ABSTRACT
Despite the importance of lipooligosaccharides (LOSs) in the pathogenicity of campylobacteriosis, little is known about the genetic and phenotypic diversity of LOS in Campylobacter coli. In this study, we investigated the distribution of LOS locus classes among a large collection of unrelated C. coli isolates sampled from several different host species. Furthermore, we paired C. coli genomic information and LOS chemical composition for the first time to investigate possible associations between LOS locus class sequence diversity and biochemical heterogeneity. After identifying three new LOS locus classes, only 85% of the 144 isolates tested were assigned to a class, suggesting higher genetic diversity than previously thought. This genetic diversity is at the basis of a completely unexplored LOS structural heterogeneity. Mass spectrometry analysis of the LOSs of nine isolates, representing four different LOS classes, identified two features distinguishing C. coli LOS from that of Campylobacter jejuni. 2-Amino-2-deoxy-d-glucose (GlcN)–GlcN disaccharides were present in the lipid A backbone, in contrast to the β-1′-6-linked 3-diamino-2,3-dideoxy-d-glucopyranose (GlcN3N)–GlcN backbone observed in C. jejuni. Moreover, despite the fact that many of the genes putatively involved in 3-acylamino-3,6-dideoxy-d-glucose (Quip3NAcyl) were apparently absent from the genomes of various isolates, this rare sugar was found in the outer core of all C. coli isolates. Therefore, regardless of the high genetic diversity of the LOS biosynthesis locus in C. coli, we identified species-specific phenotypic features of C. coli LOS that might explain differences between C. jejuni and C. coli in terms of population dynamics and host adaptation.
IMPORTANCE Despite the importance of C. coli to human health and its controversial role as a causative agent of Guillain-Barré syndrome, little is known about the genetic and phenotypic diversity of C. coli LOSs. Therefore, we paired C. coli genomic information and LOS chemical composition for the first time to address this paucity of information. We identified two species-specific phenotypic features of C. coli LOS, which might contribute to elucidating the reasons behind the differences between C. jejuni and C. coli in terms of population dynamics and host adaptation.
INTRODUCTION
Campylobacteriosis is the most common bacterial foodborne disease in developed countries, with over 200,000 human cases being reported annually in the European Union alone (1). The true burden of the disease in the population is likely underestimated, as many infections result in mild gastroenteritis (1). Approximately 80% of reported infections are caused by Campylobacter jejuni, and 7 to 18% of cases are attributed to C. coli. Therefore, C. coli is among the five most important bacterial etiological agents of human gastroenteritis (2, 3).
As in other Gram-negative bacteria, Campylobacter species cell surface glycoconjugates, including lipooligosaccharides (LOSs), play an important role in serum and bile resistance; resistance to phagocytic killing; and adhesion to, invasion of, and survival in host cells (4–8). Current knowledge on LOS diversity has been based primarily on work on C. jejuni and its role in promoting severe clinical symptoms (9–12). C. jejuni LOS is a potent Toll-like receptor 4 (TLR4) agonist, and the subsequent immune response is affected by changes in LOS structure and composition (10–14). Additionally, due to molecular mimicry between human gangliosides and certain LOS structures, C. jejuni has been identified as one of the causative agents of Guillain-Barré syndrome (GBS) (15). Contrarily, the little knowledge on C. coli LOS variability has limited our understanding of the pathogenesis of GBS in patients infected with C. coli, as it remains unclear whether C. coli is able to mimic human ganglioside structures (16–18).
Valuable insights into the genetic origins of significant strain-variable traits have been gained by studying the effects of C. jejuni LOS genotypes on phenotype (19–24). However, so far, only two studies have addressed the variation in gene composition of the C. coli LOS biosynthesis locus. Until now, nine genetic classes composed of a variable combination of 10 to 20 genes have been described in C. coli (25, 26), but no chemical analysis of their LOS structures was executed. A couple of decades ago, the LOS structure of a single C. coli strain was described (27). Additionally, three other studies have explored the chemical composition of C. coli LOSs in a few strains (28–30), but no genetic information on the strains is available to our knowledge.
In this study, we investigated the diversity and distribution of LOS locus classes among a large collection of unrelated C. coli isolates sampled from several different host species. We expanded the current C. coli LOS classification by describing three additional LOS locus classes (25, 26). Moreover, by analyzing genomic data with the LOS chemical composition of selected isolates, we identified possible associations between gene content in the LOS biosynthesis locus and observed differences in LOS phenotype. Despite the extensive introgression between C. coli and C. jejuni (31, 32), only negligible levels of recombination were detected in LOS biosynthesis genes, which might explain the distinctive species-specific chemical features observed here.
MATERIALS AND METHODS
Bacterial isolates, cultivation, and DNA extraction.
In total, 144 C. coli isolates, including 90 isolates from swine, 34 from humans, 18 from poultry, and 2 from wild birds, were chosen for LOS locus screening. The selection comprised 133 C. coli isolates collected between 1996 and 2012 from Finnish human patients, chickens, and pigs reared in Finland and wild birds sampled in the Helsinki region in previous studies (25, 33–39). This collection was supplemented with 11 C. coli isolates from the Campynet (CNET) collection (hosted by DSMZ GmbH [https://www.dsmz.de/]). Isolate selection was based on genotype (pulsed-field gel electrophoresis [PFGE] or multilocus sequence typing [MLST]), host, country of origin, and year of isolation to encompass the greatest possible diversity. Cultivation and DNA isolation were carried out as previously described (25), unless otherwise stated.
PCR.
The length of LOS biosynthesis loci was determined by amplifying the region between orthologue 10 (LOS biosynthesis glycosyltransferase [waaV]) and orthologue 16 (uncharacterized glycosyltransferase) (identifications numbers according to Richards and colleagues [26]). PCRs were set up as follows: 25-μl reaction mixtures contained 0.5 U Phusion high-fidelity DNA polymerase (Thermo Scientific), 200 μM each deoxynucleoside triphosphate (dNTP) (Thermo Scientific), 0.4 μM each primer (ORF3F2 and waaV-R) (Table 1), 1× Phusion GC buffer (Thermo Scientific), 700 μM MgCl2 (Thermo Scientific), and 50 ng of the template. Cycling conditions were as follows: 1 cycle at 98°C for 30 s followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 62.4°C for 30 s, extension at 72°C for 6 min, and a final elongation step at 72°C for 6 min. The size of the LOS locus was estimated by gel electrophoresis with 1-kb-plus (Thermo Scientific) and long-range (Thermo Scientific) molecular weight markers. Specific primers for each class were designed based on the previously described C. coli LOS locus classes (classes I to IX) (25, 26). Primer pairs and their amplicon sizes for each LOS class are shown in Table 1, and a graphic representation of the primer annealing positions within the LOS locus is shown in Fig. S2 in the supplemental material. Since global alignment using progressiveMauve (40) revealed that LOS locus classes IV and V (26) differ by only 3 single nucleotide polymorphisms (which resulted in the fragmentation of orthologue 1959 in class V), here, the two LOS locus classes are considered a single class, named class IV/V. The specificity of each primer pair was verified in silico. All primers were designed based on specific features characterizing each LOS locus class using, when possible, multiple-sequence alignments of homologous sequences to improve sensitivity and specificity. Preliminary gradient PCR was performed for each primer pair to select the most stringent conditions to minimize artifacts. Additionally, the same results were obtained when primers from PCR numbers 2 to 12 were tested on genomic DNA or in a nested PCR using PCR 1 as the template. PCRs were carried out in a semi-high-throughput manner; thus, isolates were classified into a LOS class based on the results of all PCRs (Table 1). Isolates with an unexpected LOS size, negative for all tested orthologues, or with unexpected combinations of orthologues were classified as untypeable.
TABLE 1.
Primers used in the present study and expected sizes of the amplicons
| PCR no. | Primer | Sequence | Expected amplicon size (kb) or presence of amplicon (expected size [bp]) for LOS locus classa: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV/V | VI | VII | VIII | IX | X | XI | XII | |||
| 1 | ORF3-F2 | AAA AGC TTG TGG CTG GTG GCC TGA TCA | 7.1 | 9.9 | 7.2 | 12.6 | 13.2 | 15.3 | 18.2 | 7.1 | 11.5 | 11.4 | 11.1 |
| waaV-R | AAG AGC TTT GCA AAG CTG TAT AAA TCA GAC | ||||||||||||
| 2 | 2209-L | TTC AGG TGT TTA TGA TTT GTT TC | + (355) | − | − | − | − | − | − | − | − | − | − |
| 2209-R | GCT TGT GCC TTT GGT ATA AGG | ||||||||||||
| 3 | CstIV-F | TTC CCA GCA GCT ATA AAT GGA | − | + (190) | − | − | − | − | − | − | − | − | − |
| CstIV-R | TTT CAT CTC CAA AAT CCA TGC | ||||||||||||
| 4 | 1541-L | TGG CAA YTA TGG TTT CAA GG | − | + (327) | − | + (327) | + (327) | + (327) | − | − | − | − | − |
| 1541-R | TGC YCT TTC AAA AGC AAA AAA TTC | ||||||||||||
| 5 | 1210-L | AAT TTT GCG TGG AAT GCT TG | − | − | + (337) | − | − | − | − | − | − | − | − |
| 1210-R | GCT GAA GGC AAT TGA TGA TG | ||||||||||||
| 6 | 1790-L | CCY TAA AYA CYG CTT TTR AAA AC | − | − | − | + (328) | + (328) | + (328) | − | − | − | − | + (328) |
| 1790-R | TGC GTA TCT TGT TGA TTR CAC | ||||||||||||
| 7 | 1920-L | CCA AGC CAG ATT TTC CAA GA | − | − | − | + (229) | − | + (229) | − | − | − | + (229) | + (229) |
| 1920-R | TCG TTA TAG AAA TCA CTT GCC AAT | ||||||||||||
| 8 | 2344-L | AAA GAA AGA GAA GCC AAA GGA G | − | − | − | − | − | + (348) | − | − | − | − | − |
| 2344-R | TCT TGG TTT AAT TTT CGC ATA TTC | ||||||||||||
| 9 | 1790R | TGC GTA TCT TGT TGA TTR CAC | − | − | − | + (2,252) | − | + (4,933) | − | − | − | − | − |
| 1920L | CCA AGC CAG ATT TTC CAA GA | ||||||||||||
| 10 | 38_3454 | ACG CCT AGC GTG TAA ACC AT | − | − | − | − | − | − | + (1,046) | − | − | − | − |
| 38_2031 | ATC GTC CTA TAG CTA CGG GTG A | ||||||||||||
| 11 | CstV-F | TTC CTT TGC AAC ACG AAA TAA | − | − | − | − | − | − | − | + (449) | − | − | − |
| CstV-R | GTT TTG GAG CTA GCG GAA TA | ||||||||||||
| 12 | 45_8 | GTG CTT GAG CGC AAT CTT CT | − | − | − | − | − | − | − | − | + (1,036) | + (1,036) | − |
| 45_1 | GAG GGG CCT TAT GGA GCA AA | ||||||||||||
The amplicon sizes for PCR 1 are expressed in kilobases, while all others are expressed in base pairs.
Genome sequencing and annotation.
For ascertaining the LOS locus classes, 35 isolates were chosen for genome sequencing (see Table S1 in the supplemental material) using either HiSeq or MiSeq. For HiSeq, next-generation sequencing (NGS) library preparation, enrichment, sequencing, and sequence analyses were performed by the Institute for Molecular Medicine Finland (FIMM Technology Center, University of Helsinki, Helsinki, Finland). MiSeq sequencing was performed by the Institute of Life Science, Swansea University (Swansea, United Kingdom). Reads were filtered and assembled by using SPAdes Assembler v. 3.3.0 (41). Primary annotation of all the genomes was performed by using Rapid Annotation Using Subsystems Technology (RAST) (42). Sequences were manually curated by using Artemis (43), and LOS locus classes were aligned and compared with ACT (44).
Orthologue clustering and phylogenetic analysis.
A database including all the translated coding sequences for C. jejuni and C. coli LOS biosynthesis was assembled according to the orthologue nomenclature described previously by Richards and colleagues (26). A reciprocal all-versus-all BLASTp search was performed (threshold E value of ≤1e−10) (45), and orthologous groups were determined by orthAgogue and MCL (ignoring E values, percent match lengths of ≥80%, and inflation values of 5 [46, 47]). The groups of orthologues (GOs) were then aligned by using MUSCLE and back-translated to the nucleotide sequence by using Translatorx Perl script (48–50). Maximum likelihood phylogenetic reconstruction of each GO was performed in MEGA6.06 (51), using the Kimura two-parameter model as the nucleotide substitution model and a discrete gamma distribution (4 categories) to model evolutionary rate differences among sites. A total of 100 bootstrap runs were performed and summarized in a 95% consensus tree.
LOS silver staining.
LOS profiles were assessed by silver staining as described previously (52), with some modifications. In brief, the absorbance of the biomass obtained from a culture grown for 16 h in nutrient broth 2 (Oxoid) (100 rpm, microaerobic atmosphere, and 37°C) was adjusted to an optical density at 600 nm (OD600) of 0.5. Cells were digested with 20 mg/ml proteinase K (Thermo Scientific) and incubated at 55°C for 1 h, followed by boiling for 10 min. Samples were then diluted 1:5 in loading buffer and resolved in 15% SDS-PAGE gels. Gels were silver stained for visualization as previously described (53).
Capillary electrophoresis-mass spectrometry (CE-MS) and EA-OTLC-MS analyses.
Biomass was produced in broth as indicated above, and LOS was prepared with a rapid method applying microwave irradiation as previously described (54). In short, the lyophilized biomass was suspended in 50 μl of 20 mM ammonium acetate buffer (pH 7.5) containing DNase (100 μg/ml) and RNase (200 μg/ml) and heated by direct microwave irradiation. Proteinase K was then added to a final concentration of 60 μg/ml and heated under the same conditions. Solutions were allowed to cool at room temperature and subsequently dried by using a SpeedVac (vacuum centrifuge concentrator; Savant). LOS samples were washed three times with methanol (100 μl), with vigorous stirring. Insoluble residues were collected by centrifugation and resuspended in 30 μl water for electrophoresis-assisted open tubular liquid chromatography–electrospray mass spectrometry (EA-OTLC-MS) analysis. A sheath solution (isopropanol-methanol [2:1]) was delivered at a flow rate of 1.0 μl/min. Separation was performed by using 30 mM morpholine in deionized water (pH 9.0). A separation voltage of 20 kV, together with a pressure of 50,000 Pa, was applied for the EA-OTLC-MS analysis. The electrospray ionization (ESI) voltage applied to the sprayer was set at −5.2 kV. Data acquisition was performed for an m/z range of 600 to 2,000 at a 2-s/spectrum scan rate.
Statistical analysis.
Fisher's exact test was used to assess host-LOS locus class associations. P values of ≤0.05 were considered significant.
Accession numbers.
The whole-genome sequences of C. coli are publicly available on the RAST server (http://rast.nmpdr.org) with a guest account (login and password “guest”) under accession numbers 195.91, 195.96 to 195.119, 195.124 to 195.126, 195.128 to 195.130, 195.133, 195.134, and 6666666.94320.
RESULTS
PCR typing method for C. coli LOS locus diversity.
We explored the genetic diversity of the LOS biosynthesis loci in 144 C. coli isolates (see Table S1 in the supplemental material) using a PCR typing scheme based on previously reported LOS locus class definitions (25, 26). Isolates were classified into putative LOS locus classes according to their PCR profile and LOS locus size as shown in Table 1. The LOS PCR typing scheme was validated by genome sequencing of 35 isolates (see isolates marked in yellow in Table S1 in the supplemental material). Typing results are summarized in Table 2. We were able to classify 68% of the isolates into one of the nine previously reported LOS locus classes (25, 26). Most of the isolates were assigned to LOS locus class II (17%), with the remaining isolates being assigned to LOS classes IV/V (15%), III (13%), VI (13%), VIII (7%), I (2%), VII (1%), and IX (0.7%). The final 46 (out of 144 [∼32%]) isolates remained untypeable by this method.
TABLE 2.
Distribution of LOS classes among hosts
| LOS class | Total no. of isolates (%) | No. of isolates from host: |
|||
|---|---|---|---|---|---|
| Human | Swine | Poultry | Wild birds | ||
| I | 3 | 2 | 0 | 1 | 0 |
| II | 24 (17) | 7 | 13 | 4 | 0 |
| III | 18 (13) | 4 | 13 | 0 | 1 |
| IV/V | 22 (15) | 3 | 16 | 3 | 0 |
| VI | 18 (13) | 1 | 15 | 2 | 0 |
| VII | 2 (1) | 1 | 1 | 0 | 0 |
| VIII | 10 (7) | 7 | 1 | 2 | 0 |
| IX | 1 | 1 | 0 | 0 | 0 |
| X | 22 (15) | 3 | 18 | 1 | 0 |
| XI | 1 | 0 | 1 | 0 | 0 |
| XII | 1 | 0 | 0 | 0 | 1 |
| Untypeable | 22 (15) | 5 | 12 | 5 | 0 |
Six untypeable isolates, with a LOS locus length of ∼11.5 kbp, were sequenced (isolates 45, 63, 114, 125, 149, and 153). All isolates belong to a novel LOS locus class, class X. This new class shares 12 (out of 15) orthologues with other LOS locus classes (see below) and is characterized by the presence of three unique genes (see Fig. S2 in the supplemental material). A BLASTp search of the NCBI database revealed sequence similarity with (i) a hypothetical protein of Helicobacter sp. strain strain MIT 05-5293 (E value, 1e−98; identity, 45%), (ii) a hypothetical protein of Helicobacter hepaticus (E value, 3e−108; identity, 53%), and (iii) UDP-N-acetylglucosamine 2-epimerase of H. hepaticus (E value, 3e−165; identity, 63%). Following this finding, primers were designed (Table 1) for LOS locus class X, which further identified 15% of the isolates (Table 2). The genomes of isolates 138 and 99, which have a LOS with a size similar to that of class X but with a different PCR profile (see Table S1 in the supplemental material), were also sequenced. Analysis of these genomes revealed two additional LOS locus classes, defined as classes XI (isolate 138) and XII (isolate 99). In total, we were able to assign a LOS locus class to 85% of the isolates in our collection by incorporating these additional classes. The LOS profile diversity was high, suggesting that further LOS locus classes may be described in the future.
Origin of the novel LOS locus classes X, XI, and XII.
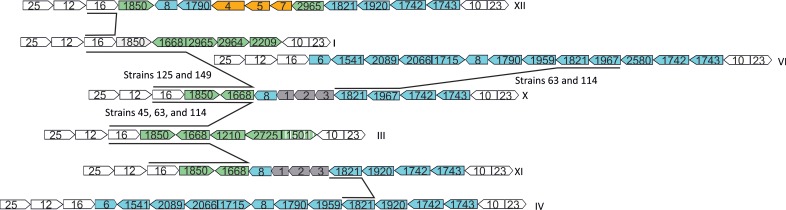
As for C. jejuni, C. coli exhibits mosaic LOS loci (22), with several classes containing similar orthologous loci. LOS locus classes X and XI are very similar to each other, diverging at only a single locus (orthologue 1967 instead of 1920) (Fig. 1). Additionally, these two classes also have gene contents and organizations similar to those of LOS locus classes I, III, IV/V, VI, and VII (Fig. 1). To infer evolutionary relationships between these classes, phylogenetic analyses were performed for each shared GO. Phylogenetic reconstruction revealed LOS class I and LOS class III as the two possible origins for the region encompassing orthologue 16 to orthologue 1668 in LOS locus class X (Fig. 1). Specifically, in the phylogenetic tree of orthologues 16, 1850, and 1668, C. coli isolates 45, 63, and 114 are monophyletic with strains from LOS locus class III, while C. coli isolates 125 and 149 formed a separate clade with LOS locus class I isolates (see Fig. S1A to C in the supplemental material). Orthologues 8 and 1821 in LOS class X and both classes IV/V and VI share the same origin. Contrarily, the origin of the region including orthologues 1967, 1742, and 1743 is less clear. In the phylogenetic tree of orthologue 1967 (see Fig. S1D in the supplemental material), C. coli isolates 63 and 114 are grouped with LOS locus class VI isolates, while the other strains form separate clades. In addition, the star-like phylogeny inferred for orthologues 1742 and 1743 hampered any kind of conclusion. These results suggest that extensive recombination and gene reorganization between LOS locus classes took place, masking the origin of common shared loci. Except for orthologue 1920, LOS locus class XI orthologues are closely related to those found in LOS locus class X (see Fig. S1 in the supplemental material). LOS locus class XII shares orthologues with LOS locus classes I, IV/V, VII, and IX. However, in our phylogenetic analysis, LOS locus class XII orthologues are distantly related to those found in other LOS classes, forming a separate branch in the phylogenetic trees. Additionally, LOS locus class XII is characterized by the presence of a set of unique genes having the best BLASTp hit against the NCBI nr database with (i) methyltransferase type 12 of Helicobacter hepaticus (E value, 6e−75; identity, 58%), (ii) a hypothetical protein of Anaerovibrio lipolyticus (E value, 5e−102; identity, 65%), and (iii) phosphoserine phosphatase of Helicobacter sp. strain MIT 05-5293 (E value, 3e−92; identity, 63%) (Fig. 1). Proposed functions for each open reading frame (ORF) of the newly identified LOS locus classes reported here are described in Table S2 in the supplemental material.
FIG 1.
LOS locus classes related to classes X, XI, and XII. Arrows represent ORFs. Genes in white are common to all LOS classes. Genes in green are present in classes I and/or III. Genes in blue are present in classes IV/V and VI. Gray genes are common among classes X and XI. Orange genes are particular to class XII. Striped genes are fragmented. Lines connect closely related orthologues. Strains are identified if more than one origin was observed in the LOS locus class (see the text). Gene size is not drawn to scale.
Cluster analysis of the LOS locus classes.
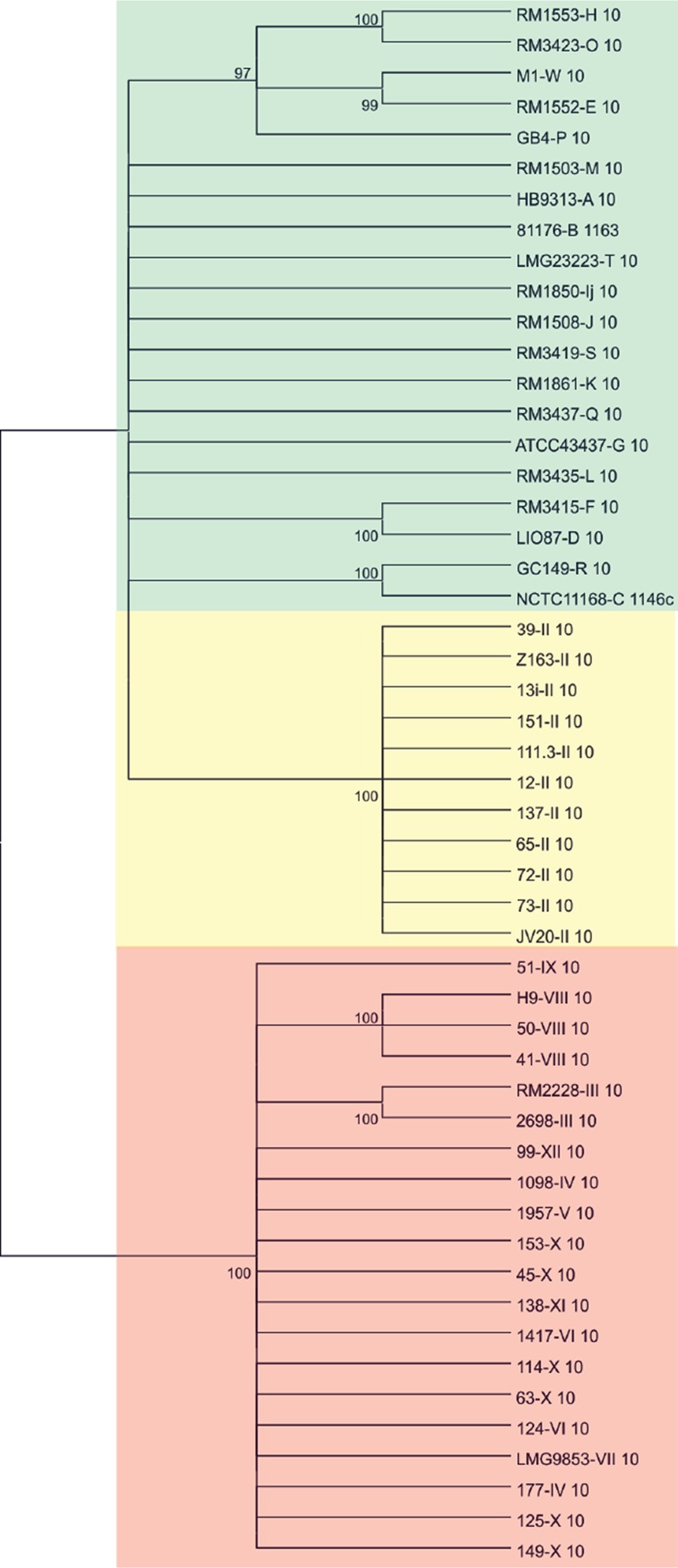
Both species share a total of 19 LOS orthologues (26), and with previously reported evidence of introgression between C. coli and C. jejuni in mind (31, 32), we attempted to quantify the level of interspecies recombination in C. coli LOS diversity. We compared individual gene descriptions of the LOS loci rather than the original gene family ontologies used by Richards and colleagues (26). Out of the 19 shared orthologues, 16 gene locus descriptions split into species-specific clusters, while only 3 were common in both species (orthologues 10, 16, and 1821). Interspecies gene transfer was investigated by comparing the topology of individual gene trees with the overall population structure (25). Evidence of interspecies gene transfer was observed only for orthologue 10 (26) (lipooligosaccharide biosynthesis glycosyltransferase [waaV]), where all C. coli loci of LOS locus class II formed a monophyletic clade with C. jejuni genes (Fig. 2). Thus, interspecies recombination is likely to have a limited effect on the LOS locus diversity observed in C. coli.
FIG 2.
Consensus cladogram representing the evolutionary relationship among orthologues belonging to GO 10 (nomenclature from Richards et al. [26]). C. jejuni strains are highlighted in green. C. coli strains, with the exception of LOS locus class II strains, are shown in red. C. coli LOS locus class II strains are highlighted in yellow. The 95% bootstrap consensus tree was built from 100 replicates. The strain LOS locus class is indicated after the strain identification.
Host-LOS locus class association.
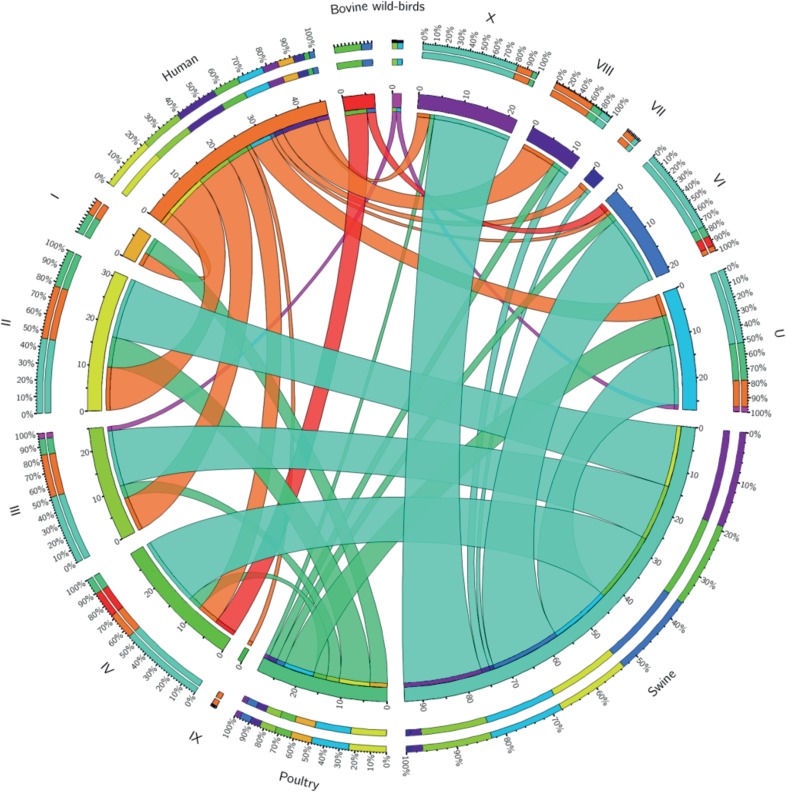
The nonrandom distribution of LOS locus classes between hosts was investigated further by supplementing data from our isolate collection with data reported previously by Richards and colleagues (26). The distribution of LOS locus classes by source of isolation is represented in Fig. 3. All LOS locus classes, except for class XII, were present among strains isolated from humans. More than one-half (57%) of the clinical isolates were of LOS locus classes II, III, and VIII, while LOS locus classes VI, VII, and X were less commonly found in clinical cases. Most pig isolates were of LOS locus class X but were also frequently found among LOS locus classes II, III, IV/V, and VI. Only one pig isolate belonged to LOS locus class VIII, and no pig strain was found to belong to class I, IX, or XII. Poultry isolates were also found among all LOS locus classes, except for classes VII, IX, and XII. Most poultry isolates were classified as LOS locus class II.
FIG 3.
Host-LOS locus class association. A Circos diagram shows the distribution of LOS locus classes of C. coli strains isolated from different hosts, from both our collection and that of Richards and colleagues (26). Ribbon ends represent links between host and LOS locus class, while the width of the ribbon correlates with the percentage of strains belonging to a specific LOS locus class in a certain host. Segments in the outer ring indicate the percentage of strains representing a certain LOS locus class or host, while the inner ring indicates the number of strains. Human strains are shown in orange, bovine strains are shown in red, poultry strains are shown in green, and swine strains are shown in cyan.
There was a positive association (P < 0.05) of class VIII with human clinical infections, while class VI was negatively associated with clinical cases. Swine isolates were positively associated with classes VI and X but negatively associated with classes I and VIII. Poultry isolates were positively associated only with LOS locus class I. Bovine and wild bird isolates were underrepresented in the data set. However, some association was observed in bovine (class IV/V) and wild bird (class XII) isolates. Isolates classified as LOS locus classes II and III were equally distributed among human, pig, and poultry isolates.
Chemical analysis of C. coli LOS composition.
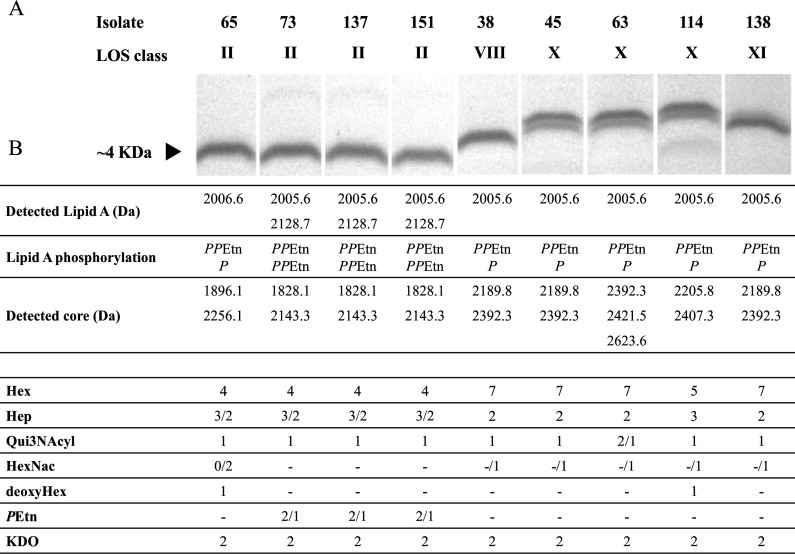
The LOS phenotypes of nine selected isolates were investigated. This selection included strains from classes overrepresented in clinical isolates, classes II and VIII, as well as isolates from two of the newly described LOS classes (classes X and XI), which are uncommon in clinical isolates. Silver-stained SDS-PAGE gels of LOS extracts provided migration profiles for the selected isolates (Fig. 4A). A complementary mass spectroscopy approach was used (CE-MS and EA-OTLC-MS) to explore inter- and intra-LOS-class structural diversity. Example spectra are shown in Fig. S3 in the supplemental material. The oligosaccharide (OS) composition of each of the nine isolates was predicted based on the fragment ions and components of the previously reported C. coli OS (27). The size and composition of the lipid A group were defined for each glycoform by tandem mass spectrometry. For example, the fragment ion at m/z 1,063.2 (doubly charged ion) in C. coli isolate 137 (see Fig. S3 in the supplemental material), which was produced from the glycoform detected as a triply charged ion at m/z 1,422.8, corresponds to a lipid A species consisting of a 2-amino-2-deoxy-d-glucose (GlcN) disaccharide backbone carrying two negatively charged pyrophosphoethanolamine (PPEtn) groups and six fatty acid chains, with a calculated mass of ∼2,128 Da. Additionally, the fragment ion at m/z 1,001.7 corresponds to a second lower-mass lipid A species (∼2,006 Da), as it carries P and PPEtn instead. All analyzed C. coli isolates exhibited a hexa-acylated lipid A species containing four tetradecanoic (14:0) and two hexadecanoic (16:0) acid chains, modified with two phosphate residues (55–57). Only GlcN disaccharides were detected in C. coli isolates, in contrast to the hybrid backbone of β-1′-6-linked 3-diamino-2,3-dideoxy-d-glucopyranose (GlcN3N) and GlcN observed in C. jejuni (55, 57). Thus, C. coli synthesizes a lipid A molecule with two ester- and two amide-linked acyl chains, while C. jejuni has a lipid A species containing mainly three amide-linked acyl chains and one ester-linked acyl chain. The lower-mass lipid A species was detected in all samples, while LOS locus class II isolates (except for isolate 65) (see Fig. S3 in the supplemental material) had an additional lipid A species, as exemplified by strain 137 in Fig. S3 in the supplemental material.
FIG 4.
C. coli LOS biochemical profiles. (A) Silver-stained LOS. (B) Proposed chemical composition based on results of MS and tandem MS analyses of intact LOS (see Fig. S3 in the supplemental material).
As in C. jejuni, C. coli exhibited a conserved inner core consisting of two l-glycero-d-manno-heptose (Hep) residues attached to a 3-deoxy-d-manno-octulosonic residue (Kdo), which is linked to lipid A through a Kdo linker (20, 57). In the variable outer core region, at least one residue with an average molecular mass of 231 Da was detected in all isolates. Based on available structures, this residue was predicted to be Quip3NAcyl (where Quip3NAcyl represents 3-acylamino-3,6-dideoxy-d-glucose in which the N-acyl residue was 3-hydroxybutanoyl). Although more than one OS was detected in all isolates by MS (Fig. 4B), only isolates from LOS locus classes X and XI exhibited visible high-Mr and low-Mr LOSs on SDS-PAGE gels (Fig. 4A). Intra-LOS class diversity was observed for both LOS class II and class X. Isolate 65 displayed a LOS composition similar to those of other LOS class II isolates but with the addition of two hexosamines (HexNAc) and one deoxyhexose (deoxyHex) and the absence of phosphoethanolamine (PEtn) residues (Fig. 4B). Likewise, isolates 45 and 63 shared similar LOS compositions, with the exception of a variable Quip3NAcyl residue in isolate 63. In contrast, isolate 114 exhibited a very different LOS composition from those of other isolates of the same class, including the presence of a third Hep and a deoxyHex residue as well as a reduced number of hexoses (Fig. 4B). The LOSs of isolates 38, 45, and 138 have similar core sizes and proposed compositions, yet they are classified into three different LOS locus classes. However, our biochemical analysis was not able to identify saccharide sequences, stereochemistry, absolute configurations (d or l), anomeric configurations (α or β), and linkage positions. Thus, further studies would be required to determine whether these three different LOS classes indeed produce the same LOS structure.
Genetic and phenotypic diversity of C. coli LOS locus class II.
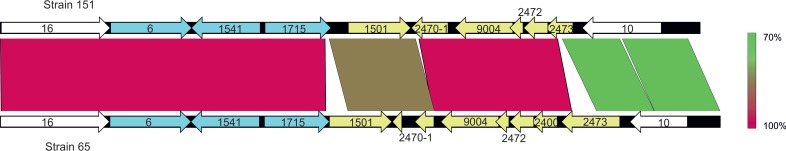
The four strains of LOS locus class II shared 99.64% DNA sequence similarity and 99.39% to 99.98% pairwise alignment identity. Isolate 65 was the most dissimilar among strains within LOS locus class II due to large fragment deletions. Deletions resulted in the shorter orthologues 2400 and 2473 being truncated into one pseudogene (Fig. 5). Orthologues 2470 and 2471 were also truncated as one pseudogene (reannotated orthologue 2470-1), as evidenced by isolate 151. The remainder of the class II isolates had an insertion of 68 nucleotides (nt) in orthologue 2470-1, disrupting the orthologue (Fig. 5). Despite the differences observed in orthologue 2470-1, isolates 73, 137, and 151 were predicted to have identical LOS chemical compositions.
FIG 5.
Comparison of nucleotide sequences of LOS locus class II strains 151 and 65. Genes in white are common to all LOS classes. Genes in blue are present in LOS locus classes IV/V, VI, and VII. Yellow genes are particular to LOS locus class II. Lines between orthologues represent sequence similarity.
Amino acid sequences of orthologues 6, 1541, 1501, 2472, and 10 were identical (100%) in all four class II strains, while orthologues 9004 and 16 exhibited a single amino acid difference in isolate 65. All isolates, with the exception of isolate 65, exhibited differences in the C terminus of orthologue 1715 and had variable numbers of Hep and/or PEtn residues observed. However, no GC homopolymeric tracts or other possible genetic signals associated with phase variation were identified within the LOS loci.
Genetic and phenotypic diversity of C. coli LOS locus class X.
In LOS locus class X, the overall sequence identity among strains was 99.31%, with the percent identity ranging from 98.96% to 99.94% in pairwise alignments and with strain 45 being the most distantly related. Although some minor gaps were observed, single point mutations were largely responsible for the diversity observed at the nucleotide level. The largest insertion (69 nt) was seen in strain 63 between orthologues 2 and 3. Between strains, 100% amino acid identity was observed for orthologues 16, 8, and 2, while one or two amino acid substitutions were present in orthologues 1668, 1, 1821, 1967, and 1743. The most prominent difference was observed in orthologue 1742 in the form of a deleted A base at position 668, resulting in premature translational termination in isolates 114 and 63. Furthermore, several single amino acid substitutions were detected in orthologue 1742 in strain 45, while 100% identity was observed between isolates 63 and 114. In spite of the dissimilar LOS compositions, the only difference observed within the LOS locus between isolates 63 and 114 was in 8 amino acids at the C terminus of orthologue 3.
DISCUSSION
Campylobacter LOS is a fundamental feature involved in the pathogenesis of gastroenteritis and postinfection sequelae (10–14, 58, 59). However, despite the burden imposed by C. coli and the importance of this structure in campylobacteriosis, little is known about LOS diversity in this species (26–29, 60). Therefore, we sought to contribute to the paucity of information by investigating the variability and distribution of C. coli LOS locus genetic classes in a large collection of isolates and by coupling genomic and LOS chemical composition data for the first time.
We developed a PCR methodology that was able to classify 85% of the isolates into a LOS class (25, 26). Among them, we described three additional LOS locus classes, named classes X, XI, and XII, which accounted for 17% of the isolates in our collection. The remaining untypeable isolates (15%) suggest that new classes will likely be described in the future and that C. coli LOS biosynthesis is more diverse than previously observed (26).
This genetic diversity is at the basis of a completely unexplored LOS structural heterogeneity, which might contribute substantially to the population dynamics of C. coli, including host specificity. We combined our 144 isolates with 33 C. coli isolates studied previously (26) to investigate the nonrandom distribution of LOS locus classes among different hosts. All hosts were significantly associated with at least one LOS locus class. In particular, isolates possessing LOS locus classes VI and X were predominantly isolated from swine, which have a very high prevalence of C. coli (up to 99%) (61). Both of these classes were rarely detected in human isolates, which is supported by a previous source attribution study in Scotland in which pigs are a relatively unimportant source of C. coli infections of humans (61). The majority of isolates from human cases in our study were assigned to LOS locus classes II and III, which were also found in swine and poultry isolates. However, human isolates were overrepresented among members of LOS locus class VIII, which was rarely detected in the sources included in this study. This indicates the presence of other, unknown potential reservoirs contributing to human infections, which corroborates data from a previous study where 54% of strains from human C. coli infections were attributed to sources other than poultry and pig (61). In opposition to previously reported findings (26), we did not observe partitioning between strains of bovine and poultry sources, and LOS locus classes previously shown to be associated with bovine hosts were populated by isolates of poultry and swine origins. Due to the limited number of isolates available from alternative sources, the host-LOS class associations found in this study may not necessarily represent the true C. coli population structure in various hosts. However, our findings suggest that generalist isolates possessing LOS locus classes II and III might be more successful at colonizing multiple species and, as seen in generalist lineages of the C. jejuni sequence type 45 (ST-45) and ST-21 clonal complexes, are largely responsible for human infections (32).
Mosaic C. coli LOS classes appear to have arisen by the insertion and/or deletion of genes or gene cassettes through homologous recombination, as previously described for C. jejuni (22). In spite of substantial genome-wide introgression between agricultural C. coli and C. jejuni isolates (25, 31), very limited interspecies recombination was detected among LOS biosynthesis loci. Only orthologue 10 (waaV) of C. coli LOS locus class II may have originated as result of recombination with C. jejuni. These results confirmed data from previous studies (31) and are supported by the species-specific features detected in the chemical composition of C. coli LOS.
GlcN disaccharide backbones, which are the most common structures among members of the family Enterobacteriaceae (57), were predicted for the lipid A species of all analyzed C. coli strains. This result is in contrast to the hybrid GlcN3N-GlcN backbone observed in C. jejuni. The gnnA and gnnB genes, located outside the LOS biosynthesis locus, are associated with the synthesis of GlcN3N-substituted lipid A (9, 62). The inactivation of either of these genes in C. jejuni resulted in the replacement of an N-linked acyl chain with an O-linked acyl chain and increased LOS biological activity in humans (9). C. coli contains the gnnA and gnnB genes in similar genomic locations, and these genes have approximately ∼70% BLASTp score ratios against C. jejuni orthologues (9). However, C. coli gnnA and gnnB are separated by a putative cobalamin-independent methionine synthase II gene in the same gene orientation. We therefore suggest three possible explanations for the absence of GlcN3N in the C. coli lipid A backbone: (i) single or multiple mutations in the putative active sites of GnnA and GnnB have rendered one or both enzymes inactive, as observed in functional studies of other bacteria (62, 63); (ii) gnnB-gnnA operon transcription might be hampered by the presence of the putative methionine synthase II (9); or (iii) GnnA and GnnB may be involved in the biosynthesis of alternative glycoconjugates in C. coli (62). Nevertheless, the replacement of an N-linked acyl chain with an O-linked acyl chain in C. coli might have an impact on host-bacterium interactions and adaptation (9).
A second species-specific feature common to all our analyzed isolates was the presence of at least one putative Quip3NAcyl residue. Quip3N is an unusual deoxysugar, which has been observed in the O-antigens of various Gram-negative bacteria and in the S layers of glycoprotein glycans of some Gram-positive bacteria (64–66). Although rarely studied, Quip3N has also been found in the OSs of LOS class E, H, and P isolates in C. jejuni exclusively as an N-acetyl derivative (Quip3NAc) (54, 67–69). Conversely, Quip3N has been reported in C. coli only as an N-acyl derivative with two possible substituents: 3-hydroxybutanoyl or 3-hydroxy-2,3-dimethyl-5-oxoprolyl (30). The presence of Quip3NAcyl in C. coli was first described by Seltmann and Beer (30), and later on, it was reported for several C. coli isolates (28). However, the molecular basis behind the biosynthesis of this sugar and the associated glycoconjugate in C. coli remains unknown. The dTDP-d-Quip3NAc biosynthesis pathway, to our knowledge, has been described only for the Gram-positive organism Thermoanaerobacterium thermosaccharolyticum (70). This pathway involves five enzymes: a thymidylyltransferase (RmlA), a 4,6-dehydratase (RmlB), a 3,4-isomerase (QdtA), a transaminase (QdtB), and a transacetylase (QdtC). Genome comparison of T. thermosaccharolyticum and C. coli identified homologues of rmlA (GO 1743), rmlB (GO 1742), qdtA (GOs 1920 and 1967), and qdtB (GO 8) in a subset of strains. However, no homologue for qdtC was found in C. coli. This may be expected, as C. coli Quip3N is an N-acyl derivative instead of the N-acetyl derivative found in T. thermosaccharolyticum (27, 30). Moreover, these results are in agreement with data from previous studies in which C. jejuni isolates carrying the above-mentioned orthologues in the LOS locus were found to express Quip3NAc in their LOSs (26, 54, 67–69). Despite the presence of this sugar in all C. coli isolates investigated in this study, as described above, the putative dTDP-d-Quip3NAc biosynthesis genes are present in only a subset of strains, all belonging to LOS classes IV/V, VI, VII, X, and XI (see Fig. S2 in the supplemental material). Furthermore, truncation of orthologue 1742 due to a single-base deletion should have resulted in the loss of Quip3NAcyl in isolates 114 and 63, which was not the case. Cross talk between different glycosylation pathways was previously observed in C. jejuni (67, 71). Thus, due to Quip3NAcyl being predicted to be ubiquitously found in C. coli LOS structures, we hypothesize that the synthesis of this residue might be carried out by genes in conserved glycosylation pathways. Because of the structural similarity between Quip3NAc and bacillosamine precursors, it is tempting to speculate that the pgl system may play a role in the biosynthesis of Quip3NAc in C. coli.
In all C. coli isolates, phenotypic variation was observed to affect at least one sugar residue, as strains exhibited different numbers of Hep, Quip3NAcyl, HexNAc, or PEtn residues (Fig. 4B). Phenotypic variation in C. jejuni has been associated mainly with phase variation of genes containing repeats of GC homopolymeric tracts (23). However, no GC tracts were detected in the LOS loci of the chemically analyzed C. coli isolates. Further inspection of all the LOS locus sequences generated in this and previous studies (25, 26) revealed that G tracts are uncommon in C. coli LOS. Only isolates of LOS classes IV/V and VI had G tracts longer than 5 bases in their LOS biosynthesis locus. It is therefore unlikely that the observed phenotypic variation in our analyzed samples was caused by slipped-strand mispairing due to homopolymeric tracts within the LOS locus. These data suggest that other mechanisms, such as posttranscriptional regulation or epigenetic methylation of DNA, might be responsible for the phenotypic variation in LOS composition in C. coli.
Among LOS locus class II isolates, strain 65 exhibited the most divergent composition. Orthologue 1715 (wlaTB) has been associated with a HexNAc residue in C. jejuni 81116 (67), and the diversity observed in the C terminus of this orthologue may be responsible for the absence of HexNAc residues in isolates 73, 137, and 151. However, further research is required to confirm the exact role of orthologue 1715 in LOS biosynthesis. Similarly to strains of LOS locus class II, strains of LOS locus class X with minor genetic dissimilarities resulted in major differences in LOS chemical composition.
Isolates 65 and 114 also contained a deoxyHex residue in the LOS. No orthologues potentially involved in deoxyHex synthesis were identified within the LOS region in isolate 65, suggesting that genes outside the LOS locus may play a larger role in LOS biosynthesis than previously thought. Deoxyhexoses, such as 6-deoxy-β-l-altrose, fucose, or rhamnose, have been frequently detected in the O-chain of the lipopolysaccharide (LPS) of several Gram-negative species (72, 73). Nevertheless, in the genus Campylobacter, these sugars have been described as components of C. jejuni capsule (74) and C. fetus LPS (75).
In conclusion, the genetic and biochemical diversity of C. coli is greater than expected. C. coli LOS is characterized by a lipid A species consisting of GlcN-GlcN disaccharides and an outer core substituted with at least one Quip3NAcyl residue. Our results hint at cross talk between different glycosylation pathways, which generally has not been considered to play a role in LOS diversity. The relevance of these characteristic features for the ecology and virulence of C. coli is yet to be explored.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ann-Katrin Llarena for her comments and Marja-Liisa Hänninen for providing the strains.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00347-16.
REFERENCES
- 1.EFSA, ECDC. 2015. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J 13:3991. doi: 10.2903/j.efsa.2015.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillespie IA, O'Brien SJ, Frost AF, Adak GK, Horby P, Swan AV, Painter MJ, Neal KR. 2002. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg Infect Dis 8:937–942. doi: 10.3201/eid0809.010817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gürtler M, Alter T, Kasimir S, Fehlhaber K. 2005. The importance of Campylobacter coli in human campylobacteriosis: prevalence and genetic characterization. Epidemiol Infect 133:1081–1087. doi: 10.1017/S0950268805004164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young KT, Davis LM, DiRita VJ. 2007. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol 5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 5.Karlyshev AV, Ketley JM, Wren BW. 2005. The Campylobacter jejuni glycome. FEMS Microbiol Rev 29:377–390. doi: 10.1016/j.fmrre.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Iwata T, Chiku K, Amano K, Kusumoto M, Ohnishi-Kameyama M, Ono H, Akiba M. 2013. Effects of lipooligosaccharide inner core truncation on bile resistance and chick colonization by Campylobacter jejuni. PLoS One 8:e56900. doi: 10.1371/journal.pone.0056900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Javed MA, Cawthraw SA, Baig A, Li J, McNally A, Oldfield NJ, Newell DG, Manninga G. 2012. Cj1136 is required for lipooligosaccharide biosynthesis, hyperinvasion, and chick colonization by Campylobacter jejuni. Infect Immun 80:2361–2370. doi: 10.1128/IAI.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naito M, Frirdich E, Fields JA, Pryjma M, Li J, Cameron A, Gilbert M, Thompson SA, Gaynor EC. 2010. Effects of sequential Campylobacter jejuni 81-176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J Bacteriol 192:2182–2192. doi: 10.1128/JB.01222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Mourik AL, Steeghs L, van Laar J, Meiring HD, Hamstra HJ, van Putten JP, Wösten MM. 2010. Altered linkage of hydroxyacyl chains in lipid A of Campylobacter jejuni reduces TLR4 activation and antimicrobial resistance. J Biol Chem 285:15828–15836. doi: 10.1074/jbc.M110.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephenson HN, John CM, Naz N, Gundogdu O, Dorrell N, Wren BW, Jarvis GA, Bajaj-Elliott M. 2013. Campylobacter jejuni lipooligosaccharide sialylation, phosphorylation, and amide/ester linkage modifications fine-tune human Toll-like receptor 4 activation. J Biol Chem 288:19661–19672. doi: 10.1074/jbc.M113.468298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuijf ML, Samsom JN, van Rijs W, Bax M, Huizinga R, Heikema AP, van Doorn PA, van Belkum A, van Kooyk Y, Burgers PC, Luider TM, Endtz HP, Nieuwenhuis EE, Jacobs BC. 2010. TLR4-mediated sensing of Campylobacter jejuni by dendritic cells is determined by sialylation. J Immunol 185:748–755. doi: 10.4049/jimmunol.0903014. [DOI] [PubMed] [Google Scholar]
- 12.Bax M, Kuijf ML, Heikema AP, van Rijs W, Bruijns SC, García-Vallejo JJ, Crocker PR, Jacobs BC, van Vliet SJ, van Kooyk Y. 2011. Campylobacter jejuni lipooligosaccharides modulate dendritic cell-mediated T cell polarization in a sialic acid linkage-dependent manner. Infect Immun 79:2681–2689. doi: 10.1128/IAI.00009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huizinga R, van Rijs W, Bajramovic JJ, Kuijf ML, Laman JD, Samsom JN, Jacobs BC. 2013. Sialylation of Campylobacter jejuni endotoxin promotes dendritic cell-mediated B cell responses through CD14-dependent production of IFN-β and TNF-α. J Immunol 191:5636–5645. doi: 10.4049/jimmunol.1301536. [DOI] [PubMed] [Google Scholar]
- 14.Huizinga R, Easton AS, Donachie AM, Guthrie J, van Rijs W, Heikema A, Boon L, Samsom JN, Jacobs BC, Willison HJ, Goodyear CS. 2012. Sialylation of Campylobacter jejuni lipo-oligosaccharides: impact on phagocytosis and cytokine production in mice. PLoS One 7:e34416. doi: 10.1371/journal.pone.0034416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuki N, Susuki K, Koga M, Nishimoto Y, Odaka M, Hirata K, Taguchi K, Miyatake T, Furukawa K, Kobata T, Yamada M. 2004. Carbohydrate mimicry between human ganglioside GM1 and Campylobacter jejuni lipooligosaccharide causes Guillain-Barré syndrome. Proc Natl Acad Sci U S A 101:11404–11409. doi: 10.1073/pnas.0402391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Funakoshi K, Koga M, Takahashi M, Hirata K, Yuki N. 2006. Campylobacter coli enteritis and Guillain-Barré syndrome: no evidence of molecular mimicry and serological relationship. J Neurol Sci 246:163–168. doi: 10.1016/j.jns.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Bersudskya M, Rosenbergb P, Rudenskyc R, Wirguin I. 2000. Lipopolysaccharides of a Campylobacter coli isolate from a patient with Guillain-Barre syndrome display ganglioside mimicry. Neuromuscul Disord 10:182–186. doi: 10.1016/S0960-8966(99)00106-6. [DOI] [PubMed] [Google Scholar]
- 18.van Belkum A, Jacobs B, van Beek E, Louwen R, van Rijs W, Debruyne L, Gilbert M, Li J, Jansz A, Mégraud F, Endtz H. 2009. Can Campylobacter coli induce Guillain-Barré syndrome? Eur J Clin Microbiol Infect Dis 28:557–560. doi: 10.1007/s10096-008-0661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert M, Karwaski M, Bernatchez S, Young NM, Taboada E, Michniewicz J, Cunningham A, Wakarchuk WW. 2002. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. J Biol Chem 277:327–337. doi: 10.1074/jbc.M108452200. [DOI] [PubMed] [Google Scholar]
- 20.Dzieciatkowska M, Brochu D, van Belkum A, Heikema AP, Yuki N, Houliston RS, Richards JC, Gilbert M, Li J. 2007. Mass spectrometric analysis of intact lipooligosaccharide: direct evidence for O-acetylated sialic acids and discovery of O-linked glycine expressed by Campylobacter jejuni. Biochemistry 46:14704–14714. doi: 10.1021/bi701229k. [DOI] [PubMed] [Google Scholar]
- 21.Chiu CP, Watts AG, Lairson LL, Gilbert M, Lim D, Wakarchuk WW, Withers SG, Strynadka NC. 2004. Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat Struct Mol Biol 11:163–170. doi: 10.1038/nsmb720. [DOI] [PubMed] [Google Scholar]
- 22.Parker CT, Gilbert M, Yuki N, Endtz HP, Mandrell RE. 2008. Characterization of lipooligosaccharide-biosynthetic loci of Campylobacter jejuni reveals new lipooligosaccharide classes: evidence of mosaic organizations. J Bacteriol 190:5681–5689. doi: 10.1128/JB.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linton D, Gilbert M, Hitchen PG, Dell A, Morris HR, Wakarchuk WW, Gregson NA, Wren BW. 2000. Phase variation of a β-1,3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol Microbiol 37:501–514. doi: 10.1046/j.1365-2958.2000.02020.x. [DOI] [PubMed] [Google Scholar]
- 24.Revez J, Hänninen M. 2012. Lipooligosaccharide locus classes are associated with certain Campylobacter jejuni multilocus sequence types. Eur J Clin Microbiol Infect Dis 31:2203–2209. doi: 10.1007/s10096-012-1556-3. [DOI] [PubMed] [Google Scholar]
- 25.Skarp-de Haan CP, Culebro A, Schott T, Revez J, Schweda E, Hänninen M, Rossi M. 2014. Comparative genomics of unintrogressed Campylobacter coli clades 2 and 3. BMC Genomics 15:129. doi: 10.1186/1471-2164-15-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richards VP, Lefébure T, Pavinski Bitar PD, Stanhope MJ. 2013. Comparative characterization of the virulence gene clusters (lipooligosaccharide [LOS] and capsular polysaccharide [CPS]) for Campylobacter coli, Campylobacter jejuni subsp. jejuni and related Campylobacter species. Infect Genet Evol 14:200–213. doi: 10.1016/j.meegid.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aspinall GO, McDonald AG, Pang H, Kurjanczyk LA, Penner JL. 1993. Lipopolysaccharide of Campylobacter coli serotype O:30. Fractionation and structure of liberated core oligosaccharide. J Biol Chem 268:6263–6268. [PubMed] [Google Scholar]
- 28.Beer W, Adam M, Seltmann G. 1986. Monosaccharide composition of lipopolysaccharides from Campylobacter jejuni and Campylobacter coli. J Basic Microbiol 26:201–204. doi: 10.1002/jobm.3620260405. [DOI] [PubMed] [Google Scholar]
- 29.Naess V, Hofstad T. 1984. Chemical studies of partially hydrolysed lipopolysaccharides from four strains of Campylobacter jejuni and two strains of Campylobacter coli. J Gen Microbiol 130:2783–2789. [DOI] [PubMed] [Google Scholar]
- 30.Seltmann G, Beer W. 1985. Vorkommen von 3-Amino-3,6-didesoxy-d-glucose in einem Lipopolysaccharid von Campylobacter coli. J Basic Microbiol 25:551–552. doi: 10.1002/jobm.3620250823. [DOI] [PubMed] [Google Scholar]
- 31.Sheppard SK, Didelot X, Jolley KA, Darling AE, Pascoe B, Meric G, Kelly DJ, Cody A, Colles FM, Strachan NJ, Ogden ID, Forbes K, French NP, Carter P, Miller WG, McCarthy ND, Owen R, Litrup E, Egholm M, Affourtit JP, Bentley SD, Parkhill J, Maiden MC, Falush D. 2013. Progressive genome-wide introgression in agricultural Campylobacter coli. Mol Ecol 22:1051–1064. doi: 10.1111/mec.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheppard SK, Maiden MC. 2015. The evolution of Campylobacter jejuni and Campylobacter coli. Cold Spring Harb Perspect Biol 7:a018119. doi: 10.1101/cshperspect.a018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olkkola SH, Juntunen P, Heiska H, Hyytiäinen H, Hänninen M. 2010. Mutations in the rpsL gene are involved in streptomycin resistance in Campylobacter coli. Microb Drug Resist 16:105–110. doi: 10.1089/mdr.2009.0128. [DOI] [PubMed] [Google Scholar]
- 34.Juntunen P, Olkkola S, Hänninen M. 2011. Longitudinal on-farm study of the development of antimicrobial resistance in Campylobacter coli from pigs before and after danofloxacin and tylosin treatments. Vet Microbiol 150:322–330. doi: 10.1016/j.vetmic.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Juntunen P, Heiska H, Olkkola S, Myllyniemi AL, Hänninen ML. 2010. Antimicrobial resistance in Campylobacter coli selected by tylosin treatment at a pig farm. Vet Microbiol 146:90–97. doi: 10.1016/j.vetmic.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 36.Lehtopolku M, Kotilainen P, Haanperä-Heikkinen M, Nakari U, Hänninen M, Huovinen P, Siitonen A, Eerola E, Jalava J, Hakanen AJ. 2011. Ribosomal mutations as the main cause of macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother 55:5939–5941. doi: 10.1128/AAC.00314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kärenlampi R, Rautelin H, Schönberg-Norio D, Paulin L, Hänninen M. 2007. Longitudinal study of Finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl Environ Microbiol 73:148–155. doi: 10.1128/AEM.01488-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llarena AK, Skarp-de Haan CP, Rossi M, Hänninen M. 2015. Characterization of the Campylobacter jejuni population in the barnacle geese reservoir. Zoonoses Public Health 62:209–221. doi: 10.1111/zph.12141. [DOI] [PubMed] [Google Scholar]
- 39.Hänninen M, Pajarre S, Klossner M, Rautelin H. 1998. Typing of human Campylobacter jejuni isolates in Finland by pulsed-field gel electrophoresis. J Clin Microbiol 36:1787–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin A, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:1–15. doi: 10.1186/1471-2164-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 44.Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, Parkhill J, Rajandream MA. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ekseth OK, Kuiper M, Mironov V. 9 October 2013. orthAgogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics doi: 10.1093/bioinformatics/btt582. [DOI] [PubMed] [Google Scholar]
- 48.Edgar RC, Sjolander K. 2004. A comparison of scoring functions for protein sequence profile alignment. Bioinformatics 20:1301–1308. doi: 10.1093/bioinformatics/bth090. [DOI] [PubMed] [Google Scholar]
- 49.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:1–19. doi: 10.1186/1471-2105-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abascal F, Zardoya R, Telford MJ. 30 April 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 16 October 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Revez J, Rossi M, Ellström P, de Haan CP, Rautelin H, Hänninen M. 2011. Finnish Campylobacter jejuni strains of multilocus sequence type ST-22 complex have two lineages with different characteristics. PLoS One 6:e26880. doi: 10.1371/journal.pone.0026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsai C, Frasch CE. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem 119:115–119. doi: 10.1016/0003-2697(82)90673-X. [DOI] [PubMed] [Google Scholar]
- 54.Dzieciatkowska M, Liu X, Heikema AP, Houliston RS, van Belkum A, Schweda EK, Gilbert M, Richards JC, Li J. 2008. Rapid method for sensitive screening of oligosaccharide epitopes in the lipooligosaccharide from Campylobacter jejuni strains isolated from Guillain-Barré syndrome and Miller Fisher syndrome patients. J Clin Microbiol 46:3429–3436. doi: 10.1128/JCM.00681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moran AP, Zähringer U, Seydel U, Scholz D, Stütz P, Rietschel ET. 1991. Structural analysis of the lipid A component of Campylobacter jejuni CCUG 10936 (serotype O:2) lipopolysaccharide. Eur J Biochem 198:459–469. doi: 10.1111/j.1432-1033.1991.tb16036.x. [DOI] [PubMed] [Google Scholar]
- 56.Moran AP, Rietschel ET, Kosunen TU, Zähringer U. 1991. Chemical characterization of Campylobacter jejuni lipopolysaccharides containing N-acetylneuraminic acid and 2,3-diamino-2,3-dideoxy-d-glucose. J Bacteriol 173:618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moran AP. 1997. Structure and conserved characteristics of Campylobacter jejuni lipopolysaccharides. J Infect Dis 176:S115–S121. doi: 10.1086/513781. [DOI] [PubMed] [Google Scholar]
- 58.Stahl M, Ries J, Vermeulen J, Yang H, Sham HP, Crowley SM, Badayeva Y, Turvey SE, Gaynor EC, Li X. 2014. A novel mouse model of Campylobacter jejuni gastroenteritis reveals key pro-inflammatory and tissue protective roles for Toll-like receptor signaling during infection. PLoS Pathog 10:e1004264. doi: 10.1371/journal.ppat.1004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bereswill S, Fischer A, Plickert R, Haag L, Otto B, Kühl AA, Dashti JI, Zautner AE, Muñoz M, Loddenkemper C, Groß U, Göbel UB, Heimesaat MM. 2011. Novel murine infection models provide deep insights into the “ménage à trois” of Campylobacter jejuni, microbiota and host innate immunity. PLoS One 6:e20953. doi: 10.1371/journal.pone.0020953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aspinall GO, McDonald AG, Pang H, Kurjanczyk LA, Penner JL. 1993. An antigenic polysaccharide from Campylobacter coli serotype O:30. Structure of a teichoic acid-like antigenic polysaccharide associated with the lipopolysaccharide. J Biol Chem 268:18321–18329. [PubMed] [Google Scholar]
- 61.Roux F, Sproston E, Rotariu O, MacRae M, Sheppard SK, Bessell P, Smith-Palmer A, Cowden J, Maiden MC, Forbes K, Strachan NJ. 2013. Elucidating the aetiology of human Campylobacter coli infections. PLoS One 8:e64504. doi: 10.1371/journal.pone.0064504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sweet CR, Ribeiro AA, Raetz CR. 2004. Oxidation and transamination of the 3″-position of UDP-N-acetylglucosamine by enzymes from Acidithiobacillus ferrooxidans. J Biol Chem 279:25400–25410. doi: 10.1074/jbc.M400596200. [DOI] [PubMed] [Google Scholar]
- 63.Jansonius JN. 1998. Structure, evolution and action of vitamin B6-dependent enzymes. Curr Opin Struct Biol 8:759–769. doi: 10.1016/S0959-440X(98)80096-1. [DOI] [PubMed] [Google Scholar]
- 64.Altman E, Schäffer C, Brisson JR, Messner P. 1995. Characterization of the glycan structure of a major glycopeptide from the surface layer glycoprotein of Clostridium thermosaccharolyticum E207-71. Eur J Biochem 229:308–315. doi: 10.1111/j.1432-1033.1995.tb20470.x. [DOI] [PubMed] [Google Scholar]
- 65.Ovchinnikova OG, Rozalski A, Liu B, Knirel YA. 2013. O-antigens of bacteria of the genus Providencia: structure, serology, genetics, and biosynthesis. Biochemistry (Mosc) 78:798–817. doi: 10.1134/S0006297913070110. [DOI] [PubMed] [Google Scholar]
- 66.Veremeichenko S, Zdorovenko GM. 2004. Structure and properties of the lipopolysaccharide of Pseudomonas fluorescens IMV 2366 (biovar III). Microbiology 73:312–319. [PubMed] [Google Scholar]
- 67.Holden KM, Gilbert M, Coloe PJ, Li J, Fry BN. 2012. The role of WlaRG, WlaTB and WlaTC in lipooligosaccharide synthesis by Campylobacter jejuni strain 81116. Microb Pathog 52:344–352. doi: 10.1016/j.micpath.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 68.Godschalk PC, Gilbert M, Jacobs BC, Kramers T, Tio-Gillen AP, Ang CW, Van den Braak N, Li J, Verbrugh HA, Van Belkum A, Endtz HP. 2006. Co-infection with two different Campylobacter jejuni strains in a patient with the Guillain-Barré syndrome. Microb Infect 8:248–253. doi: 10.1016/j.micinf.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 69.Aspinall GO, Lynch CM, Pang H, Shaver RT, Moran AP. 1995. Chemical structures of the core region of Campylobacter jejuni O:3 lipopolysaccharide and an associated polysaccharide. Eur J Biochem 231:570–578. doi: 10.1111/j.1432-1033.1995.tb20734.x. [DOI] [PubMed] [Google Scholar]
- 70.Pföstl A, Zayni S, Hofinger A, Kosma P, Schäffer C, Messner P. 2008. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-α-d-glucose. Biochem J 410:187–194. doi: 10.1042/BJ20071044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bernatchez S, Szymanski CM, Ishiyama N, Li J, Jarrell HC, Lau PC, Berghuis AM, Young NM, Wakarchuk WW. 2005. A single bifunctional UDP-GlcNAc/Glc 4-epimerase supports the synthesis of three cell surface glycoconjugates in Campylobacter jejuni. J Biol Chem 280:4792–4802. doi: 10.1074/jbc.M407767200. [DOI] [PubMed] [Google Scholar]
- 72.Ma B, Simala-Grant JL, Taylor DE. 2006. Fucosylation in prokaryotes and eukaryotes. Glycobiology 16:158R–184R. doi: 10.1093/glycob/cwl040. [DOI] [PubMed] [Google Scholar]
- 73.Knirel Y. 2011. Structure of O-antigens, p 41–115. In Knirel YA, Valvano MA (ed), Bacterial lipopolysaccharides. Springer, Vienna, Austria. [Google Scholar]
- 74.Hanniffy OM, Shashkov AS, Moran AP, Prendergast MM, Senchenkova SN, Knirel YA, Savage AV. 1999. Chemical structure of a polysaccharide from Campylobacter jejuni 176.83 (serotype O:41) containing only furanose sugars. Carbohydr Res 319:124–132. doi: 10.1016/S0008-6215(99)00129-9. [DOI] [PubMed] [Google Scholar]
- 75.Moran AP, O'Malley DT, Kosunen TU, Helander IM. 1994. Biochemical characterization of Campylobacter fetus lipopolysaccharides. Infect Immun 62:3922–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.