction 37 to 41 were pooled as P3, respectively. The P1, P2, and P3 samples were lyophilized prior to quadrupole time of flight liquid chromatography mass spectrometry (Q-TOF-LC/MS) analysis.
Methods in supplemental materials.
DNA manipulation and plasmid construction, protein purification, DNA sequencing, electrophoretic mobility shift assay, DNase I footprinting, primer extension, and thermal shift assay methods are explained in the supplemental material.
RESULTS
GanB degrades arabinose-free galactan with its endo-β-1,4-galactosidase activity.
To study the galactan utilization in B. subtilis, the characteristics and function of GanB encoded by the last gene of the cycB-ganPQAB operon were primarily investigated. GanB (also known as GalA or YvfO) is annotated as arabinogalactan endo-β-1,4-galactosidase, which belongs to glycoside hydrolase (GH) family 53. Analysis of the GanB protein sequence with the SignalP 4.1 program (29) predicted a signal peptide of 26 amino acid residues (data not shown). To further characterize the activity of GanB, the N-terminal His6-tagged mature protein was produced in E. coli strain JM109(pHWG1119) and purified by immobilized-metal affinity chromatography (IMAC). The purification of His6-GanB from the crude extract yielded over 90% pure His6-GanB, with a molecular mass of 46 kDa and a protein content of 0.6 mg/ml. To characterize GanB, different substrates and methods were applied in the enzyme assay (see Table S3 in the supplemental material). To begin with, AZCL galactan, which is a dyed substrate, was used for a semiquantitative assay. By doing this, the optimal temperature of activity was determined to be 50°C and the optimal pH was 6.5 (data not shown). Also, the specific activity of GanB toward galactan that originated from lupin was determined by measuring the increase of reducing sugars during the enzymatic reaction and resulted in 75 ± 5 U/mg GanB activity.
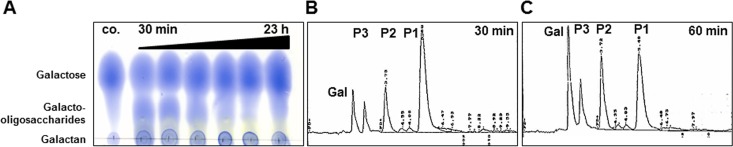
Given that galactan is degraded by GanB, in order to find out the possible mode of action of GanB, arabinogalactan from larch wood and lupin galactan were qualitatively tested by TLC analysis. After degradation of lupin galactan by GanB, galactose and galacto-oligosaccharides were produced. The latter products diminished after a longer period of degradation, and the proportion of galactose increased (Fig. 2A). No product was released from arabinogalactan (data not shown). This showed that arabinose-free galactan is the natural substrate for GanB. In order to identify the galacto-oligosaccharides generated during the degradation of lupin galactan by GanB, the reaction mixture was further analyzed by HPLC (Fig. 2B and C). Two products, i.e., galactose and β-d-Gal-(1→4)-β-d-Gal (here Gal2 or galactobiose), were identified using commercial standards (Fig. 2B and C). Additionally, two further unidentified main products (shown as P2 and P1 in Fig. 2) were obtained with a higher degree of polymerization (DP) and retention times of 3.78 and 5.49, respectively (Fig. 2B and C). The proportion of these galacto-oligosaccharides with higher DP decreased after a longer incubation of lupin galactan with GanB (Fig. 2B and C).
FIG 2.
(A) Analysis of the products of lupin galactan degradation by GanB after 30 min and up to 23 h using thin-layer chromatography. As a control (co.), a mixture of galactose (20 μg) and galactan (5 μg) was applied. (B and C) HPLC profiles of the products of lupin galactan degradation by purified GanB after 30 min (B) and 60 min (C). The separation of the galacto-oligosaccharides was performed by HPLC with pulsed amperometric detection as described in Materials and Methods.
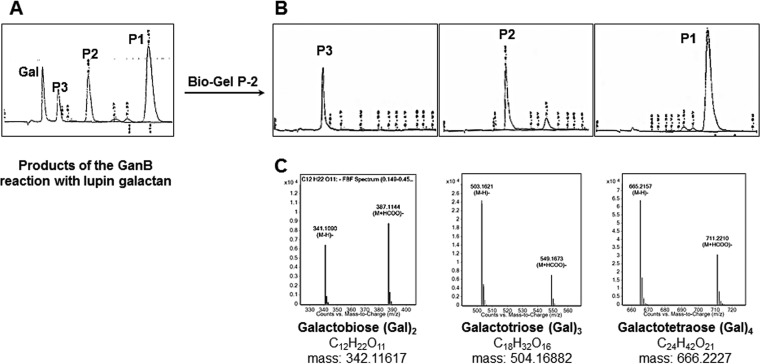
In order to characterize the unknown galacto-oligosaccharides generated during the galactan degradation, a preparative approach was applied. First, the reaction mixture of the degradation of 4 mg lupin galactan was separated on a Bio-Gel P-2 chromatography column, and products with different DP were isolated (labeled as P1, P2, and P3 fractions) (Fig. 3A and B). Afterward, their molecular masses were determined by Q-TOF-LC/MS with a negative-ion [M-H]− mode (Fig. 3C). Mass spectrometry analysis revealed that P1 was a tetrasaccharide with a molecular weight of 666.2227, P2 was a trisaccharide with a molecular weight of 504.1688, and P3 was a disaccharide with a molecular weight of 342.1161 (Fig. 3C). The latter substance has also been identified as β-d-Gal-(1→4)-β-d-Gal with an authentic standard by HPLC. These experiments show that GanB cleaves the high-molecular-weight arabinose-free galactan polymer producing galacto-oligomers with a DP of up to 4 and provide evidence for the endo-type mode of action.
FIG 3.
(A) HPLC profile of products from the preparative degradation of galactan with GanB. (B and C) HPLC chromatograms of the galacto-oligosaccharides P1 to P3 that were isolated after Bio-Gel P-2 chromatography (B), along with their mass spectra peaks elucidated by Q-TOF-LC/MS (C).
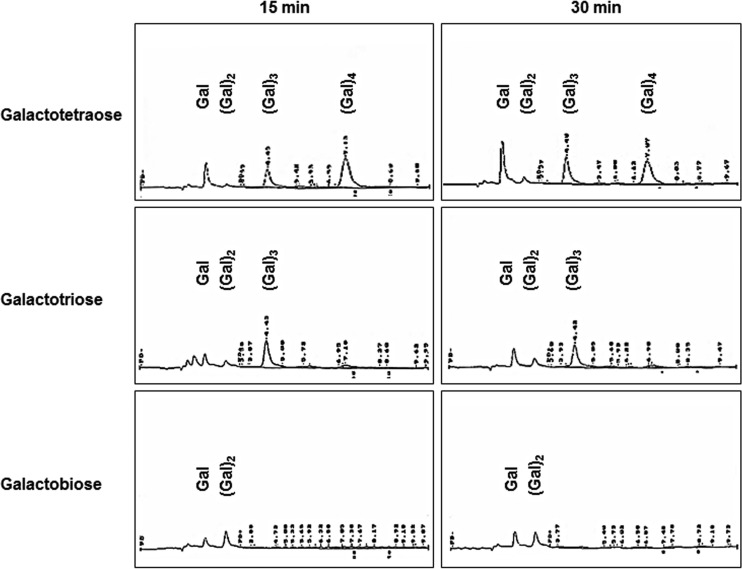
GanB was also subjected to substrates other than high-molecular-weight galactan in order to determine the GanB mode of action. First, the isolated galactotetraose, galactotriose, and galactobiose served as GanB substrates, and the degradation products were analyzed by HPLC (Fig. 4). In all of these cases, the release of galactose was detected, suggesting that GanB somewhat shows an exo-mode of action. However, GanB was unable to act on p-nitrophenyl-β-d-galactopyranoside (pNP-β-Gal), indicating that a solely exo-mode could not be detected (see Table S3 in the supplemental material). All in all, GanB shows both endo- and exo-activity, with galactan as a substrate generating galactotetraose as its main product.
FIG 4.
HPLC profile of the degradation of galactotetraose, galactotriose, and galactobiose by GanB after reaction times of 15 and 30 min to demonstrate the exo-mode of action of GanB. The separation of the galacto-oligosaccharides was performed by HPLC with pulsed amperometric detection as described in Materials and Methods.
Galactotetraose and galactotriose are transported by galactan ABC transport system (GanSPQ-MsmX).
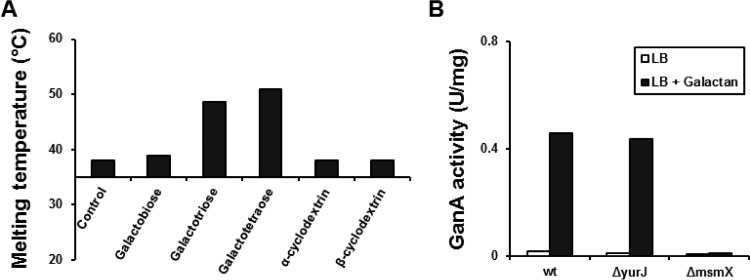
As shown before, degradation of galactan by GanB generated galacto-oligosaccharides with a DP of up to 4. To clarify the pathway of galactan utilization, the next step was to investigate the substrate specificity of the galactan transporter. The galactan transporter is an ABC transport system, consisting of GanP and GanQ as the transmembrane proteins and CycB as the substrate-binding lipoprotein. The substrate specificity of the transporter depends on CycB, which has been shown to be a cyclodextrin-binding lipoprotein (21). Analysis of the CycB protein sequence indicated an LAAC lipoprotein signal peptide motif (30) with a cleavage site between amino acid positions 22 and 23 (data not shown). To carry out the in vitro studies, the mature CycB was fused to the His6 tag at its N terminus, and the His6-CycB was overproduced in E. coli using the plasmid pHWG1151. SDS-PAGE analysis of the crude extract of induced JM109(pHWG1151) showed the production of the 44-kDa His6-CycB protein. Purified His6-CycB (2.5 mg/ml) was used for the thermal shift assay (Fig. 5A). In this assay, CycB was subjected to three substrates, namely, galactobiose, galactotriose, and galactotetraose, which are produced by the activity of GanB. Only galactotetraose and galactotriose were able to increase the melting temperature of CycB by their binding to His6-CycB (Fig. 5A). Additionally, α- and β-cyclodextrins were tested with the thermal shift assay. Although cyclodextrin was supposed to be the ligand of CycB (21), no affinity of cyclodextrin to His6-CycB was observed under the experimental conditions (Fig. 5A). Therefore, we renamed CycB to GanS here. Altogether, this experiment shows that galactotetraose and galactotriose are the main ligands for the GanS protein and may be mainly transported by the galactan ABC transporter into the cytoplasm.
FIG 5.
(A) Thermal shift assay using purified His6-CycB (later renamed GanS) in the presence of galactobiose, galactotriose, galactotetraose, α-cyclodextrin, and β-cyclodextrin with a final concentration of 1 mM. The melting temperature of His6-CycB was measured without the substrate (control). (B) Finding the ATP-binding protein of the galactan ABC transporter. The wild-type strain BKE32550 (ΔyurJ) and BKE38810 (ΔmsmX) were cultivated in LB with or without 0.2% lupin galactan with a starting OD600 of 0.05 and were induced after 2 h of incubation at 37°C. The β-galactosidase activity of GanA was measured in crude extracts after 6 h of induction.
Another important question about the uptake of the galactotetraose and galactotriose by the galactan ABC transporter was the ATP-binding protein. Since the ganSPQAB operon contained no ATP-binding protein, the genome of B. subtilis was searched for the probable ATP-binding proteins. So far, 70 ATP-binding proteins have been annotated and predicted in the genome of B. subtilis, among which 2 proteins, namely, MsmX and YurJ, are suggested to be multiple sugar-binding transporter ATP-binding proteins (18). Therefore, two strains lacking msmX (strain BKE38810) or yurJ (strain BKE32550) were cultivated in LB with or without galactan as an inducer. As a reporter system, the β-galactosidase activity of GanA in the crude extract was measured (Fig. 4B). In the wild-type strain, the β-galactosidase activity was increased by 24-fold when galactan was added to LB medium. Deletion of yurJ had no influence on the β-galactosidase activity, showing similar inducibility to the wild-type strain. On the contrary, adding galactan to LB had no influence on the β-galactosidase activity in the ΔmsmX mutant. The loss of induction of the gan operon in the ΔmsmX mutant is caused by a galacto-oligosaccharide transport deficiency. Taken together, MsmX is the ATP-binding component of the galactan ABC transport system.
GanA degrades galacto-oligosaccharides in an exo-type mode of action.
The gan operon contains another structural gene, ganA (also known as lacA or yvfN), which encodes a β-galactosidase. The activity of this enzyme toward the synthetic substrate oNP-β-Gal was shown several years ago (20). Nevertheless, by knowing the mechanism of the extracellular degradation of galactan and the uptake of the produced galacto-oligosaccharides, it was essential to understand the degradation of these galacto-oligosaccharides by GanA. Therefore, the his6-ganA gene was overexpressed in E. coli using strain JM109(pHWG1111). The production of His6-GanA was confirmed by SDS-PAGE and showed a prominent protein band with a molecular mass of 80 kDa (data not shown). The purified His6-GanA exhibited a specific activity of 80 U/mg toward the synthetic substrate pNP-β-Gal at 37°C. Using pNP-β-Gal as the substrate, no metal dependency was observed, and GanA exhibited an optimum pH and temperature of 6.5 and 55°C, respectively (data not shown). Likewise, GanA was able to cleave lactose with a specific activity of 12 U/mg.
Finally, the target substrates of the galactan utilization system were tested for GanA activity (see Table S4 in the supplemental material). At first, the high-molecular-weight substrates were used as the substrates for GanA. For this purpose, different methods were used to detect the degradation products of arabinogalactan, AZCL galactan, and lupin galactan. In all of these cases, no GanA activity was detectable (see Table S4). Conversely, the galacto-oligosaccharides, galactotetraose, galactotriose, and galactobiose, were degraded by GanA generating galactose as the end product. The HPLC degradation profile of galactotetraose is shown in Fig. 6. Altogether, these results clearly indicated that GanA degrades the transported galacto-oligosaccharides into galactose as the end product. Since galactose was the main end product of the degradation of galactotetraose, it is highly assumable that GanA act as an exo-saccharolytic enzyme. Furthermore, it should be noted that the activity for galactotetraose was approximately 20-fold higher than that for lactose with a 5 mM substrate in an activity test (data not shown).
FIG 6.
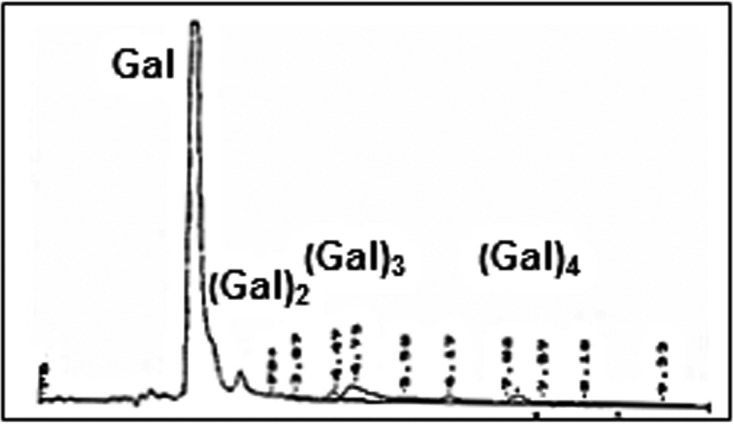
Degradation of prepared galactotetraose by GanA. The reaction was carried out for 15 min at 37°C.
Induction of the ganSPQAB operon by galactobiose and its repression by glucose.
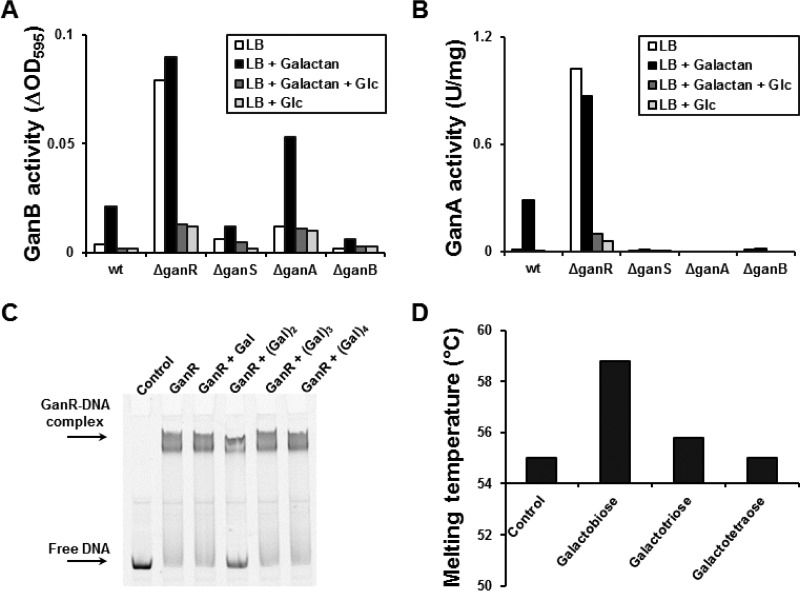
The galactan utilization operon is repressed by GanR (19); however, the possible inducer of this system has not yet been reported. In order to explore the possible inducer of GanR, wild-type B. subtilis and ΔganR, ΔganS, ΔganA, and ΔganB mutant strains were cultivated in LB containing galactan or glucose alone or in combination, and their galactanase activities (GanB) were measured in the culture medium (Fig. 7A). The wild-type strain cultivated in LB with galactan showed significantly higher GanB activity (5-fold) than the wild-type strain cultivated in LB without galactan. This shows that the presence of galactan induced ganB expression. Similar to many carbohydrate utilization systems in B. subtilis, the addition of glucose to the culture medium reduced GanB activity regardless of the presence of galactan, showing that the galactan utilization system is repressed by carbon catabolite repression. Deletion of GanR caused a high level of ganB expression where the GanB activity with or without galactan was almost identical, albeit significantly higher than the induced wild-type strain. The carbon catabolite repression remained functional in the ΔganR mutant. Deletion of ganS resulted in lower GanB activity when galactan was present in the medium, showing a lower induction of the system that was probably caused by the loss of inducer uptake. Interestingly, deletion of ganA increased GanB activity by 2.5-fold compared to that of the wild-type strain cultivated in LB with galactan. In this case, the inducibility of ganB was 4-fold. When ganB was deleted, negligible GanB activity was measured in the culture medium (Fig. 7A).
FIG 7.
(A and B) GanB (A) and GanA (B) activities after 6 h of cultivation of B. subtilis strains, i.e., wt (KM0), ΔganR (BKE34170), ΔganS (KM468), ΔganA (KM589), and ΔganB (KM588), with different sugars in LB as described in Materials and Methods. GanB activity was measured in a 1:10 diluted culture medium toward the substrate AZCL galactan. GanA activity was determined in crude extracts toward pNP-β-Gal. (C) An electrophoretic mobility shift assay was carried out using the amplified 5′-end Cy5-Pgan DNA in the absence (control) and presence of GanR. The potential inducers of GanR were added to the reaction with the final concentration of 5 mM. (D) Thermal shift assay using purified GanR-Strep tagged in the absence of substrate (control) and in the presence of galactose, galactobiose, galactotriose, or galactotetraose with a final concentration of 1 mM.
In order to confirm the results obtained by the measurement of the GanB activity, the GanA activity in the wild-type strain and in the ΔganR, ΔganS, ΔganA, and ΔganB mutants was also determined (Fig. 7B). In this experiment, the gan system was inducible by galactan, while the ganR deletion rendered the gan operon constitutive. The ganS deletion significantly reduced GanA activity, which may be due to the reduced uptake of the probable inducer of the system (Fig. 7B). The ganA deletion showed negligible β-galactosidase activity as expected. Finally, when ganB was deleted, also no β-galactosidase activity was detected (Fig. 7B). Taken together, these results indicate that the inducer of the system is a galacto-oligosaccharide generated by GanB activity. Since deletion of ganA did not affect the induction of the system and even increased GanB activity in the presence of galactan (Fig. 7A), it is likely that the inducer of the system is one of the transported galacto-oligosaccharides.
To identify the inducer of the galactan utilization system, an electrophoretic mobility shift assay was conducted using the purified Strep-tagged GanR and different potential substrates (Fig. 7C). The 5′-end fluorescein isothiocyanate (FITC)-labeled Pgan DNA was amplified from the promoter region located upstream of ganS. From the addition of GanR to the Pgan DNA in the reaction mixture, a DNA-protein complex formed that migrated slower than the free Pgan DNA. Next, galactose or different galacto-oligosaccharides were added to the DNA-protein complex. Only galactobiose was able to dissociate the GanR from its target Pgan DNA, showing that the galactobiose is the inducer of the galactan utilization system (Fig. 7C). Lactose and isopropyl-β-d-thiogalactopyranoside (IPTG) also had no influence on the DNA-protein complex (data not shown). To confirm this result, thermal shift assay was also carried out using the purified GanR and the galacto-oligosaccharides. Indeed, the presence of galactobiose increased the melting temperature of GanR (Fig. 7D). Altogether, galactobiose was found as the inducer of the gan system using in vivo and in vitro experiments.
GanR inhibits the Pgan transcription by binding to the −35 box.
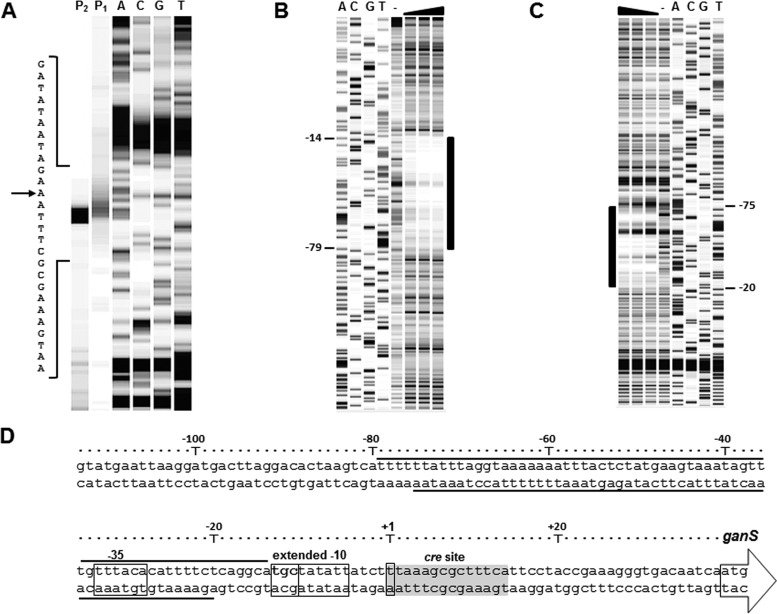
As the final step in this study, the promoter region of the ganSPQAB (Pgan) was characterized. The transcription start site of the gan operon was identified by primer extension method (Fig. 8A). The results of the primer extension experiment indicated that the transcription of ganSPQAB starts 38 bp upstream of the translation start codon of ganS (Fig. 8A). By the identification of the +1 position, the typical −10 (TATAtT) and −35 (TTtACA) boxes of the σA-type (housekeeping) promoters were found in the Pgan DNA sequence, each with a mismatch to the consensus sequence (small letters) (Fig. 8D). Interestingly, a TGN motif was found next to the −10 box showing that Pgan likely had an extended −10 box (Fig. 8D). Given that the Pgan is repressed by GanR, DNA footprinting was carried out using purified Strep-tagged GanR (see Fig. S1 in the supplemental material). Digestion of the Pgan DNA by DNase I indicated a protected region between nucleotides −14 and −79 with respect to the transcription start site at the coding strand (Fig. 8B), whereas the noncoding strand was protected between bp −20 and −75 from the transcription start site (Fig. 8C). These results show that GanR represses the Pgan transcription initiation via the steric hindrance mechanism and by prevention of the binding of the RNA polymerase.
FIG 8.
Characterization of the core and regulatory elements of Pgan. (A) Identification of the transcription start site of Pgan was carried out using primer extension reactions. The generated cDNA probes (P1 and P2 repeats) were compared with the sequencing reaction using the dideoxy chain-termination method (A, C, G, T). (B and C) The DNase I footprinting reactions for the Pgan coding strand DNA (B) and noncoding strand DNA (C) were compared to the dideoxy chain-termination reactions (A, C, G, and T). The DNase I footprinting reactions were performed without GanR as the negative control (−) or with different amounts of GanR (1.95, 3.90, and 7.79 μM). The bar shows the protected DNA region in each DNase I footprinting reaction. (D) The DNA sequence of the Pgan region. The promoter core elements (−35 box and extended −10 box) and the transcription start site (+1) are shown by rectangles. The protected DNA region is shown by solid lines. The arrow shows the start codon of ganS. The predicted cre site is highlighted in gray.
DISCUSSION
Galactan is an abundant molecule in the cell wall of plants and obviously a suitable substrate for the soil bacteria as an important source of galactose. For the first time, the galactanase activity of B. subtilis was detected by its cultivation in a medium containing soybean. This galactanase was able to degrade β-1,4-galactan into the predominant product galactotetraose (17). The galactanase-encoding gene, galA (later renamed ganB), was discovered in B. subtilis after its importance for the degradation of galactan by E. coli was proven in vivo (18). In this study, we provided in vitro evidence that GanB, which belongs to GH family 53 of the β-1,4-galactanases, indeed acts as a β-galactanase. Moreover, GanB exhibits an endo- and exo-mode of action producing galactotetraose as the main product along with galactotriose, galactobiose, and galactose (Fig. 3). Similar patterns of product release were reported from the β-galactanase GanA from Geobacillus stearothermophilus (31) and the β-galactosidase from Bacillus licheniformis (BLGAL) (32). Nevertheless, there is a difference in the substrate preference of these enzymes. While GanB is able to degrade galactotetraose, galactotriose, and galactobiose, suggesting an additional exo-mode of activity (Fig. 4), BLGAL is not able to hydrolyze galactotriose and galactobiose to any appreciable extent although their amino acid sequences are 80% identical. This sequence identity between GanB and BLGAL even includes three homologous tryptophan side chains that are involved in galactotriose binding (32). Compared to galactanases from Bacilli species, fungal β-galactanases perform a more complete degradation of galactan, producing mostly galactose and galactobiose as end products (32, 33). The liberation of galactotriose as the sole product in an exo-fashion was reported from an endo-β-galactanase from Bifidobacterium longum (34). These results support the idea that different types of product formation exist among GH family 53 galactanases.
After degradation of galactan by the extracellular GanB, the released galactotetraose, galactotriose, and galactobiose must be transported into the cell. The galacto-oligosaccharide transport system is one of the 10 carbohydrate-specific ABC transport systems in B. subtilis (35) with two transmembrane proteins, GanP and GanQ, and a substrate-binding protein, CycB. The annotation of CycB originates from a previous report on the ability of CycB to bind β- or γ-cyclodextrin (21). Here, we provided evidence that CycB is clearly a galacto-oligosaccharide-binding protein, and it showed no affinity toward β-cyclodextrin as a substrate under our experimental condition. Therefore, we renamed cycB to ganS as a substrate-binding lipoprotein. The next unknown component of the galacto-oligosaccharides transport system was the ATP-binding protein. Previously, Shipkowski and Brenchley (18) proposed that one of the two MalK homologs, namely, YurJ and MsmX proteins, is the possible ATPase of the galactan ABC transporter. Our results indeed showed that MsmX is the ATPase of the galactan ABC transporter. In addition to the galactan ABC transport system, MsmX energizes the AraNPQ (arabino-oligosaccharides) (36) and MdxEFG (maltodextrin) (37) ABC transport systems.
The transported galactotetraose and galactotriose are further degraded in the cell. The ganB gene is flanked by ganA, which encodes a GH family 42 β-galactosidase. Interestingly, lactose, which is probably the natural substrate for β-galactosidases (38), is weakly degraded by GanA (see Table S4 in the supplemental material). Similar to B. subtilis GanA, GanB of G. stearothermophilus shows no detectable activity toward lactose (31). In B. subtilis, the physiological role of GanA is to hydrolyze the transported short β-1,4-galacto-oligosaccharides into galactose, while it is unable to degrade the galactan polymer similar to GanB of G. stearothermophilus (31). The arrangement of GH family 53- and GH family 42-encoding enzymes within the galactan utilization cluster was already reported for Geobacillus stearothermophilus (31). It seems a common feature that the degradation of galactan in nature is carried out by an extracellular endo-β-galactanase that cleaves the high-molecular-weight polysaccharide inside the chain and produces short oligomers. Afterward, the intracellular β-galactosidase enzyme GH family 42 hydrolyzes these short galacto-oligosaccharides into galactose. Galactose can then be metabolized into UDP-glucose via the Leloir pathway (23).
Transcriptome analysis of B. subtilis shows that the ganSPQAB genes form an operon (39), which is repressed by GanR (19). Prior to this study, the inducer of the system was not reported. Here, we found out that galactobiose is the inducer of the operon. This was in line with in vivo findings that the presence of the substrate-binding lipoprotein, GanS, and the extracellular GanB is necessary for the induction of the system. In contrast, the deletion of ganA even increased the inducibility of the gan operon and raised its maximal expression level in the presence of galactan (see Fig. 7A). This suggests that GanB extracellularly produces galactobiose from galactan, and GanS takes up galactobiose while GanA activity results in the degradation of galactobiose. Interestingly, the maximal activity of Pgan in the induced ΔganA mutant was higher than that in the induced wild-type strain. This is probably due to the deficiency of the ΔganA mutant to consume the galactobiose as a carbon source. Conversely, this maximal Pgan activity in the ΔganA strain remained less than that in the ΔganR mutant for two reasons. First, GanB mainly generates galactotetraose and, only after longer incubation times, galactobiose. Second, GanS has a lower affinity toward galactobiose than galactotetraose (or galactotriose) for the uptake. Nevertheless, it seems that the amount of galactobiose in the ΔganA mutant is enough for the induction of Pgan. Finally, the characterization of the Pgan elements revealed that GanR represses the binding of the RNA polymerase with the housekeeping sigma factor by the steric hindrance mechanism. In addition, the gan operon is repressed in the presence of glucose similar to other carbohydrate utilization systems in B. subtilis (40). A potential cre (catabolite responsive element) site was found at position +1 (Fig. 8D). The cre sites are the specific binding sites of the carbon catabolite protein A (CcpA)-histidine-containing protein (HPr) complex, which acts as the general carbon catabolite repression system (41). The CcpA-HPr forms as a response to the high level of fructose 1,6-bisphosphate and glucose 6-phosphate in the cytoplasmic pool, which increases during the presence of preferred sugars, such as glucose or fructose (42). Depending on the location of the cre sites, binding of the CcpA-HPr complex results in the activation (upstream of −35 box) or repression (downstream of −10 box) of the system (40). In the case of the gan operon, the location of the cre site, which is downstream of the −10 box, results in blocking the transcription initiation of the gan operon in the presence of glucose.
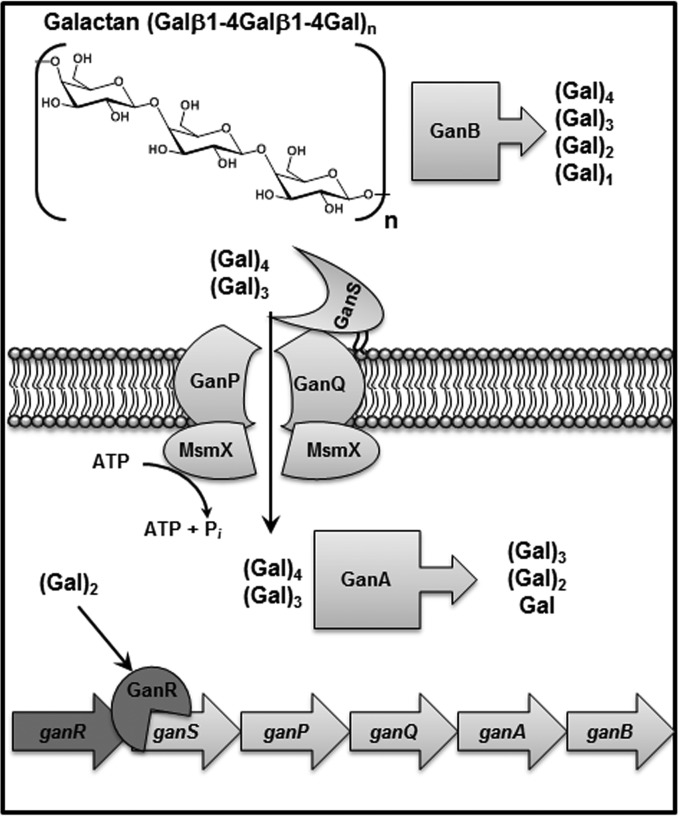
Based on the results obtained in this study, the model of the galactan utilization system by B. subtilis is demonstrated in Fig. 9. In the first step, extracellular galactan is degraded by GanB generating mainly galactotetraose as well as galactotriose and galactobiose. Next, galactotetraose and galactotriose are transported via the GanSPQ-MsmX complex and further degraded by GanA inside the cell. Binding the galactobiose to GanR dissociates the GanR-Pgan complex, resulting in expression of the gan operon.
FIG 9.
Model of the galactan degradation pathway in B. subtilis.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate Andrés Sánchez-Kopper (Institute of Biochemical Engineering, University of Stuttgart, Germany) for performing the Q-TOF-LC/MS experiments. We thank Gisela Kwiatkowski, Gisela Wajant, Silke Weber, and Annette Schneck for technical assistance throughout this study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00468-16.
REFERENCES
- 1.Keegstra K. 2010. Plant cell walls. Plant Physiol 154:483–486. doi: 10.1104/pp.110.161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. 2010. Lignin biosynthesis and structure. Plant Physiol 153:895–905. doi: 10.1104/pp.110.155119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohnen D. 2008. Pectin structure and biosynthesis. Curr Opin Plant Biol 11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert HJ. 2010. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol 153:444–455. doi: 10.1104/pp.110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochiai A, Itoh T, Kawamata A, Hashimoto W, Murata K. 2007. Plant cell wall degradation by saprophytic Bacillus subtilis strains: gene clusters responsible for rhamnogalacturonan depolymerization. Appl Environ Microbiol 73:3803–3813. doi: 10.1128/AEM.00147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva IR, Jers C, Meyer AS, Mikkelsen JD. 2016. Rhamnogalacturonan I modifying enzymes: an update. N Biotechnol 33:41–54. doi: 10.1016/j.nbt.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Nasser W, Awadé AC, Reverchon S, Robert-Baudouy J. 1993. Pectate lyase from Bacillus subtilis: molecular characterization of the gene, and properties of the cloned enzyme. FEBS Lett 335:319–326. doi: 10.1016/0014-5793(93)80410-V. [DOI] [PubMed] [Google Scholar]
- 8.Soriano M, Diaz P, Pastor FI. 2006. Pectate lyase C from Bacillus subtilis: a novel endo-cleaving enzyme with activity on highly methylated pectin. Microbiology 152:617–625. doi: 10.1099/mic.0.28562-0. [DOI] [PubMed] [Google Scholar]
- 9.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Cordani JJ, Connerton IF, Cummings NJ, Daniel RA, Denziot F, Devine KM, Dusterhoft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, et al. . 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 10.Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A, Vallenet D, Wang T, Moszer I, Medigue C, Danchin A. 2009. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155:1758–1775. doi: 10.1099/mic.0.027839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itoh T, Ochiai A, Mikami B, Hashimoto W, Murata K. 2006. A novel glycoside hydrolase family 105: the structure of family 105 unsaturated rhamnogalacturonyl hydrolase complexed with a disaccharide in comparison with family 88 enzyme complexed with the disaccharide. J Mol Biol 360:573–585. doi: 10.1016/j.jmb.2006.04.047. [DOI] [PubMed] [Google Scholar]
- 12.Mølgaard A, Kauppinen S, Larsen S. 2000. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Structure 8:373–383. doi: 10.1016/S0969-2126(00)00118-0. [DOI] [PubMed] [Google Scholar]
- 13.Pages S, Valette O, Abdou L, Belaich A, Belaich JP. 2003. A rhamnogalacturonan lyase in the Clostridium cellulolyticum cellulosome. J Bacteriol 185:4727–4733. doi: 10.1128/JB.185.16.4727-4733.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inácio JM, de Sá-Nogueira I. 2008. Characterization of abn2 (yxiA), encoding a Bacillus subtilis GH43 arabinanase, Abn2, and its role in arabino-polysaccharide degradation. J Bacteriol 190:4272–4280. doi: 10.1128/JB.00162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leal TF, de Sá-Nogueira I. 2004. Purification, characterization and functional analysis of an endo-arabinanase (AbnA) from Bacillus subtilis. FEMS Microbiol Lett 241:41–48. doi: 10.1016/j.femsle.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Inácio JM, Correia IL, de Sá-Nogueira I. 2008. Two distinct arabinofuranosidases contribute to arabino-oligosaccharide degradation in Bacillus subtilis. Microbiology 154:2719–2729. doi: 10.1099/mic.0.2008/018978-0. [DOI] [PubMed] [Google Scholar]
- 17.Labavitch JM, Freeman LE, Albersheim P. 1976. Structure of plant cell walls. Purification and characterization of a beta-1,4-galactanase which degrades a structural component of the primary cell walls of dicots. J Biol Chem 251:5904–5910. [PubMed] [Google Scholar]
- 18.Shipkowski S, Brenchley JE. 2006. Bioinformatic, genetic, and biochemical evidence that some glycoside hydrolase family 42 beta-galactosidases are arabinogalactan type I oligomer hydrolases. Appl Environ Microbiol 72:7730–7738. doi: 10.1128/AEM.01306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniel RA, Haiech J, Denizot F, Errington J. 1997. Isolation and characterization of the lacA gene encoding beta-galactosidase in Bacillus subtilis and a regulator gene, lacR. J Bacteriol 179:5636–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Errington J, Vogt CH. 1990. Isolation and characterization of mutations in the gene encoding an endogenous Bacillus subtilis β-galactosidase and its regulator. J Bacteriol 172:488–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamionka A, Dahl MK. 2001. Bacillus subtilis contains a cyclodextrin-binding protein which is part of a putative ABC-transporter. FEMS Microbiol Lett 204:55–60. doi: 10.1111/j.1574-6968.2001.tb10862.x. [DOI] [PubMed] [Google Scholar]
- 22.Quentin Y, Fichant G, Denizot F. 1999. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J Mol Biol 287:467–484. doi: 10.1006/jmbi.1999.2624. [DOI] [PubMed] [Google Scholar]
- 23.Chai Y, Beauregard PB, Vlamakis H, Losick R, Kolter R. 2012. Galactose metabolism plays a crucial role in biofilm formation by Bacillus subtilis. mBio 3:e00184-12. doi: 10.1128/mBio.00184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anagnostopoulos C, Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J Bacteriol 81:741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruand C, Le Chatelier E, Ehrlich SD, Janniere L. 1993. A fourth class of theta-replicating plasmids: the pAMβ1 family from Gram-positive bacteria. Proc Natl Acad Sci U S A 90:11668–11672. doi: 10.1073/pnas.90.24.11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 27.Placier G, Watzlawick H, Rabiller C, Mattes R. 2009. Evolved beta-galactosidases from Geobacillus stearothermophilus with improved transgalactosylation yield for galacto-oligosaccharide production. Appl Environ Microbiol 75:6312–6321. doi: 10.1128/AEM.00714-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravaud S, Robert X, Watzlawick H, Haser R, Mattes R, Aghajari N. 2007. Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization. J Biol Chem 282:28126–28136. doi: 10.1074/jbc.M704515200. [DOI] [PubMed] [Google Scholar]
- 29.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 30.Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol Mol Biol Rev 64:515–547. doi: 10.1128/MMBR.64.3.515-547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tabachnikov O, Shoham Y. 2013. Functional characterization of the galactan utilization system of Geobacillus stearothermophilus. FEBS J 280:950–964. [DOI] [PubMed] [Google Scholar]
- 32.Ryttersgaard C, Le Nours J, Lo Leggio L, Jorgensen CT, Christensen LL, Bjornvad M, Larsen S. 2004. The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. J Mol Biol 341:107–117. doi: 10.1016/j.jmb.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 33.Ryttersgaard C, Lo Leggio L, Coutinho PM, Henrissat B, Larsen S. 2002. Aspergillus aculeatus beta-1,4-galactanase: substrate recognition and relations to other glycoside hydrolases in clan GH-A. Biochemistry 41:15135–15143. doi: 10.1021/bi026238c. [DOI] [PubMed] [Google Scholar]
- 34.Hinz SW, Pastink MI, van den Broek LA, Vincken JP, Voragen AG. 2005. Bifidobacterium longum endogalactanase liberates galactotriose from type I galactans. Appl Environ Microbiol 71:5501–5510. doi: 10.1128/AEM.71.9.5501-5510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saier MH Jr, Goldman SR, Maile RR, Moreno MS, Weyler W, Yang N, Paulsen IT. 2002. Transport capabilities encoded within the Bacillus subtilis genome. J Mol Microbiol Biotechnol 4:37–67. [PubMed] [Google Scholar]
- 36.Ferreira MJ, Sá-Nogueira I. 2010. A multitask ATPase serving different ABC-type sugar importers in Bacillus subtilis. J Bacteriol 192:5312–5318. doi: 10.1128/JB.00832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schönert S, Seitz S, Krafft H, Feuerbaum EA, Andernach I, Witz G, Dahl MK. 2006. Maltose and maltodextrin utilization by Bacillus subtilis. J Bacteriol 188:3911–3922. doi: 10.1128/JB.00213-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juers DH, Matthews BW, Huber RE. 2012. LacZ beta-galactosidase: structure and function of an enzyme of historical and molecular biological importance. Protein Sci 21:1792–1807. doi: 10.1002/pro.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Härtig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rügheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, et al. . 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 40.Fujita Y. 2009. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci Biotechnol Biochem 73:245–259. doi: 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- 41.Aung-Hilbrich LM, Seidel G, Wagner A, Hillen W. 2002. Quantification of the influence of HPrSer46P on CcpA-cre interaction. J Mol Biol 319:77–85. doi: 10.1016/S0022-2836(02)00245-0. [DOI] [PubMed] [Google Scholar]
- 42.Ramström H, Sanglier S, Leize-Wagner E, Philippe C, Van Dorsselaer A, Haiech J. 2003. Properties and regulation of the bifunctional enzyme HPr kinase/phosphatase in Bacillus subtilis. J Biol Chem 278:1174–1185. doi: 10.1074/jbc.M209052200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.