ABSTRACT
Listeria monocytogenes is a significant foodborne human pathogen that can cause severe disease in certain high-risk individuals. L. monocytogenes is known to produce high-molecular-weight, phage tail-like bacteriocins, or “monocins,” upon induction of the SOS system. In this work, we purified and characterized monocins and found them to be a new class of F-type bacteriocins. The L. monocytogenes monocin genetic locus was cloned and expressed in Bacillus subtilis, producing specifically targeted bactericidal particles. The receptor binding protein, which determines target cell specificity, was identified and engineered to change the bactericidal spectrum. Unlike the F-type pyocins of Pseudomonas aeruginosa, which are related to lambda-like phage tails, monocins are more closely related to TP901-1-like phage tails, structures not previously known to function as bacteriocins. Monocins therefore represent a new class of phage tail-like bacteriocins. It appears that multiple classes of phage tails and their related bacteriocins have coevolved separately in parallel.
IMPORTANCE Phage tail-like bacteriocins (PTLBs) are structures widespread among the members of the bacterial kingdom that are evolutionarily related to the DNA delivery organelles of phages (tails). We identified and characterized “monocins” of Listeria monocytogenes and showed that they are related to the tail structures of TP901-1-like phages, structures not previously known to function as bacteriocins. Our results show that multiple types of envelope-penetrating machines have coevolved in parallel to function either for DNA delivery (phages) or as membrane-disrupting bacteriocins. While it has commonly been assumed that these structures were coopted from phages, we cannot rule out the opposite possibility, that ancient phages coopted complex bacteriocins from the cell, which then underwent adaptations to become efficient at translocating DNA.
INTRODUCTION
Two types of high-molecular-mass (>106-Da), phage tail-like bacteriocins (PTLBs), the R type and the F type, are known to exist in the bacterial kingdom (1, 2). R-type bacteriocins (RTBs) are contractile nanotube machines evolutionarily related to Myoviridae tails (e.g., that of bacteriophage T4) (3), type 6 secretion systems (4, 5), insecticidal protein injector complexes (6, 7), and structures involved in bacterium-marine animal interactions (8). RTBs are composed of a tube surrounded by a contractile sheath (9). At one end of the sheath is a complex baseplate structure to which six receptor binding proteins (RBPs) are attached. RTBs kill target bacteria by first binding to a receptor on the cell surface via RBPs. This event triggers sheath contraction, which then drives the tube structure through the cell envelope. The result is a dissipation of the membrane potential and cell death. Contact with a single RTB particle is sufficient to kill a cell (10). Far less studied are the F-type bacteriocins (FTBs). These high-molecular-weight bactericidal protein structures are evolutionarily related to the noncontractile structures of Siphoviridae (e.g., bacteriophage lambda) tails and are exemplified by the F-type pyocins of Pseudomonas aeruginosa (11–14). FTBs are structurally less complex than RTBs and consist of a flexible noncontractile tube attached to a baseplate structure to which RBPs are attached. As with RTBs, contact with a single FTB particle can result in cell death; however, the mechanism of cell killing is not well understood. There is no core or tube to be driven into the cell to form a channel as is the case with RTBs. However, given the similar potency, it might be expected that FTBs also disrupt the cell membrane potential via creation of a pore.
RTBs and FTBs share many common features. Both are produced intracellularly upon induction, usually of the SOS response. After intracellular assembly, the particles are released into the medium by cell lysis, and they can go on to kill competing target cells in the medium. Sister cells of the producer strain are typically not sensitive to the bactericidal activity of their own bacteriocin. Production of these structures can be viewed as a form of altruism; cells must sacrifice themselves by lysis to release the bacteriocins and thereby provide sister cells a competitive advantage by killing other (sensitive) bacteria competing for the same niche. The target bacteria are usually other strains within the same species, but sometimes they are members of other species or genera. An example of an R-type bacteriocin providing its producer a competitive advantage in vivo is described by Morales-Soto and Forst (15).
Several strains of Listeria monocytogenes and Listeria innocua were shown to produce PTLBs termed “monocins” upon induction of the SOS response, and these have been used for bacterial typing (16–18). We found that these structures are not related to known PTLBs but rather to the tail structures of TP901-1-like phages. We identified the monocin genetic locus, cloned and functionally expressed the monocin genes in Bacillus subtilis, and analyzed the bactericidal spectra of the resulting monocins. By engineering the monocin RBP, it was possible to retarget strain specificity. This is the first example of an FTB that has been cloned, expressed, and engineered to retarget its killing spectrum. These studies indicate that multiple classes of phage organelles which function as DNA delivery machines and PTLBs have independently diverged and evolved in parallel.
MATERIALS AND METHODS
Primers, plasmid constructs, and strain constructs are shown in Tables S1 to S3 in the supplemental material.
Bacterial growth.
Listeria strains were propagated in nutrient broth (Difco) or on 1.5% nutrient agar plates at 30°C. Bacillus strains were maintained on Trypticase soy broth (TSB) plates supplemented with chloramphenicol (5 μg/ml [final concentration]).
Molecular biology.
All enzymes were obtained from New England BioLabs (Ipswich, MA) unless stated otherwise. PCRs were carried out using Phusion polymerase. Ligation of the hyper-spank promoter (Phyper-spank) into the vector backbone pDG630 was carried out using T4 DNA ligase. The monocin gene cluster and its variants were cloned into the hyper-spank promoter plasmid by using Gibson assembly. B. subtilis genomic DNA was isolated using a MasterPure Gram-positive DNA purification kit (Illumina, San Diego, CA).
Bacteriocin killing assays.
Spot killing assays were performed as previously described (19, 20). Briefly, Listeria strains were grown to an optical density (OD) of ∼1.0. One hundred microliters of culture was added to a 0.5% nutrient agar overlay and poured onto a 1.5% nutrient agar plate. After the overlay was set, 3- to 4-μl drops of diluted monocin preparations were spotted onto the plate and allowed to dry/adsorb. The plates were incubated at 30°C overnight, and zones of clearing indicated bactericidal activity. For assay of activity against the target panel, a positive hit was scored if clearing was noted for a dilution of at least 1/125. The survival assay protocol was also performed as previously described (19), with the exceptions that the plates and cultures contained brain heart infusion (BHI) medium and all incubations were carried out at 30°C.
Monocin purification.
Listeria cultures were grown in nutrient broth at 30°C with shaking at 250 rpm; when the OD at 600 nm (OD600) reached ∼0.2, mitomycin C (Sigma) was added to a final concentration of 3 μg/ml. The cultures were then allowed to incubate overnight, after which lysis was typically noted by a clearing of the culture opacity and the presence of cellular debris. The cell debris was removed by centrifugation at 23,000 × g for 60 min. The supernatant was collected, and 10% (wt/vol) polyethylene glycol 8000 (PEG; OmniPure) was added along with 10% (vol/vol) 5 M NaCl. The PEG was allowed to dissolve, and the mixture was incubated overnight at 4°C. The PEG-precipitated material was then collected by centrifugation at 16,000 × g for 45 min. The pellet was resuspended in a 1/10 culture volume in TN50 (10 mM Tris, 50 mM NaCl, pH 7.5). Monocins were collected by high-speed ultracentrifugation at 90,000 × g for 3 to 4 h, and the pellets were resuspended in 1/50 of the original culture volume.
Bacillus subtilis monocin expression strains were grown in TSB at 37°C to an OD600 of approximately 0.2 to 0.4. They were then induced with either 5 mM H2O2 or 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside), depending on the construct, shifted to 28°C, and allowed to express the monocins overnight. Purification of monocins was similar to that from Listeria for the strains that contained the monocin lysis cassette. For constructs in which the lysis cassette was deleted, the cells were collected by centrifugation and resuspended in a 1/10 culture volume of TN50, lysozyme was added (1 mg/ml), and the paste was sonicated using a Biologics Inc. V/T sonicator at half power with three 30-s pulses. The sonicate was centrifuged at 23,000 × g to remove debris, and the supernatant was PEG precipitated and ultracentrifuged as described above.
To identify monocin proteins, purified samples were electrophoresed for a distance of 0.5 cm by 10% SDS-PAGE (Invitrogen), followed by Coomassie blue staining. The single band containing all monocin proteins was excised, digested in-gel with trypsin, and subjected to tandem mass spectrometry (MS-MS) analysis conducted at the University of California, Davis, Proteomics Core.
Cloning and expressing the L. monocytogenes strain 35152 monocin (M35152) in Bacillus subtilis.
Primers, plasmids, and strains are listed in Tables S1 to S3 in the supplemental material. The monocin gene cluster (ftbA to ftbR) was PCR amplified from genomic DNA isolated from Listeria monocytogenes strain 35152 by using primers oGL-054 and oGL-057. The PCR product was cloned into the Bacillus subtilis integration vector pDG630 between the AscI and NotI sites and ligated with T4 DNA ligase to create plasmid pGL-031. The monocin gene cluster was flanked by amyE sequences to promote homologous recombination into the amyE locus. Also carried within the plasmid pGL-031 is the cat gene for selection of recombinants.
pGL-031 was linearized and transformed into the Bacillus subtilis host strain BDG9 (21). Chloramphenicol-resistant colonies were selected, and DNAs were isolated and screened by PCR to verify proper integration. The strain was termed sGL-064.
Deletion of lysis genes.
To remove the predicted holin and lysin genes (ftbQ and ftbR) from the monocin gene cluster, the region containing ftbA to ftbP was PCR amplified from L. monocytogenes 35152 genomic DNA by using primers oGL-054 and oUC-001. The PCR product and the vector DG630 were both digested with the restriction enzymes AscI and NotI and ligated together using T4 DNA ligase. The resultant construct was named pUC-001. After integration into BDG9 as described above, the resulting integrant strain was termed sUC-001.
Introducing Phyper-spank.
To generate a version of pDG630 with an inducible promoter driving the expression of the monocin genes, the B. subtilis hyper-spank promoter along with the lacI gene was PCR amplified from plasmid pDR111 (a gift from David Rudner, Harvard Medical School) by using primers oGL-084 and oGL-085. The PCR product was digested with AscI and NotI and ligated into vector pDG630 (21) which had also been digested with the same restriction enzymes. The resultant construct was named pGL-034. The monocin gene cluster from ftbD to ftbP was PCR amplified using primers oGL-086 and oGL-087. The PCR product was then cloned into HindIII-digested pGL-034 by using Gibson assembly. The manufacturer's standard protocol was used. The resultant construct was named pGL-036. After integration into BDG9, the resulting strain was termed sGL-071. Similarly, to place Phyper-spank upstream of ftbF, we used primers oGL-086 and oGL-090 to create pGL-039, which was transformed into B. subtilis to create sGL-077.
Construction of Bacillus subtilis strain Δ8.
Bacillus subtilis knockouts were made following the methods described by Tanaka et al. (22). Strain Δ6 was described by Westers et al. (23) and is a prophage deletion strain. In order to manipulate strain Δ6 further, we removed the cat gene, a remnant of the pks operon knockout (23). First, the upp::kan marker was amplified from Bacillus subtilis strain TF8A λPr-neo::Δupp by use of primers oDG1013 and oDG1014. The PCR product was cloned into pETcoco1 linearized with NotI. The plasmid was then linearized with SpeI, transformed into Δ6, and selected for Kanr. The strain was termed BDG243. To delete the cat gene, the phleomycin cassette with two flanking regions from Δ6 was amplified from pUC18 (22) in a sewing PCR by using the primers oDG1001 and oDG1002 (left flank), oDG999 and oDG1000 (phleomycin cassette), and oDG1003 and oDG1004 (right flank), and the three pieces were combined by amplification with the two outside primers oDG1001 and oDG1004. The PCR product was transformed into BDG243 and selected on phleomycin plates, followed by screening for kanamycin sensitivity. The resultant strain was designated BDG247. The phleomycin marker was deleted by growing BDG247 in LB without selection for 4 h and plating the strain on kanamycin, and colonies were picked and screened for phleomycin sensitivity. The resultant strain, BDG252, is a markerless knockout strain that is useful for making further modifications. To delete hag, the 5′-flanking region of hag was amplified with primers oDG1019 and oDG1020, the 3′ flank was amplified with oDG1021 and oDG1022, and the phleomycin cassette was amplified with oDG999 and oDG1000. The three PCR products were combined and a sewing reaction performed with oDG1019 and oDG1022. The product was transformed into BDG252, selected on phleomycin, and then screened for kanamycin sensitivity to create BDG253. The phleomycin marker was again deleted by growing BDG253 in LB without selection for 4 h, plating the strain on kanamycin, and screening colonies for phleomycin sensitivity to create strain BDG255. To delete spoIIga, the 5′ flank was amplified with primers oDG1023 and oDG1024, the 3′ flank was amplified with oDG1025 and oDG1026, and the phleomycin cassette was amplified with oDG999 and oDG1000. The three products were combined in a sewing reaction using primers oDG1023 and oDG1026. After transformation, selection on phleomycin, and screening for phleomycin resistance, the resulting strain was named BDG256. The phleomycin marker was deleted, again by growing BDG256 in LB without selection for 4 h, plating it on kanamycin, and screening colonies for phleomycin sensitivity, to create strain BDG257, also known as the Δ8 strain.
Construction of M35152-A118.
A118 was obtained from Richard Calendar (University of California, Berkeley, CA) and was from a stock of the originally sequenced phage. The 35152 monocin gene cluster of ftbD to the 5′ portion of ftbP was PCR amplified using primers oGL-086 and oGL-112. The A118 phage tail fiber gene was PCR amplified from phage genomic DNA by using the forward primer oGL-120 and the reverse primer oGL-162, oGL-163, oGL-164, or oGL-165 to include three, two, one, or no downstream chaperone. The monocin PCR product and each of the A118 PCR products were cloned in a three-piece Gibson assembly into HindIII-digested pGL-034. The constructs were named pGL-075, pGL-076, pGL-077, and pGL-078, and the resulting integrants were named sGL-364, sGL-158, sGL-365, and sGL-366, respectively.
EM.
Monocins were purified by sucrose density gradient centrifugation. Two microliters of purified monocin was placed onto a carbon-coated copper electron microscopy (EM) grid. After 1 min, the sample was blotted from the edge of the grid with filter paper to leave a thin layer of sample, which was subsequently stained with 2% uranyl acetate for 30 s. The stain solution was blotted with filter paper. This staining and blotting step was repeated five times. The sample grid was air dried and inserted into an FEI Tecnai F20 transmission electron microscope. Images were recorded on a TVIPS 16-megapixel charge-coupled device (CCD) camera.
RESULTS
Identification of the monocin genetic locus.
Listeria monocytogenes strain ATCC 35152 and Listeria innocua strain ATCC 33090 were both reported to produce monocins (16). To confirm these observations, we induced expression of the monocins by using mitomycin C, harvested the particles by ultracentrifugation, and then tested the resulting preparations for bactericidal activity against Listeria strains by the spot method (see Materials and Methods) (Table 1). The preparations were designated M35152 and M33090, respectively, and were found to have differing bactericidal spectra. M35152 has a broad spectrum that consists mainly of serotype 4b strains (Table 1). M33090 has a much narrower spectrum, forming spots only on L. monocytogenes 19111 and Listeria ivanovii 19119 (data not shown). Consistent with other PTLBs, neither preparation showed bactericidal activity against its own natural producer strain.
TABLE 1.
Bactericidal activities of monocins on a panel of Listeria strains
| Strain | Other designation | Source | Serotype | Sensitivity to monocin: |
Phage A118 plaque formation | ||
|---|---|---|---|---|---|---|---|
| M35152 (from Listeria) | M35152 (from Bacillus) | M35152-A118 (from Bacillus) | |||||
| 15313 | ATCC | 1/2a | No | No | Yes | Yes | |
| 35152 | ATCC | 1/2a | No | No | Yes | Yes | |
| 19111 | ATCC | 1/2a | Yes/noa | No | Yes | Yes | |
| DP-L4056 | 10403s cured of phage | Daniel Portnoy | 1/2a | No | No | Yes | Yes |
| DP-L3817 | 1993 Halifax | Daniel Portnoy | 1/2a | No | No | Yes | No |
| DP-L3633 | EGDe | Daniel Portnoy | 1/2a | No | No | Yes | No |
| DP-L1171 | Daniel Portnoy | 1/2b | No | No | Yes | No | |
| DP-L3293 | LO28 | Daniel Portnoy | 1/2c | No | No | Yes | No |
| DP-L188 | ATCC 19113 | Daniel Portnoy | 3 | No | No | No | No |
| 23074 | ATCC | 4b | Yes | Yes | No | No | |
| DP-L185 | F2397 | Daniel Portnoy | 4b | Yes | Yes | No | No |
| DP-L186 | ScottA | Daniel Portnoy | 4b | Yes | Yes | No | No |
| DP-L1173 | Daniel Portnoy | 4b | Yes | Yes | No | No | |
| DP-L1174 | Daniel Portnoy | 4b | Yes | Yes | No | No | |
| DP-L1168 | Daniel Portnoy | 4b | Yes | Yes | No | No | |
| DP-L1169 | Daniel Portnoy | 4b | Yes | Yes | No | No | |
| 33030 | ATCC (L. innocua) | Yes | Yes | No | No | ||
Strain 19111 was originally scored as sensitive, but a reexamination of this result indicated that it may be incorrect (see Discussion).
Monocin preparations were analyzed by mass spectrometry (MS) to identify proteins present in the samples that had similarity to components of phage tail-like structures. Although genome sequences for strains 35152 and 33090 are not available, numerous other Listeria genomes have been sequenced. Querying these genomes revealed open reading frames (ORFs) whose gene products matched the monocin peptides. The most abundant protein in the M35152 preparation corresponded to the lmaA gene, or antigen A gene, carried by numerous L. monocytogenes strains. Antigen A is a protein originally found to elicit an immune response in Listeria infections (see Discussion). BLAST searches showed that it has high sequence similarity to the major tail protein of TP901-1-like Siphoviridae bacteriophages of Gram-positive bacteria, including the Listeria phage A118 (24). The M35152 antigen A sequence has identical or nearly identical homologues in several known Listeria monocytogenes genomes. We chose strain 1/2a F6854 as a reference and used the gene numbering system of that strain for this work. Antigen A corresponds to gene 0131. The MS data for monocin preparations identified several other peptide matches that correspond to ORFs carried nearby in the genome, including ORFs 0130, 0132, 0135, 0136, 0138, and 0140. Several of these also have sequence similarities to phage tail proteins (Table 2).
TABLE 2.
Genes, predicted amino acid lengths, and putative functions of the monocin gene cluster
| ORFa | Gene designation | Product length (aa) | Putative product function | Phage A118 homologue (% aa identity) | Comment |
|---|---|---|---|---|---|
| 0125 | ftbA | 231 | Transcriptional regulator | ||
| 0126 | ftbB | 150 | Toxin/antitoxin | gp35 (46) | |
| 0127 | ftbC | 117 | Toxin/antitoxin, lambda C1, HTH | ||
| 0128 | ftbD | 142 | Antigen D (unknown function) | gp61 (46) | |
| 0129 | ftbE | 138 | Antigen C/transcriptional regulator | gp66 (39) | |
| 0130 | ftbF | 129 | Antigen B (unknown function) | ||
| 0131 | ftbG | 170 | Antigen A major phage tail | gp12 (15) | Strong similarity to several putative TP901-1-like prophage major tail proteins among diverse bacteria |
| 0132 | ftbH | 100 | Hypothetical phage-related protein | ||
| 0133 | ftbI | 130 | Hypothetical phage-related protein | ||
| 0134 | ftbJ | 622 | Tape measure | gp16 (19) | Less than half the length of the A118 tape measure |
| 0135 | ftbK | 272 | Phage tail component | gp17 (36) | |
| 0136 | ftbL | 378 | Phage tail component | gp18 (33) | |
| 0137 | ftbM | 99 | Hypothetical phage-related protein | ||
| 0138 | ftbN | 191 | Hypothetical phage-related protein | Potential peptidase | |
| 0139 | ftbO | 159 | Hypothetical phage-related protein | ||
| 0140 | ftbP | 178 | Receptor binding | gp20 | 50% identity between aa 36 to 68 and A118 gp20 |
| 0141 | ftbQ | 140 | Holin | ||
| 0142 | ftbR | 242 | Lysin |
The ORF numbering system is based on strain F6854.
A close inspection of this genomic region revealed that ORFs 0130 to 0140 encode components analogous to a Siphoviridae tail structure module (Fig. 1). In addition to the above-described genes, ORF 0134 encodes a putative tape measure protein, and ORFs 0137 and 0139 encode putative proteins that share sequence similarity to known phage tail proteins. These ORFs are all transcribed on one strand, with few or no intergenic spaces. Just downstream of the structural genes, ORFs 0141 and 0142 encode putative holin and lysin proteins, which, in general, are responsible for timed cell lysis for particle release (25). Upstream of ORF 0130, we annotated five putative regulatory genes. ORF 0125 has sequence similarity to transcriptional regulator genes, ORFs 0126 and 0127 have sequence similarity to phage regulatory protein genes, and ORFs 0128 and 0129 have sequence similarities to antigen D and antigen C, respectively (see Discussion), and some similarity to transcriptional regulators (Table 2). ORFs 0125 to 0127 are transcribed in the opposite direction from the structural genes, while ORFs 0128 and 0129 are transcribed in the same direction as the structural genes, suggesting that an operator region lies between the diverging transcripts. No genes encoding phage capsid, capsid assembly, or portal proteins were found in this region or nearby in the genome. Also not present were any genes encoding DNA replication or packaging machinery or integration/excision proteins or any other ORFs often associated with temperate prophages. Thus, this region is consistent with a locus encoding an F-type bacteriocin. The locus on the genome is found between nucleotides 107301 and 118941 with the strain F6854 annotation. This corresponds to nucleotides 119740 to 131380 for EGD-e and 116542 to 128182 for 10403s. Because some of the genes of this locus share sequence similarity to phage genes, it has been annotated a prophage insertion and designated LambdaLm01 or the lma locus by some genome sequencing groups characterizing other Listeria strains. We chose to rename these genes ftbA to ftbR (for F-type bacteriocin) since we now know the functions of the products of these genes.
FIG 1.
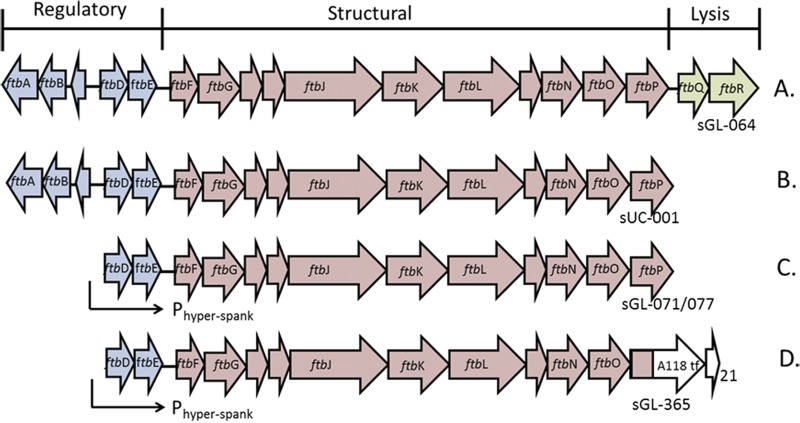
Gene clusters encoding monocins. The blue genes represent putative regulatory protein genes, the pink genes encode structural proteins, and the green genes form the lysis cassette encoding holin and lysin. (A) Entire wild-type gene cluster. (B) Holin/lysin deletion construct (sUC-001). (C) Construct under the control of the inducible Phyper-spank promoter (sGL-071). (D) Monocin gene cluster with the tail fiber fusion plus the downstream chaperone (sGL-365).
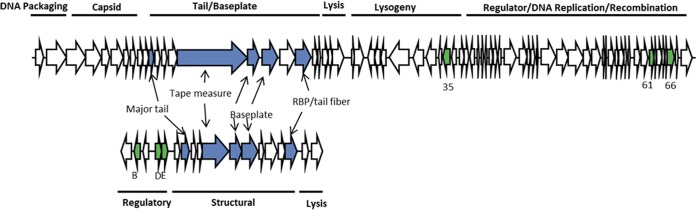
Viewing the monocin gene cluster as a whole, its closest Listeria phage tail relatives are those of A118-like phages (Fig. 2; Table 2). However, the similarity is relatively weak. The ftbK and ftbL genes encode predicted baseplate components that are the two most similar structural proteins, with 33 to 36% amino acid identities. The tape measure protein is only weakly similar to (19% identity) and half the size of the A118 counterpart. The receptor binding protein encoded by ftbP shares only a small region of similarity with the A118 tail fiber. The major tail protein actually shares more sequence similarity with other putative TP901-1 phage counterparts than with the A118 counterpart. BLAST searches revealed several major tail protein homologues encoded within the genomes of several Gram-positive bacterial species, most of which appear to be part of uncharacterized prophages. For example, the major tail protein shares 73% amino acid sequence similarity to a tail protein of Paenibacillus larvae (GenBank accession no. WP_023484977) and 72% similarity to that of Enterococcus avium 14025 (GenBank accession no. WP_016180823). Five putative structural genes, ftbH, ftbI, ftbM, ftbN, and ftbO, share no recognizable sequence similarity to their A118 counterparts and very little similarity to any other phage genes in any bacteria, although we predict that ftbM, ftbN, and ftbO encode baseplate components, based on their position within the cluster. Three putative regulatory proteins, encoded by ftbB, ftbD, and ftbE, are more similar to their A118 counterparts than any of the structural proteins, with sequence identities ranging from 39 to 49%; however, the genes are not clustered together in the A118 genome as they are in the monocin cluster. The lysis cassette is not related by sequence similarity to the A118 lysis genes.
FIG 2.
Comparison of the monocin gene cluster and bacteriophage A118. The monocin structural genes are analogous to the Siphoviridae phage tail module. The monocin regulatory genes also have A118 homologues, although they are not in the same context (also see Table 2).
Expressing the 35152 monocin (M35152) in Bacillus subtilis.
The monocin gene cluster from ftbA to ftbR was cloned and integrated into the genome of Bacillus subtilis strain BDG9 (see Materials and Methods) to generate strain sGL-064. Monocin was produced from sGL-064 by induction of the SOS response with addition of hydrogen peroxide, which we found to be more reliable than mitomycin C for induction in B. subtilis. Monocin particles recovered from cell lysates were tested by spot assay on a panel of target strains (Table 1). The bactericidal spectrum of the recombinant monocin corresponded to that of the wild type, with the exception of one strain (see below and Discussion). For subsequent work, we chose ATCC 23074 (serotype 4b) as the standard assay strain for studying M35152.
The entire M35152 gene cluster was sequenced by primer walking. The locus was found to be highly similar to that of F6854, with six of the predicted proteins being identical, seven having only 1 or 2 amino acid substitutions, three having 6 or 7 amino acid substitutions, and the most divergent (FtbP) having 25 amino acid substitutions. The monocin locus has a GC content of 37%, the same as that of the L. monocytogenes genome.
In order to produce monocins intracellularly for ease of production and potentially higher yields, we deleted ftpQ and ftbR (encoding holin and lysin, respectively) (Fig. 1B). The resultant strain, sUC-001, produced monocins primarily intracellularly upon SOS induction, with only minor (<5%) leakage into the culture medium in older cultures (data not shown).
Expression of M35152 from an inducible promoter.
Yields of monocins from B. subtilis were still quite low, showing bactericidal activity only when spotted undiluted on a lawn. We suspected that the natural monocin SOS regulatory circuit was not functioning well in Bacillus. To bypass the need for SOS induction and to increase yields, we eliminated the three upstream putative regulatory genes (ftbA, ftbB, and ftbC) and placed the IPTG-inducible B. subtilis Phyper-spank promoter upstream of ftbD to drive transcription of all of the remaining monocin genes (Fig. 1C). We also generated a construct in which Phyper-spank was placed upstream of ftbF, since we believed that ftbD and ftbE encode regulatory proteins and that we could completely bypass natural monocin gene regulation by driving transcription of only the structural components. Both constructs also included lacI downstream of cat to provide reversible repression. The resulting Bacillus integrants were designated sGL-071 and sGL-077. Monocins could now be induced from these strains by addition of IPTG (see Materials and Methods). By spot assay, functional M35152 was produced by both strains; however, the yields were considerably different. When Phyper-spank was positioned upstream of ftbF (sGL-077), very little monocin was produced upon IPTG induction, and it was barely detectable by spot assay. However, Phyper-spank positioned in front of ftbD (sGL-071) gave robust, inducible yields: spot assays with 5-fold dilutions of purified monocin showed bactericidal activity at dilutions of 1/625 to 1/3,125 (Fig. 3A).
FIG 3.
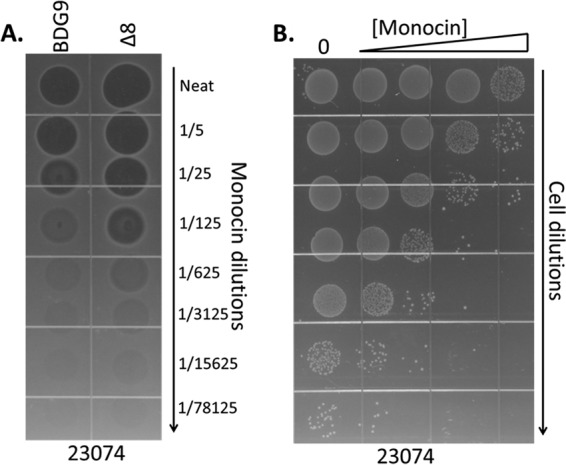
Bactericidal activity of M35152 on strain 23074. (A) Lawn spot assay of monocin activity to compare materials produced in the B. subtilis BDG9 and Δ8 backgrounds. 23074 was the target strain used to make the lawn. Clearing in the lawn indicates bactericidal activity. (B) Liquid survival assay showing killing activity. The target strain was 23074, and it was spotted directly onto an agar plate.
Improved B. subtilis expression strain.
To further improve monocin yields, we constructed a modified B. subtilis production strain deleted of prophage genes, sporulation functions, and flagellum synthesis genes. The hypothesis was that potential prophage production and flagellum synthesis could interfere with monocin production and that prevention of sporulation would allow cells to remain in a productive, vegetative state longer. We started with B. subtilis strain Δ6, which has a series of prophage element deletions, including deletions of prophage 1, prophage 3, SPβ, PBSX, and Skin (23). We further modified Δ6 by knocking out flagellum production (Δhag) and sporulation (ΔspoIIga) genes to generate strain Δ8 (see Materials and Methods). The M35152 gene cluster, minus the holin/lysin genes, was transformed/integrated into Δ8 and regulated with Phyper-spank upstream of ftbD as in sGL-071. The resulting strain, sGL-157 (M35152), had improved monocin production (typically 5- to 10-fold) over that of its BDG9-based counterpart (Fig. 3A).
Monocins have potent bactericidal activity.
M35152 was produced from sGL-157 in sufficient amounts to perform quantitative survival assays to calculate the number of killing units (KU; the theoretical activity required to kill a single cell) in a given preparation (Fig. 3B). A 1-liter shake flask preparation can produce ∼1015 KU of active material. The specific activity is ∼1014 KU/mg of protein. Assuming that the molecular mass of monocins is similar to that of F-type pyocins, i.e., 2 × 106 to 3 × 106 Da (11), a monocin KU corresponds roughly to 2 or 3 purified recombinant monocin particles. This is close to the theoretical maximum potency of a single PTLB particle resulting in cell death, an observation also noted with both R- and F-type pyocins of Pseudomonas aeruginosa.
Generating a recombinant RBP by using the tail fiber of listeriophage A118.
Previous studies with other PTLBs demonstrated that it is possible to retarget the bactericidal spectrum of an RTB by making fusions between the native RBP and heterologous RBPs from phages, either tail fibers or tail spikes (19–21, 26, 27). It appears almost universal that the N-terminal domain of such proteins forms the attachment to the baseplate of the RTB (or phage) and that the C-terminal domain interacts with a receptor on the target cell surface. The RBP (tail fiber; gp20) of phage A118 has a small region of amino acid similarity with the monocin product of ftbP, located at approximately amino acids 30 to 45. Based on this observation and its position within the gene cluster, we predicted ftbP to encode the monocin RBP. We designed constructs to fuse the putative baseplate binding function of the monocin RBP (amino acids 1 to 40) to the receptor binding function of the A118 tail fiber (gp20; amino acids 210 to 357).
In addition to the tail fiber fusion, we included 3 small ORFs located downstream of the A118 gp20 ORF. Many bacteriophage tail fibers are known to require additional proteins, often referred to as chaperones, for proper folding, and these are usually encoded just downstream of the tail fiber (19, 28). Bacteriophage A118 possesses three small ORFs, ORF 21 (105 amino acids), ORF 22 (52 amino acids), and ORF 23 (121 amino acids), in this position, any one of which might encode a protein that has this role. These 3 ORFs have little sequence similarity to any other known protein. To determine which, if any, might be required, we designed 4 constructs to include the ORF(s) downstream of the monocin/A118 tail fiber fusion. One construct included all three ORFs, one included ORFs 21 and 22, one included just ORF 21, and one construct had none of the putative chaperone ORFs. These were kept in the same context with respect to the end of the A118 tail fiber (i.e., same spacing and ribosome binding sites) (Fig. 4).
FIG 4.
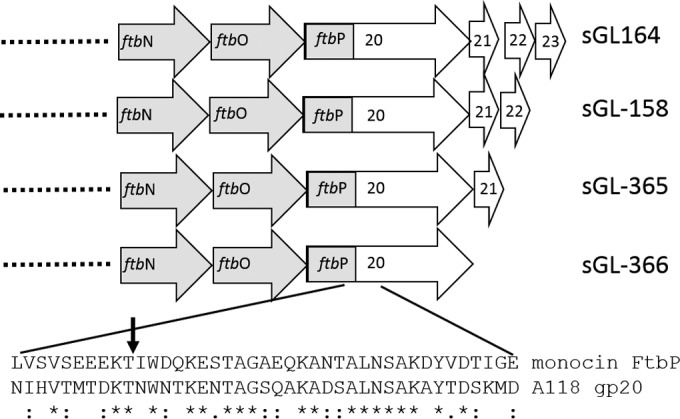
M35152-A118 constructs made in this study, showing the distal portion of the monocin cluster. The fusion site between FtbP and gp20 is shown with an arrow and includes amino acids 1 to 40 of FtbP and 210 to 357 of gp20. gp21, gp22, and gp23 were left in their natural context in relation to gp20.
These constructs were integrated into B. subtilis Δ8 as described above, generating strains sGL-366 (no downstream ORFs), sGL-365 (ORF 21), sGL-158 (ORFs 21 and 22), and sGL-364 (ORFs 21 to 23) (Fig. 4). Monocins were induced from each strain as described above and tested for activity by spot assay on Listeria monocytogenes 19111, a strain known to serve as a host for phage A118 and therefore predicted to be sensitive to a monocin equipped with the A118 RBP (Table 1; Fig. 5). When no putative downstream ORFs were present, no monocin activity was detected. When ORF 21 or both ORF 21 and ORF 22 were included in the expression construct, robust activity was seen. When all three downstream ORFs were present, there was a decreased monocin yield compared to that with ORF 21 or ORF 21 plus ORF 22 (Fig. 5B). The monocins produced from strains sGL-365 and sGL-158 were termed M35152-A118.
FIG 5.
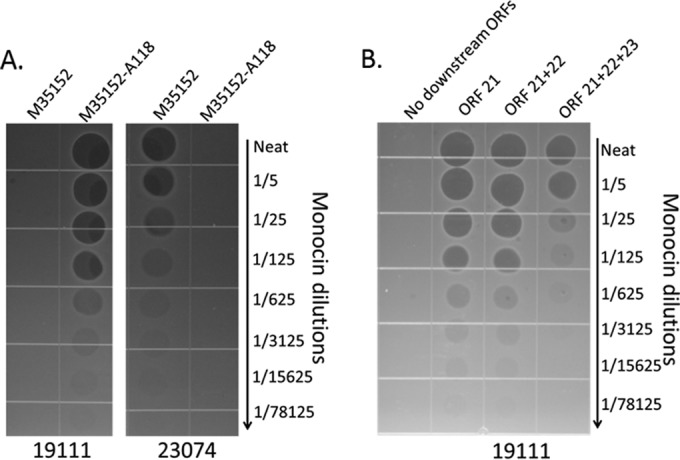
Bactericidal activity of the recombinant monocin M35152-A118. (A) Comparison of wild-type M35152 and M35152-A118 on two different L. monocytogenes strains, showing the switch in bactericidal spectrum upon engineering of the RBP. M35152 killed strain 23074 but not strain 19111, whereas M35152-A118 killed 19111 but not 23074. (B) Comparison of activities of the 4 putative downstream ORF constructs, showing that ORF 21 is required to generate functional M35152-A118.
While this work was being completed, it was shown directly that gp20 is the A118 RBP (29). It was also shown that gp21 is part of the A118 phage baseplate structure. To see if gp21 is also part of the structure of M35152-A118, ultracentrifugation-purified preparations of M35152-A118 were analyzed by mass spectrometry. gp21 was found to be associated with the monocin structure along with the RBP fusion protein (see Discussion).
Both M35152 and M35152-A118 produced in Bacillus subtilis were tested for bactericidal activity against a larger panel of Listeria strains (Table 1). M35152 killed all strains of the 4b serotype. M35152 targeted 1/2a strains as well as 1/2b and 1/2c strains, consistent with the fact that A118 infects these strains. One serotype 3 strain was tested and was insensitive to both monocins. One strain, serotype 1/2a strain ATCC 19111, seemed to be variably sensitive to material produced from Listeria, but the recombinant M35152 monocin did not show any signs of activity against this strain (see Discussion).
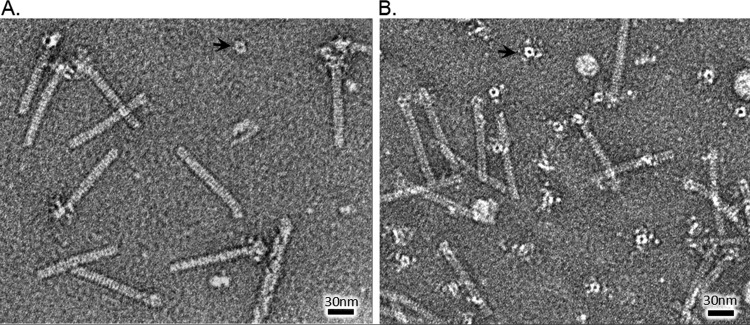
Representative electron micrographs of the monocins produced from sGL-157 and sGL-158 are shown in Fig. 6. The monocins are fully formed and consist of a rod structure of about 100 nm in length and a large baseplate complex. They are consistent with the monocin particles previously described for Listeria (17). The rod is made up of stacked disks, and the baseplate appears to be hexagonal, both of which are features resembling those of the TP901-1-like phage tail.
FIG 6.
Transmission electron micrographs of monocins produced in B. subtilis. (A) M35152 from strain sGL-157. (B) M35152-A118 from strain sGL-158. Arrows indicate detached tube disks.
DISCUSSION
Monocins now represent the best-characterized F-type bacteriocins (FTBs). We cloned and expressed the Listeria FTB gene cluster in Bacillus subtilis and identified the receptor binding protein (RBP), which can be engineered to alter the bactericidal spectrum. Like those of other PTLBs, the monocin genetic locus resembles a phage tail module, with a regulatory region, structural component genes, and a lysis cassette. Based on sequence similarity and gene order, the closest common ancestor of monocins is the tail structure of A118-like phages, which are TP901-1-like Siphoviridae phages. However, sequence similarity between A118 and monocins is low, suggesting that the divergence is ancient.
Both monocins and phage A118 are most closely related to lactococcal Siphoviridae phages (tails) TP901-1, p2, and Tuc2009 (30–33). Negatively stained EM images confirm that monocins likely have a similar structure (Fig. 6). The tail structures of these phages possess a complex baseplate in which the RBPs are arranged in an outer ring with six units, each composed of three “tripods,” each of which in turn is a trimer of the RBP, for a total of 54 RBP subunits (34, 35). This high density of RBPs would allow very high avidity of the monocin for the receptor, such that contact between a monocin and its cell target is likely to result in irreversible binding, and therefore in a bactericidal event. Indeed, we did observe potency consistent with approximately one particle killing one target cell in the survival assay. The cellular receptor for wild-type monocins has yet to be determined; however, the receptor for phage A118 (and likely M35152-A118) is known to be teichoic acid (29). Work is under way to determine if natural monocins also utilize teichoic acids or perhaps some other cell surface structure as a receptor.
The successful fusion of the A118 RBP (gp20) to FtbP confirms that ftbP encodes the monocin RBP and that a change in this protein alone changes the target cell specificity. Consistent with this is the observation that FtbP is the most divergent protein encoded among monocin gene clusters. This was noted with other PTLBs and is likely the result of selective pressure to acquire different target specificities. During the course of this work, it was verified experimentally by others that A118 gp20 is the phage's RBP and that gp21 is part of the A118 baseplate (29). We show here that fully functional production of M35152-A118 also requires the presence of gp21. Given its position in relation to gp20, we originally hypothesized that it might be a “chaperone” or tail fiber assembly protein required for the formation of trimers of gp20. If so, we might predict that it would not be part of the structure. However, given that it is part of both the phage A118 and M35152-A118 structures, this assessment is probably incorrect. gp21 likely is associated with the C terminus of gp20, since this is the portion transferred to the monocin, but its role remains unknown.
One strain of L. monocytogenes (ATCC 19111) was originally scored as sensitive to monocins produced by hydrogen peroxide-induced Listeria strain 35152. A reexamination of this result by looking at serial dilutions of monocin preparations from Listeria showed that it is likely to be incorrect. We believe that this variability is probably due to some other antibacterial entity induced in strain 35152 and is not a property of M35152 itself. Indeed, we conducted a study to identify new monocins from other Listeria strains and found that many strains variably produced particles likely to be phages that are coinduced with monocins (data not shown). It was often difficult to distinguish if a bactericidal activity was due to a monocin or due to “killing from without” by a temperate phage. By cloning the monocin into B. subtilis, we could examine the bactericidal properties of this structure without interference from contaminating antibacterial entities, such as phages. M35152 produced from Bacillus consistently and potently killed 4b strains, and M35152-A118 consistently killed 1/2a strains as well as 1/2b and 1/2c strains. Note that the killing spectrum of M35152-A118 is broader than the host range of phage A118, likely due to phage defense mechanisms that do not affect bacteriocin function.
This work sheds light on some confusing issues with this gene cluster. Gohmann et al. described antigens that elicit an immune response during Listeria infection (36). One of the proteins was named antigen A (lmaA), and it is encoded by ftbG. Here we show that the gene product of ftbG is actually the monocin major tail protein. There must be some amount of monocin induction during Listeria infection that elicits an immune response and antibody production. This group went on to clone surrounding genes, whose products they labeled antigens B, C, and D, without any evidence to indicate that they encoded actual antigens other than their proximity to lmaA (37); these correspond to ftbF, ftbE, and ftbD, respectively. These B, C, and D “antigen” designations have been inherited and annotated in several new genomes. Whether there is a biological significance to this remains to be seen, but researchers should be aware of the origin of these gene designations. Many other genome sequencing groups have annotated the monocin gene cluster a prophage insertion, LambdaLm01. We now know that this locus encodes a functional bacteriocin. Another level of confusion involves the lysis cassette. Previous work identified and cloned a putative monocin lysin gene (17). However, that particular gene shares little sequence similarity to the lysin gene identified in the FTB gene cluster in the present study; instead, it is nearly identical to a lysin associated with phage phiLM4 (GenBank accession no. ABG75905), prophage FSL F8-0076 (GenBank accession no. KES26481), and several L. monocytogenes prophages. It is likely that this early work misidentified a prophage lysin as the monocin lysin.
Another interesting note is that Hain et al. examined the lma locus, which is widespread among pathogenic Listeria species, and speculated that it may play a role in pathogenesis (38). Deletion of lmaB and lmaD (ftbF and ftbE) resulted in somewhat attenuated virulence in a mouse model; however, a mechanism for this observation remained unclear. To complicate matters, it was also recently shown that excision of A118-like prophage sequences (without infectious particle production) promotes virulence by restoring comK (39). Perhaps there is cross talk between phage/monocin regulatory proteins involved in this process. Considering the observation that monocin antibodies are produced during Listeria infection, one might assume that monocins are produced and released, possibly inside macrophages. Further studies are needed to determine whether there is any pathogenic function of monocin production during an L. monocytogenes infection.
Both R-type and F-type pyocins kill cells by a dissipation of the membrane potential. Given the high potency of monocins, we expect that they function similarly. The mechanics of achieving this membrane disruption for the two types of PTLB bactericidal activity appear to be quite different. R-type pyocin employs a contractile mechanism by which a core is inserted through the cell envelope, presumably to create a channel (9). F-type bacteriocins, on the other hand, have no sheath proteins to drive contractility. Recent data suggest that the tape measure protein of Siphoviridae phages becomes inserted into the inner membrane and is involved in DNA translocation (40). It is possible that PTLB tape measure proteins also insert into the inner membrane but that, instead of facilitating DNA translocation, they are involved in pore formation. Since both R- and F-type pyocins possess tape measure proteins, their mechanisms of killing may be very similar, with the difference being the mechanism by which the tape measure protein is delivered.
PTLBs appear to be widespread among the members of the bacterial kingdom, with examples from Rhizobium lupini (41), Xenorhabdus nematophilus (42), Xenorhabdus bovienii (43), Erwinia carotovora (44), Yersinia enterocolitica (45), Erwinia amylovora (46), Budvicia aquatic (47), Pragia fontium (47), Pseudomonas fluorescens (48), Stenotrophomonas maltophilia (49), Clostridium difficile (21), Pseudomonas aeruginosa (1), and Pseudomonas syringae (50). Those that have been studied in detail do not appear to share a single common PTLB ancestor, suggesting that each arose independently. Even on a functional level, contractile PTLBs are more similar to specific contractile phages than to each other. For example, R-type pyocins are related to PS17-like tails (51), RTBs of C. difficile are related to phiC2-like phages (21), and the Pseudomonas syringae RTB is related to a Mu-like tail structure (50). The same can be said for noncontractile PTLBs. The FTBs of P. aeruginosa are related to lambda-like phage tails (1), and we now show that F-type monocins are related to A118 and TP901-1 phage tails.
A common hypothesis is that PTLBs arose from “defective” prophages which lost genes encoding head proteins and replication machinery, while the remaining tail genes underwent adaptations, such as internal tube charge, that allowed them to function more efficiently as bacteriocins (9). One observation used to support the phage origin of PTLBs is that PS17 tails have bactericidal activity, albeit 100-fold lower than that of R-type pyocin (52). However, detailed structural comparisons of Myoviridae phage tails, type VI secretion systems, and insecticidal contractile complexes revealed a common ancestry for envelope-penetrating complexes, without clear evidence that they originated in either phages or bacteria (3, 30, 53). These recent observations might support the alternate hypothesis that Myoviridae phages evolved by acquisition of a contractile nanotube module, perhaps even one that functioned as a bacteriocin. Monocins are clearly related to TP901-1-like phage tails. However, they share little sequence similarity to their closest known Listeria phage relatives, suggesting that the divergence was quite ancient, and one might not a priori assume that they are descendants of degenerate prophages. It is possible that TP901-1-like phage tails and monocins share an ancestor that predates Siphoviridae phages. However, the picture is further blurred by the observation that horizontal gene transfer plays a major role in phage evolution (54), and probably also in PTLB evolution. The monocin major tail protein shares as much as 73% sequence similarity with a putative phage protein in Paenibacillus larvae and with other TP901-1-like prophages in other bacterial species, suggesting that there is still potential for genetic exchange between the two entities.
Monocins have very potent bactericidal activity and can be produced at high concentrations in a food-grade production organism. Two monocins, the natural monocin M35152 and the engineered monocin M35152-A118 created in this work, together kill serotype 4b and 1/2a strains, which are among the most significant foodborne isolates (55). These structures may potentially be deployed in food safety applications to precisely kill pathogenic Listeria strains and to reduce the incidence of illness.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel Portnoy and Richard Calendar for supplying strains and phages. We are grateful to the AvidBiotics scientific advisory board, especially Jeffery F. Miller, Martin Blaser, and John Mekalanos, for helpful suggestions and reviews of this work. We also thank Brett Phinney and the staff at the UC Davis proteomic core for assistance with the mass spectrometry analysis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00489-16.
REFERENCES
- 1.Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol 38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 2.Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499–510. doi: 10.1016/S0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 3.Leiman PG, Shneider MM. 2012. Contractile tail machines of bacteriophages. Adv Exp Med Biol 726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- 4.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell AB, Peterson SB, Mougous JD. 2014. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurst MRH, Glare TR, Jackson TA. 2004. Cloning Serratia entomophila antifeeding genes: a putative defective prophage active against the grass grub Costelytra zealandica. J Bacteriol 186:5116–5128. doi: 10.1128/JB.186.15.5116-5128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang G, Dowling AJ, Gerike U, Ffrench-Constant RH, Waterfield NR. 2006. Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J Bacteriol 188:2254–2261. doi: 10.1128/JB.188.6.2254-2261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ, Newman DK. 2014. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 343:529–533. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge P, Scholl D, Leiman PG, Yu X, Miller JF, Zhou ZH. 2015. Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states. Nat Struct Mol Biol 22:377–382. doi: 10.1038/nsmb.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uratani Y, Hoshino T. 1984. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J Bacteriol 157:632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuroda K, Kageyama M, Maeda T, Fujime S. 1979. Physicochemical properties of pyocin F1. J Biochem 85:21–28. [DOI] [PubMed] [Google Scholar]
- 12.Kuroda K, Kageyama M. 1979. Biochemical properties of a new flexuous bacteriocin, pyocin F1, produced by Pseudomonas aeruginosa. J Biochem 85:7–19. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda K, Kageyama M. 1981. Comparative study of F-type pyocins of Pseudomonas aeruginosa. J Biochem 89:1721–1736. [DOI] [PubMed] [Google Scholar]
- 14.Kuroda K, Kageyama M. 1983. Biochemical relationships with three F-type pyocins, pyocin F1, F2, and F3, and phage KF1. J Biochem 94:1429–1441. [PubMed] [Google Scholar]
- 15.Morales-Soto M, Forst S. 2011. The xnp1 P2-like tail synthesis gene cluster encodes xenorhabdicin and is required for interspecies competition. J Bacteriol 193:3624–3632. doi: 10.1128/JB.00092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zink R, Loessner MJ, Glas I, Scherer S. 1994. Supplementary Listeria-typing with defective Listeria phage particles (monocins). Lett Appl Microbiol 19:99–101. doi: 10.1111/j.1472-765X.1994.tb00915.x. [DOI] [PubMed] [Google Scholar]
- 17.Zink R, Loessner MJ, Scherer S. 1995. Characterization of cryptic prophages (monocins) in Listeria and sequence analysis of a holin/lysin gene. Microbiology 141:2577–2584. doi: 10.1099/13500872-141-10-2577. [DOI] [PubMed] [Google Scholar]
- 18.Bannerman E, Boerlin P, Bille J. 1996. Typing of Listeria monocytogenes by monocin and phage receptors. Int J Food Microbiol 31:245–262. doi: 10.1016/0168-1605(96)01003-3. [DOI] [PubMed] [Google Scholar]
- 19.Williams S, Gebhart D, Martin DW, Scholl D. 2008. Re-targeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol 74:3868–3876. doi: 10.1128/AEM.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholl D, Cooley M, Williams S, Gebhart D, Martin D, Bates A, Mandrell R. 2009. An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the foodborne pathogen, Escherichia coli O157:H7. Antimicrob Agents Chemother 53:3074–3080. doi: 10.1128/AAC.01660-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gebhart D, Williams SR, Bishop-Lilly KA, Govoni GR, Willner KM, Butani A, Sozhamannan S, Martin D, Fortier LC, Scholl D. 2012. Novel high-molecular-weight, R-type bacteriocins of Clostridium difficile. J Bacteriol 194:6240–6247. doi: 10.1128/JB.01272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka K, Henry CS, Zinner JF, Jolivet E, Cohoon MP, Xia F, Bidnenko V, Ehrlich SD, Stevens RL, Noirot P. 2013. Building the repertoire of dispensable chromosome regions in Bacillus subtilis entails major refinement of cognate large-scale metabolic model. Nucleic Acids Res 41:687–699. doi: 10.1093/nar/gks963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westers H, Dorenbos R, van Dijl JM, Kabel J, Flanagan T, Devine KM, Jude F, Seror SJ, Beekman AC, Darmon E, Eschevins C, de Jong A, Bron S, Kuipers OP, Albertini AM, Antelmann H, Hecker M, Zamboni N, Sauer U, Bruand C, Ehrlich DS, Alonso JC, Salas M, Quax WJ. 2003. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol Biol Evol 20:2076–2090. doi: 10.1093/molbev/msg219. [DOI] [PubMed] [Google Scholar]
- 24.Loessner MJ, Inman RB, Lauer P, Calendar R. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol Microbiol 35:324–340. doi: 10.1046/j.1365-2958.2000.01720.x. [DOI] [PubMed] [Google Scholar]
- 25.Young R. 2013. Phage lysis: do we have the hole story yet? Curr Opin Microbiol 16:790–797. doi: 10.1016/j.mib.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scholl D, Gebhart D, Williams S, Bates A, Mandrell R. 2012. Genome sequence of E. coli O104:H4 leads to rapid development of a targeted antimicrobial agent against this emerging pathogen. PLoS One 7:e33637. doi: 10.1371/journal.pone.0033637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gebhart D, Lok S, Clare S, Tomas M, Stares M, Scholl D, Donskey CJ, Lawley TD, Govoni GR. 2015. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. mBio 6:e02368-14. doi: 10.1128/mBio.02368-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashemolhosseini S, Stierhof YD, Hindennach I, Henning U. 1996. Characterization of the helper proteins for the assembly of tail fibers of coliphages T4 and lambda. J Bacteriol 178:6258–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bielmann R, Habann M, Eugster MR, Lurz R, Calendar R, Klumpp J, Loessner MJ. 2015. Receptor binding proteins of Listeria monocytogenes bacteriophages A118 and P35 recognize serovar-specific teichoic acids. Virology 477:110–118. doi: 10.1016/j.virol.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 30.Veesler D, Cambillau C. 2011. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol 75:423–433. doi: 10.1128/MMBR.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bebeacua C, Lai L, Vegge CS, Brøndsted L, van Heel M, Veesler D, Cambillau C. 2013. Visualizing a complete Siphoviridae member by single-particle electron microscopy: the structure of lactococcal phage TP901-1. J Virol 87:1061–1068. doi: 10.1128/JVI.02836-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bebeacua C, Tremblay D, Farenc C, Chapot-Chartier MP, Sadovskaya I, van Heel M, Veesler D, Moineau S, Cambillau C. 2013. Structure, adsorption to host, and infection mechanism of virulent lactococcal phage p2. J Virol 87:12302–12312. doi: 10.1128/JVI.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins B, Bebeacua C, Mahony J, Blangy S, Douillard FP, Veesler D, Cambillau C, van Sinderen D. 2013. Structure and functional analysis of the host recognition device of lactococcal phage tuc2009. J Virol 87:8429–8440. doi: 10.1128/JVI.00907-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veesler D, Spinelli S, Mahony J, Lichière J, Blangy S, Bricogne G, Legrand P, Ortiz-Lombardia M, Campanacci V, van Sinderen D, Cambillau C. 2012. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc Natl Acad Sci U S A 109:8954–8958. doi: 10.1073/pnas.1200966109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cambillau C. 2015. Bacteriophage module reshuffling results in adaptive host range as exemplified by the baseplate model of listerial phage A118. Virology 484:86–92. doi: 10.1016/j.virol.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 36.Gohmann S, Leimeister-Wachter M, Schiltz E, Goebel W, Chakraborty T. 1990. Characterization of a Listeria monocytogenes-specific protein capable of inducing delayed hypersensitivity in Listeria-immune mice. Mol Microbiol 4:1091–1099. doi: 10.1111/j.1365-2958.1990.tb00683.x. [DOI] [PubMed] [Google Scholar]
- 37.Schaferkordt S, Chakraborty T. 1997. Identification, cloning, and characterization of the lma operon whose gene products are unique to Listeria monocytogenes. J Bacteriol 179:2707–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hain T, Ghai R, Billion A, Kuenne CT, Steinweg C, Izar B, Mohamed W, Mraheil MA, Domann E, Schaffrath S, Kärst U, Goesmann A, Oehm S, Pühler A, Merkl R, Vorwerk S, Glaser P, Garrido P, Rusniok C, Buchrieser C, Goebel W, Chakraborty T. 2012. Comparative genomics and transcriptomics of lineages I, II, and III strains of Listeria monocytogenes. BMC Genomics 13:144. doi: 10.1186/1471-2164-13-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits AA. 2012. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 150:792–802. doi: 10.1016/j.cell.2012.06.036. [DOI] [PubMed] [Google Scholar]
- 40.Cumby N, Reimer K, Mengin-Lecreulx D, Davidson AR, Maxwell KL. 2015. The phage tail tape measure protein, an inner membrane protein and a periplasmic chaperone play connected roles in the genome injection process of E. coli phage HK97. Mol Microbiol 96:437–447. doi: 10.1111/mmi.12918. [DOI] [PubMed] [Google Scholar]
- 41.Lotz W, Mayer F. 1972. Isolation and characterization of a bacteriophage tail-like bacteriocin from a strain of Rhizobium. J Virol 9:160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thaler JO, Baghdiguian S, Boemare N. 1995. Purification and characterization of xenorhabdicin, a phage tail-like bacteriocin, from the lysogenic strain F1 of Xenorhabdus nematophilus. Appl Environ Microbiol 61:2049–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morales-Soto N, Gaudriault S, Ogier JC, Thappeta KR, Forst S. 2012. Comparative analysis of P2-type remnant prophage loci in Xenorhabdus bovienii and Xenorhabdus nematophila required for xenorhabdicin production. FEMS Microbiol Lett 333:69–76. doi: 10.1111/j.1574-6968.2012.02600.x. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen HA, Tomita T, Hirota M, Kaneko J, Hayashi T, Kamio Y. 2001. DNA inversion in the tail fiber gene alters the host range specificity of carotovoricin Er, a phage-tail-like bacteriocin of phytopathogenic Erwinia carotovora subsp. carotovora Er. J Bacteriol 83:6274–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauch E, Kaspar H, Schaudinn C, Dersch P, Madela K, Gewinner C, Hertwig S, Wecke J, Appel B. 2001. Characterization of enterocoliticin, a phage tail-like bacteriocin, and its effect on pathogenic Yersinia enterocolitica strains. Appl Environ Microbiol 67:5634–5642. doi: 10.1128/AEM.67.12.5634-5642.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jabrane A, Sabri A, Compère P, Jacques P, Vandenberghe I, Van Beeumen J, Thonart P. 2002. Characterization of serracin P, a phage-tail-like bacteriocin, and its activity against Erwinia amylovora, the fire blight pathogen. Appl Environ Microbiol 68:5704–5710. doi: 10.1128/AEM.68.11.5704-5710.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smarda J, Benada O. 2005. Phage tail-like (high-molecular-weight) bacteriocins of Budvicia aquatica and Pragia fontium (Enterobacteriaceae). Appl Environ Microbiol 71:8970–8973. doi: 10.1128/AEM.71.12.8970-8973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer S, Godino A, Quesada JM, Cordero P, Jofré E, Mori G, Espinosa-Urgel M. 2012. Characterization of a phage-like pyocin from the plant growth-promoting rhizobacterium Pseudomonas fluorescens SF4c. Microbiology 158:1493–1503. doi: 10.1099/mic.0.056002-0. [DOI] [PubMed] [Google Scholar]
- 49.Liu J, Chen P, Zheng C, Huang YP. 2013. Characterization of maltocin P28, a novel phage tail-like bacteriocin from Stenotrophomonas maltophilia. Appl Environ Microbiol 79:5593–5600. doi: 10.1128/AEM.01648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hockett KL, Renner T, Baltrus DA. 2015. Independent co-option of a tailed bacteriophage into a killing complex in Pseudomonas. mBio 6:e00452. doi: 10.1128/mBio.00452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shinomiya T. 1984. Phenotypic mixing of pyocin R2 and bacteriophage PS17 in Pseudomonas aeruginosa PAO. J Virol 49:310–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shinomiya T, Shiga S. 1979. Bactericidal activity of the tail of Pseudomonas aeruginosa bacteriophage PS17. J Virol 32:958–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A 106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. 2011. Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol Mol Biol Rev 75:610–635. doi: 10.1128/MMBR.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, Kolonay JF, Rasko DA, Angiuoli SV, Gill SR, Paulsen IT, Peterson J, White O, Nelson WC, Nierman W, Beanan MJ, Brinkac LM, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Haft DH, Selengut J, Van Aken S, Khouri H, Fedorova N, Forberger H, Tran B, Kathariou S, Wonderling LD, Uhlich GA, Bayles DO, Luchansky JB, Fraser CM. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res 32:2386–2395. doi: 10.1093/nar/gkh562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.