ABSTRACT
Modulation of the intestinal microbial ecosystem (IME) is a useful target to establish probiotic efficacy in a healthy population. We conducted a randomized, double-blind, crossover, and placebo-controlled intervention study to determine the impact of Bifidobacterium bifidum strain Bb on the IME of adult healthy volunteers of both sexes. High-throughput 16S rRNA gene sequencing was used to characterize the fecal microbiota before and after 4 weeks of daily probiotic cell consumption. The intake of approximately one billion live B. bifidum cells affected the relative abundance of dominant taxa in the fecal microbiota and modulated fecal butyrate levels. Specifically, Prevotellaceae (P = 0.041) and Prevotella (P = 0.034) were significantly decreased, whereas Ruminococcaceae (P = 0.039) and Rikenellaceae (P = 0.010) were significantly increased. We also observed that the probiotic intervention modulated the fecal concentrations of butyrate in a manner dependent on the initial levels of short-chain fatty acids (SCFAs). In conclusion, our study demonstrates that a single daily administration of Bifidobacterium bifidum strain Bb can significantly modify the IME in healthy (not diseased) adults. These findings demonstrate the need to reassess the notion that probiotics do not influence the complex and stable IME of a healthy individual.
IMPORTANCE Foods and supplements claimed to contain health-promoting probiotic microorganisms are everywhere these days and mainly intended for consumption by healthy people. However, it is still debated what actual effects probiotic products may have on the healthy population. In this study, we report the results of an intervention trial aimed at assessing the modifications induced in the intestinal microbial ecosystem of healthy adults from the consumption of a probiotic product. Our results demonstrate that the introduction of a probiotic product in the dietary habits of healthy people may significantly modify dominant taxa of the intestinal microbiota, resulting in the modulation of short-chain fatty acid concentrations in the gut. The overall changes witnessed in the probiotic intervention indicate a mechanism of microbiota modulation that could have potential effects on human health.
INTRODUCTION
The prevailing notion that the deliberate intake of viable cells of certain microorganisms through food and supplements may be beneficial for health underlies the worldwide commercial success of probiotic products. Probiotics have been defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (1). Therefore, a positive consequence on consumer health is an intrinsic feature of any formulation that is considered a probiotic. Accordingly, the European Union considers the term probiotic a health claim per se (“reference to probiotic/prebiotic implies a health benefit” [2]). However, although numerous studies have demonstrated the efficacy of different probiotic preparations in a number of pathological conditions (3), the potential benefits associated with probiotic consumption by the general (healthy) population remain unclear (4). The lack of a clear cause-and-effect relationship between probiotic intake and health benefits for the general consumer is the most frequent reason stated by the European Food Safety Authority (EFSA) for the rejection of all health claims requested for probiotics.
The intestinal microbiota has been proposed as an additional organ of the human body that performs numerous functions, ranging from vitamin production and immunomodulation to improvement of nutrient bioavailability and competitive exclusion against potential detrimental microorganisms (5). Therefore, modification of the intestinal microbial ecosystem (IME) may potentially induce functional changes that affect host physiology (5) and is generally recognized by certain health agencies (e.g., Italian Ministry of Health and Health Canada Federal Department) as the primary element supporting probiotic efficacy. Studies that describe and demonstrate the ability of specific probiotics to impact the IME of healthy consumers, however, remain limited (6, 7). Contradictory results have also been obtained (8), possibly owing to differences in the microbial strain used, the number of viable cells administered, and the product formulation. Furthermore, the deep complexity and profound variability of microbiota composition among subjects can hinder the recognition of actual modifications. As a consequence, the impact of a probiotic, dietary, or pharmacologic intervention on the IME can be studied only by adopting sensitive analytical tools, such as 16S rRNA gene profiling, and an appropriate trial design.
We therefore performed an intervention trial with a crossover design and used 16S rRNA gene profiling together with short-chain fatty acid (SCFA) quantification in fecal samples from healthy adults to investigate the impact on the IME of a product containing a single Bifidobacterium bifidum strain. The strain was selected as a representative of a species that has been reported to possess numerous host interaction properties (9), including marked adhesion to enterocytes (10–12), immunomodulation (12–14), and metabolism of mucin and human milk oligosaccharides (15, 16).
MATERIALS AND METHODS
Participants.
Thirty-eight healthy human volunteers (21 women and 17 men; age, 24 to 54 years; mean, 31 years) participated in the study, named PROBIOTA-Bb: “Effect of the probiotic strain Bifidobacterium bifidum Bb on the fecal microbiota of healthy adults.” All patients provided written informed consent, and the study protocol was approved by the Ethics Committee for Research of the Università degli Studi di Milano (opinion 37/12, 19 December 2012). The procedures were carried out in accordance with the approved trial synopsis. The following inclusion criteria were adopted for the enrollment of participants: age between 18 and 55 years, good general health, and a signed consent form. The following exclusion criteria were adopted: antibiotic therapy during the 1 month prior to the first visit, intentional intake of probiotic or prebiotic products 1 month before the first visit, viral or bacterial enteritis during the 2 months before the first visit, presence of gastrointestinal disorders (e.g., diarrhea, inflammatory bowel disease, or irritable bowel syndrome), pregnancy or breastfeeding, and recent or presumed episodes of alcoholism or drug addiction. Participants in the study were prohibited from eating probiotic foods and supplements and any foods or supplements enriched in prebiotic compounds. Traditional yogurt was allowed.
Experimental design.
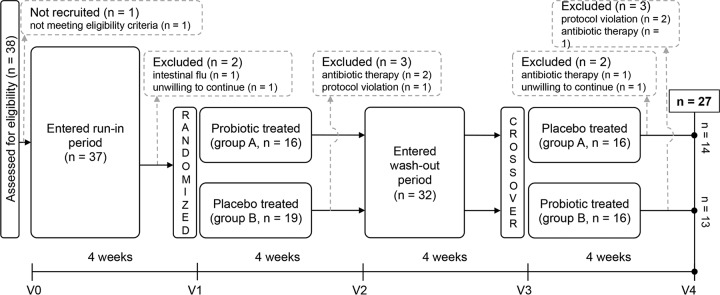
The PROBIOTA-Bb study was a randomized, double-blind, and placebo-controlled crossover trial with two parallel groups (Fig. 1). The study consisted of a 4-week prerecruitment (run-in) phase, followed by random assignment of participants to group A (n = 16) or group B (n = 19). The group A protocol included a 4-week probiotic treatment (one capsule every day for 4 weeks in addition to habitual diet), followed by a 4-week washout period and a 4-week placebo phase. Group B followed the opposite sequence, with placebo, washout, and then probiotic treatment. Participants received written and oral instructions to store the capsules at room temperature, to avoid exposure of the capsules to heat sources, and to consume one capsule every day in the morning at least 15 min before breakfast with natural (not sparkling) water (alternatively, to consume the capsule in the evening at least 3 h after the last meal of the day). No research has tested which mode of administration is better, ingesting probiotics on an empty stomach or with meals. In this study, we decided to invite volunteers to consume capsules on an empty stomach because food intake may vary enormously from meal to meal and from subject to subject, differently affecting cell transit through the stomach and therefore diversely influencing probiotic activity.
FIG 1.
Schematic of the study design and flow.
The study consisted of five visits: before the run-in period (visit V0), before and after the first treatment (V1 and V2, respectively), and before and after the second treatment (V3 and V4, respectively) (Fig. 1). During each consultation, participants completed a short food frequency questionnaire that was specifically prepared to include a section for items considered potential sources of prebiotic fibers. In addition, participants compiled a weekly Bristol stool chart to report their bowel habits.
Products used in the trial.
The probiotic preparation consisted of gelatin uncoated capsules filled with Bifidobacterium bifidum Bb isolated from the stool from a healthy adult woman and available in the culture collection of the section of Food Microbiology and Bioprocesses at the Department of Food, Environmental and Nutritional Sciences (University of Milan). Viable and total bacterial cell counts were performed approximately 1 week before the beginning of the trial on several randomly selected capsules. Viable counts were performed by serial dilutions and plating on de Man-Rogosa-Sharpe (MRS) agar plates with the addition of 0.05% cysteine-HCl. After 72 h of anaerobic incubation at 37°C, we calculated that each capsule contained 1.3 × 109 ± 0.1 × 109 CFU. Total bacterial cell counts, which were performed by cytofluorimetry (BD Accuri C6; Becton Dickinson Italia, Milan, Italy) upon SYBR green cell labeling, revealed that each capsule contained 3.8 × 109 bacterial cells per capsule. The capsules also contained maltodextrin, cellulose powder, dextrose, a separating agent (magnesium salts of edible fatty acid), and silica. Placebo capsules were identical and were filled with maltodextrin instead of dry powder probiotic bacteria. This trial was registered at www.isrctn.com/search?q=ISRCTN56945491 under trial no. ISRCTN56945491.
Collection of fecal samples and extraction of metagenomic DNA.
A fecal sample was collected from each participant in a sterile plastic pot no more than 24 h before visits V1, V2, V3, and V4. According to the recommendations for “storage conditions of intestinal microbiota matter in metagenomic analysis” (17), participants were asked to preserve the sample at room temperature until delivery to the laboratory. At delivery, stool specimens were immediately stored at −80°C until metagenomic DNA extraction, which was performed within 14 days by means of a QIAamp DNA stool minikit (Qiagen, Valencia, CA), according to the manufacturer's specifications, adopting a temperature of 95°C in step three to maximize bacterial cell lysis.
Profiling of fecal microbiota composition.
The bacterial community structure of fecal samples was determined by 16S rRNA gene profiling, as previously described (18). In brief, a DNA fragment encompassing the variable region V3 of the 16S rRNA gene was amplified from metagenomic DNA with the primers Probio_Uni (5′-CCTACGGGRSGCAGCAG-3′) and Probio_Rev (5′-ATTACCGCGGCTGCT-3′) and was sequenced by means of Ion Torrent PGM sequencing technology (Life Technologies, Carlsbad, CA). Specifically, emulsion PCR was performed using the Ion OneTouch 200 template kit version 2 DL (Life Technologies, Guilford, CT), according to the manufacturer's instructions. Amplicon library sequencing was performed on 316 Chips using the Ion sequencing 200 kit (Life Technologies). The sequencing runs were multiplexed, and barcode sequences were used to discriminate the samples. Sequence reads were then analyzed using the bioinformatic pipeline Quantitative Insights Into Microbial Ecology (QIIME) version 1.7.0 (19) with the GreenGenes database updated to version 13.5. Bacterial relative abundances in each fecal sample were reported at the taxonomic levels of phylum, class, order, family, and genus.
Quantification of fecal SCFAs.
SCFAs were quantified in the fecal samples from 25 out of 27 subjects who completed the intervention trial. The remaining two subjects were excluded from the analysis due to insufficient fecal material availability. Fecal samples were extracted according to Huda-Faujan et al. (20), with some modifications. In detail, stool specimens (100 mg) were suspended in 2 ml of 0.001% HCOOH and vortexed for 1 min. The suspension was centrifuged at 1,000 × g for 2 min at 4°C, and the supernatant was recovered. The residue was extracted again, as described above. The supernatants were combined, and the volume was adjusted to 5 ml with a solution of 0.001% HCOOH in water. All extracts were stored at −20°C. Before ultraperformance liquid chromatography–high-resolution-mass spectrometry (UPLC-HR-MS) analysis, samples were diluted 1:100 in 0.001% HCOOH and centrifuged at 3,000 × g for 1 min.
UPLC-HR-MS analysis was carried out on an Acquity UPLC separation module (Waters, Milford, MA, USA) coupled with an Exactive Orbitrap MS with an HESI-II probe for electrospray ionization (Thermo Scientific, San Jose, CA, USA). The ion source and interface conditions were as follows: spray voltage, −3.0 kV, sheath gas flow rate, 35 arbitrary units; auxiliary gas flow rate, 10 arbitrary units; temperature, 120°C; and capillary temperature, 320°C. A 1.8-μm HSS T3 column (150 by 2.1 mm; Waters) was used for separation at a flow rate of 0.2 ml/min. The eluents were 0.001% HCOOH in MilliQ-treated water (solvent A) and CH3OH:CH3CN (1:1 [vol/vol], solvent B). A 5-μl aliquot of the sample was separated by the UPLC using the following elution gradient: 0% B for 4 min, 0 to 15% B in 6 min, 15 to 20% B in 5 min, 20% for 13 min, and then return to initial conditions in 1 min. The column and samples were maintained at 30 and 15°C, respectively. The UPLC eluate was analyzed in full-scan MS in the range m/z 50 to 130. The resolution was set at 50 K, the automatic gain control (AGC) target was 1E6, and the maximum ion injection time was 100 ms. The ion with m/z 91.0038, corresponding to the formic acid dimer [2M-H]−, was used as the lock mass. The mass tolerance was 2 ppm. The MS data were processed using Xcalibur software (Thermo Scientific). Analytical-grade SCFAs were used as standards (Sigma-Aldrich, Milan, Italy). Five-point external calibration curves were adopted to quantify pyruvic, lactic, succinic, acetic, propionic, butyric, isobutyric, valeric, and isovaleric acids in fecal samples. SCFA concentrations were expressed in milligrams per kilogram of wet feces.
Statistics.
Statistical analyses were performed using the R statistical software (version 3.1.2). To measure valid outcomes, only participants with 100% compliance with the treatments and the experiment protocol were included in the analysis (per-protocol analysis). The numerical value of 0 was given to any taxon that was absent in a specific sample to allow a comparison. Because of the necessary crossover design for significant results, intention-to-treat analysis was not performed. Differences between the effects on microbiota composition of probiotic and placebo treatments were evaluated by analyzing the data with nonparametric Wilcoxon-Mann-Whitney test with Benjamini-Hochberg correction using paired data. In particular, we performed two separated statistical analyses: in the first analysis, we compared data for each taxon before versus after probiotic intake; in the second, data from the same subjects were compared before versus after placebo. This analysis allowed us to decide whether the population distributions were identical without assuming them to display a normal distribution. A nonparametric test derived from the Shapiro-Francia test was performed for the composite hypothesis of normality. The P value was computed from the formula given by Royston (21). Repeated-measures analysis of variance (ANOVA) was used when the data were consistent with the assumption of a parametric test for normal distribution. Statistical significance was set at a P value of ≤0.05, and mean differences with 0.05 < P ≤ 0.10 were accepted as trends. To group subjects into enterotypes, the tutorial by the European Molecular Biology Laboratory (EMBL) was used (http://enterotype.embl.de/). With respect to differences in the absolute quantity of SCFAs, the subjects were clustered using a Jensen-Shannon divergence (JSD) algorithm based on the SCFA concentration. Moreover, the treatment response was evaluated by performing the Wilcoxon-Mann-Whitney test with Benjamini-Hochberg correction using the paired-data test.
Accession number(s).
Sequence reads have been deposited in NCBI's Sequence Read Archive (ENA) under accession no. PRJEB11694.
RESULTS
Study compliance and questionnaire analyses.
All participants tolerated the capsules well, and no adverse events were reported. Participants maintained their usual dietary habits during the study, and no significant differences in the intake of potentially prebiotic foods were observed. The only registered modifications were related to slight seasonal differences in the availability of fruits and vegetables (the study began in June and ended in October). Participants' adherence to the study protocol was assessed based on capsule counts and fecal sample collection, and compliance was higher than 95%. In total, 38 participants were assessed for eligibility, and 35 participants were randomly assigned; 27 participants (77%) concluded the study, with 14 participants in the randomization group A (6 females and 8 males) and 13 participants in group B (7 women and 6 men). The drop-out rate of volunteers who began the first treatment was 20% (n = 7), consistent with the literature (6), and apparently justified by the strict exclusion criteria and quite long duration of the study (four months). No significant changes in stool consistency and evacuation frequency were noted according to the analysis of data reported in stool diaries (data not shown).
16S rRNA gene profiling revealed that the fecal microbiota composition was markedly varied among the participants in the PROBIOTA-Bb study.
A total of 15,845,061 filtered high-quality sequence reads were generated (average, 135,428 reads per sample), with a mean ± standard deviation (SD) length of 179 ± 4 bp. Rarefaction curves indicated that most fecal microbiota diversity had been covered (see Fig. S1A in the supplemental material). We identified a total of 93 bacterial families and 190 bacterial genera, with a minimum number of 23 families and 38 genera and a maximum of 65 families and 125 genera per fecal sample. Only 17 genera were detected in all subjects at the 4 time points (8.9% of all detected genera), and 47 genera were present in at least one sample for all subjects (approximately 25% of all detected genera).
Subsequently, microbiota profiling data were stratified by enterotyping based on the relative abundances of the bacterial genera (22). The microbiota compositions of all samples in this study were clustered in two groups (Silhouette index [SI] = 0.25; see Fig. S2 in the supplemental material) resembling the Bacteroides-dominant (Ba) and the Prevotella-dominant (Pr) enterotypes (22). During the study, 8 of 27 subjects changed enterotypes; specifically, we observed 11 shifts from one enterotype to the other (accounting for the 14.7% of all possible shifts), with 3 during the probiotic intervention, 5 in the placebo treatment, and 3 in the washout phase (Fig. S2B). Therefore, the treatments did not affect the affiliation of subjects to enterotypes. Interestingly, bacterial richness differed significantly between the two enterotype clusters: the α-diversity estimated by the Chao1 index was significantly higher in the Ba group than in the Pr group of samples (see Fig. S3 in the supplemental material).
The analyses described below were conducted to identify the effects induced by the probiotic intake on fecal microbial ecology and were restricted to the participants who received the intended interventions in accordance with the protocol (per-protocol analysis).
B. bifidum intake did not modify the α- and β-diversities of the fecal microbiota.
The effect of the probiotic intervention was first investigated with respect to the modification induced by B. bifidum intake on the richness and evenness of the operational taxonomic units (OTUs) in each sample (α-diversity) and the intersample relationship of the bacterial compositions (β-diversity). According to both parametric (repeated-measures ANOVA) and nonparametric (Wilcoxon text) statistics, the intake of B. bifidum did not significantly affect the intrasample biodiversity as measured by the Chao1 and Shannon coefficients (predictors of taxonomic richness and evenness; Fig. S1A and B in the supplemental material) or the intersample diversity determined by principal-coordinate analysis (PCoA) based on weighted and unweighted UniFrac distances (measure of β-diversity; Fig. S1C).
Probiotic intervention with B. bifidum modified the relative abundance of dominant taxa in the fecal microbiota.
The Wilcoxon test with Benjamini-Hochberg correction was used to identify the bacterial taxa (from phylum to genus) that were significantly affected by the probiotic or placebo treatments. This nonparametric statistical analysis revealed that B. bifidum Bb intake had a greater impact than placebo on the relative abundance of OTUs (see Table S1 in the supplemental material). In detail, the probiotic intervention induced the significant modification of 25 taxa, whereas only 13 taxa were different after placebo. More importantly, only 1 OTU that changed during the placebo treatment had a relative abundance above 1% (undefined members of the Bacteroidales order, from 1.10 to 0.46%; P = 0.036; Table S1), whereas probiotic intake significantly modulated the abundance of several dominant taxa of the fecal microbiota, including the families Prevotellaceae (mean value of the relative abundances from 14.18 to 11.97%; P = 0.041), Rikenellaceae (from 3.99 to 5.92%; P = 0.010), and Ruminococcaceae (from 12.21 to 15.27%; P = 0.039), and the genus Prevotella (from 14.16 to 11.96%; P = 0.034) (Fig. 2; see also Table S1).
FIG 2.
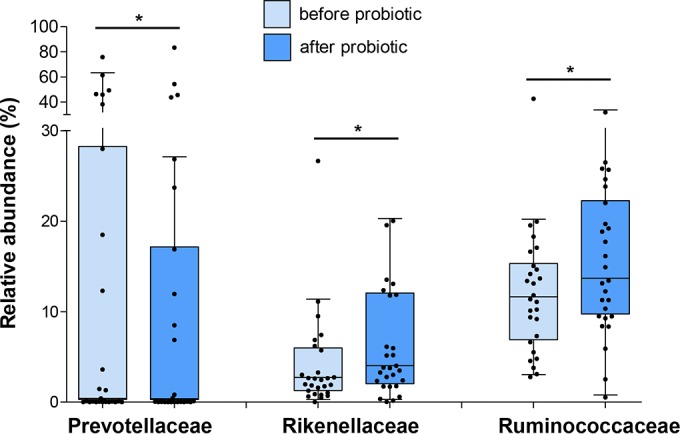
Relative abundance of dominant bacterial families in fecal samples significantly modified by the probiotic treatment. Data are shown as Tukey box plots. Statistically significant differences are according to the Wilcoxon-Mann-Whitney test with Benjamini-Hochberg correction; *, P < 0.05.
The statistical strength of the observed differences between the effects induced by the two treatments is corroborated by the comparison of OTU relative abundances before treatments (i.e., for samples collected at V1 and V3; Fig. 1). The Wilcoxon test revealed that only three OTUs with relative abundances of less than 1% were significantly different between the placebo and probiotic pretreatment phases (see Table S1 in the supplemental material).
We did not find a significant modulation of the genus Bifidobacterium by probiotic treatment, suggesting that the intake of a billion B. bifidum cells was not enough to affect the relative abundance of the entire genus. On the contrary, we found that the relative abundance of the reads associated with the B. bifidum species increased importantly upon probiotic consumption, changing from a median below the detection limit to 0.005%. In contrast, during placebo treatment, the relative abundance of B. bifidum reads decreased from 0.002% to below the detection limit.
Finally, we also observed that the impact of the probiotic intervention on enterotypes was greater on the Pr group of samples (see Table S2 in the supplemental material); this result, however, may have been merely a reflection of the change in Prevotella levels.
Probiotic intervention modulated the fecal levels of butyrate.
We quantified the concentration of SCFAs in fecal samples by UPLC-HR-MS (see Table S3 in the supplemental material). Statistical analyses (repeated-measures ANOVA and Wilcoxon test) revealed that neither the probiotic nor placebo intervention significantly affected the levels of SCFAs quantified in all fecal samples (see Table S4 in the supplemental material).
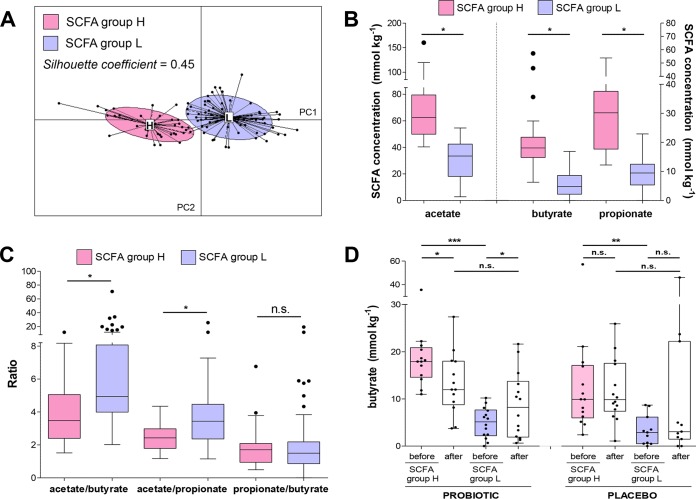
Subsequently, we clustered subjects by PCoA based on the concentrations of the most abundant fecal SCFAs, i.e., acetate, butyrate, and propionate. According to the highest Silhouette coefficient of clustering prediction (SI = 0.43), we separated the samples in two groups (Fig. 3A), which do not correspond to enterotype clusters. SCFA group H was characterized by higher concentrations of acetate, butyrate, isovalerate, propionate, succinate, and valerate than the second group (SCFA group L; Fig. 3B). Furthermore, SCFA group H exhibited significantly lower acetate/butyrate and acetate/propionate ratios than SCFA group L (Fig. 3C). Subsequently, we investigated whether the intervention affected fecal levels of taxa and SCFAs in the two groups. We did not find significant modifications of taxon relative abundance upon probiotic or placebo treatment in the SCFA groups (see Table S5 in the supplemental material). On the contrary, we determined that the probiotic (but, notably, not the placebo) treatment induced a significant change in the fecal level of butyrate in both SCFA groups (Fig. 3D; see also Table S3 in the supplemental material). Specifically, the median of butyrate levels increased from 5.2 to 8.2 mg kg−1 of wet feces in group L and dropped from 17.9 to 12.0 mg kg−1 of wet feces in group H, erasing the significant difference initially existing in the levels of this SCFA between the groups (Fig. 3D). None of the other SCFAs were significantly affected by the treatments (Table S3).
FIG 3.
Short-chain fatty acid (SCFA) cluster analysis. (A) Principal-coordinate analysis (PCoA; the first two principal components are shown); clustering was based on the fecal concentrations of acetate, butyrate, and propionate using JSD distance and the Partitioning around Medoids (PAM) algorithm. (B) Tukey box plots representing the proportion of main fecal SCFAs in groups H (displaying higher concentrations of SCFAs) and L (lower SCFA concentrations). (C) Tukey box plots of the ratios between the three main fecal SCFAs in groups H and L. (D) Tukey box plots representing the effect of the probiotic and placebo intervention on butyrate levels in SCFA groups H and L. Asterisks are according to the Wilcoxon-Mann-Whitney test (with paired data, when possible) with Benjamini-Hochberg correction to determine statistically significant differences between groups (B to D) and before and after the probiotic treatment (D). *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., no significant difference.
DISCUSSION
The PROBIOTA-Bb trial was undertaken to contribute to the elucidation of the probiotic potential of strains of the species Bifidobacterium bifidum, which is a specialized human commensal possessing a large arsenal of host interaction properties (9). Specifically, we studied the impact of B. bifidum Bb on the intestinal microbial ecology of healthy adults. Strain Bb was isolated from the feces from a healthy adult woman and, according to in silico analysis of its draft genome (our unpublished data), possesses the genetic determinants known to support the B. bifidum-host interaction, including genes encoding sortase-dependent pili (12), mucin-metabolizing enzymes (23), human-milk oligosaccharide hydrolases (24), BopA outer surface lipoprotein (10), and Tal transaldolase (25). In this study, we used capsules containing approximately one billion viable Bb cells, which corresponds to the minimal daily dosage recommended for probiotics by the Italian Ministry of Health (26).
16S rRNA gene profiling revealed considerable variation of the fecal microbiota composition of the volunteers enrolled in the PROBIOTA-Bb trial, consistent with previous studies (27, 28). These interindividual differences support the choice of the crossover design, in which individual participants serve as their own controls, resulting in the reduction of interindividual variation.
To characterize the wide interindividual variability of the fecal microbiota, we clustered subjects according to common features in the taxonomic composition. In light of the theory suggesting that intestinal microbiota variation is generally stratified and not continuous (22), we adopted the enterotype classification to cluster the data collected during the PROBIOTA-Bb study, according to the relative abundance of bacterial genera. The proposed approach for enterotyping is subject to limitations. In particular, enterotyping reflects overconfidence in the assumption of discrete enterotypes without consistent evidence to refute the simpler hypothesis of continuous variation of the microbiota (29). The existence of discrete structures for gut microbiota has not been convincingly demonstrated (30) and represents a crucial assumption in applying an appropriate prediction model. Nonetheless, we used the original tutorial to define enterotypes (http://enterotype.embl.de/enterotypes.html) in the present study, because it has been demonstrated to be useful to correlate the gut microbial community structure with host biomarkers and diet (31, 32). In our study, enterotyping clustered the microbiota taxonomic structures of the fecal samples in two groups corresponding to the most common enterotypes of healthy adult populations, namely, the Bacteroides-dominant (Ba) and the Prevotella-dominant (Pr) enterotypes (22, 33).
In our trial, the α- and β-diversities were not significantly affected by probiotic intervention. In a recent intervention study that was performed adopting the same trial design (6), the administration of capsules containing the probiotic strain Lactobacillus paracasei DG did not modify the α-diversity of participants' fecal samples, but, in contrast to the results of this study, induced a significant change in the β-diversity in terms of weighted UniFrac distances. Kim et al. (34) did not observe significant alterations in α- and β-diversities following the consumption of various probiotic products; however, a very limited number of subjects per group (only three) were used in that study. Furthermore, no alterations in α- and β-diversities were observed in the fecal microbiota of 1- to 2-year-old children following the consumption of probiotic milk containing Lactobacillus rhamnosus GG (LGG), Lactobacillus acidophilus La-5, and Bifidobacterium animalis subsp. lactis Bb-12 (35). Other intervention trials investigating the effect of probiotics on the intra- and interindividual biodiversities of the gut microbiota in healthy subjects have not been reported. Taken together, these data confirm the literature (although limited) suggesting that probiotic intake may be an insufficiently weak perturbation to modify the α- and β-diversities of the intestinal microbiota of healthy adults. This hypothesis is plausible in light of the recognized stability throughout adulthood and the reported resilience of the human intestinal microbiota to short-term dietary changes (36).
This study revealed that the intervention with strain Bb affected the relative abundances of several dominant taxa of the intestinal microbiota; specifically, the families Ruminococcaceae and Rikenellaceae increased, whereas Prevotellaceae decreased. Remarkably, Ruminococcaceae, Prevotellaceae, and Rikenellaceae (together with Lachnospiraceae and Bacteroidaceae) have been identified by metatranscriptomics as the predominant families of the active microbiota (37).
Ruminococcaceae is a family of obligate anaerobes that include bacteria (e.g., Faecalibacterium, Ruminiclostridium, and Ruminococcus spp.) that may degrade numerous polysaccharides in the lower gastrointestinal tract, such as starch, cellulose, and xylan, and produce SCFAs (38). The expansion of Ruminococcaceae in centenarians has been reported, with a positive correlation with high-fiber diets (39) and after intervention with resistant starch (40). Furthermore, in the study by Martínez et al., Ruminococcaceae were more dominant in normoweight than obese individuals and negatively correlated with markers of inflammation (41). A lower abundance of this taxon was associated with exaggerated Toll-like receptor 2 (TLR-2) responses and an increased risk of developing IgE-associated eczema in infants (42). The relative abundance of Ruminococcaceae was also lower in acute-on-chronic liver failure patients (43); in the same study, a negative correlation of Ruminococcaceae with tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6) and endotoxemia was also observed. Finally, two recent studies reported that Ruminococcaceae are diminished in the guts of inflammatory bowel disease (IBD) patients, particularly those with ileal Crohn's disease (44, 45). Therefore, a number of observations suggest that Ruminococcaceae are commonly associated with a healthy gut microbiota and may exert a protective role on host health.
A couple of studies have also suggested a potential positive role for Rikenellaceae, a Bacteroidales family significantly enhanced by probiotic treatment with strain B. bifidum Bb. Specifically, Rikenellaceae family members were depleted in patients who had chronic HIV infection relative to HIV-uninfected controls (46) and suppressed in IBD patients relative to healthy controls (45, 47). Conversely, one study reported a higher abundance of Ruminococcaceae and Rikenellaceae and a decrease in the abundance of Prevotellaceae in the terminal ileum microbiota of subjects with ankylosing spondylitis (AS) compared with healthy controls (48).
In our study, the probiotic intervention reduced the relative abundance of the family Prevotellaceae and, particularly, the genus Prevotella. The relative richness of Prevotella spp. and Prevotellaceae is frequently modulated following dietary interventions (49, 50) and, in general, by lifestyle modifications (51), suggesting that these microorganisms represent active and rapidly reacting components of the human intestinal microbiota (37). Notably, Prevotella spp. are the dominant colonizers of agrarian societies and are associated with long-term diets rich in plant carbohydrates and fibers, whereas the abundance of Bacteroides spp. is increased in individuals from urbanized societies and is associated with diets high in animal fat and proteins (33, 52). Although all subjects enrolled in the present study lived in an urbanized European area (Lombardia region), Prevotella was the dominant genus in approximately 30% of study participants (8 of 27 subjects at V1). This high prevalence of Prevotella in the population under study may potentially be explained by the higher-than-average fruit and vegetable (and thus fiber and starch) intake of Italian subjects (53).
A few studies have suggested that the abundance of Prevotellaceae/Prevotella (Bacteroidetes phylum) is inversely associated with the relative richness of Ruminococcaceae (Firmicutes phylum). For instance, increased Ruminococcaceae have been proposed to compensate for lower levels of Prevotellaceae in Parkinson's disease patients (54). Conversely to Ruminococcaceae, Prevotellaceae have been observed to be overrepresented in obese people (55). Furthermore, although typically associated with plant carbohydrate consumption, enriched abundance of Prevotella has also been linked to the high consumption of l-carnitine-containing foods, such as red meat (56).
Notably, several studies also suggested a potential role of Prevotellaceae as intestinal pathobionts. The family Prevotellaceae was, in fact, demonstrated to elicit a strong inflammatory response in the guts of mice (57) and is overrepresented in patients with ulcerative colitis (58). An increase in Prevotella spp. was also reported in the intestinal lumen microbiota of colorectal cancer patients (59) and in children diagnosed with irritable bowel syndrome (60). Finally, the species Prevotella copri was identified as strongly correlated with disease in new-onset untreated rheumatoid arthritis patients (61). However, increased abundance of P. copri has also been associated with dietary fiber-induced improvement in glucose and insulin responses (62). In addition, low Prevotellaceae levels have been reported in patients with type 1 diabetes (63) and children with autism (64). However, a subsequent study involving a larger population of autistic children reported the opposite result (i.e., a significant increase in Prevotella spp. [65]). In conclusion, the scientific literature is far from a final and unambiguous understanding of the role of a specific taxon in host health; plausibly, the same bacterial taxa of the gut microbiota may exert opposite effects on the host (health preserving versus health threatening), depending on physiological background. This variability is particularly true for Prevotella spp., which appear to be critical bacteria for healthy microbiota that have been linked to plant-rich diets but also to chronic inflammatory conditions (66). In addition, genera such as Prevotella include numerous species that possess wide genetic diversity, which may at least partially explain the observed differences in interactions between Prevotella and its host (66).
Modification of the intestinal microbiota structure may potentially lead to alterations of the gut levels of SCFAs, which are microbial metabolic products exerting a number of effects on host physiology (67). Diet (probiotics included) may modify the level of SCFAs in the intestinal lumen by affecting their uptake/utilization by host and intestinal microbes or by changing the relative abundance of specific butyrate-producing bacteria. In our study, we observed the modification of Clostridiales bacteria of the family Ruminococcaceae, which are among the primary producers of SCFAs and, in particular, butyrate in the human large intestine. We therefore quantified the concentrations of SCFAs in fecal samples by UPLC-HR-MS. We observed that subjects could be clustered into groups, H and L, according to the levels of the three most abundant SCFAs. Group H was characterized by higher concentrations of several SCFAs, including butyrate, than those in group L. Notably, we observed that the probiotic treatment induced a significant change in butyrate levels, which decreased in group H and increased in L. Notably, this modification was not observed with placebo treatment. A similar effect on fecal butyrate levels was also observed during an intervention study with the probiotic strain Lactobacillus paracasei DG (6). In light of literature suggesting that excessive intestinal butyrate may be detrimental in certain physiological conditions, such as irritable bowel syndrome (IBS) or metabolic syndrome (68–71), although considered improbable by some researchers (72), we recently postulated the potential existence of an optimal butyrate concentration range in the human intestine (6, 73). In this context, the hypothesis can be made that probiotics might be used to decrease high butyrate concentrations or increase low butyrate concentrations to maintain butyrate homeostasis in healthy people, potentially preventing disorders associated with altered butyrate levels (73).
In conclusion, in our study, the probiotic treatment modified the relative abundances of bacterial taxa that have often been associated with health conditions; in addition, the probiotic treatment modulated the fecal levels of butyrate, a microbial metabolite exerting multiple effects on human health. Therefore, the daily consumption of B. bifidum Bb cells may affect human health; however, as for most dietary interventions, the current state of knowledge does not allow us to better define the significance of any taxonomic or metabolite changes of the intestinal microbial ecosystem on the host health.
In a wider perspective, the PROBIOTA-Bb trial contributes to the field of research on probiotics in healthy populations, which is currently attracting significant attention in the context of probiotic health claim assessment by the EFSA. In particular, our study demonstrates that a single daily administration of one bacterial strain approximately at the minimal recommended dose (1 billion CFU [26]) can modify the human intestinal microbial ecology of healthy (not diseased) adults in a significant fashion. These findings emphasize the need to reassess the notion that probiotics do not influence the complex and stable intestinal microbial ecosystem of a healthy individual and the importance of a proper intervention setting coupled with the use of adequate analytical and bioinformatic tools.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Parisa Abbasi and Lorenzo Zamboni for their technical assistance.
We declare no conflicts of interest.
This work was entirely financially supported by Fondazione Cariplo (grant 2010-0678).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01753-16.
REFERENCES
- 1.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. 2014. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11:506–514. [DOI] [PubMed] [Google Scholar]
- 2.Food Safety Authority of Ireland. Guidance on the implementation of Regulation 1924/2006 no. 1924/2006 on nutrition and health claims made on foods conclusions of the standing committee on the food chain and animal health. Food Safety Authority of Ireland, Dublin, Ireland: https://www.fsai.ie/uploadedFiles/EU_guidance_ClaimsRegulation.pdf. [Google Scholar]
- 3.Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB, Sherman PM, Mayer EA. 2013. An update on the use and investigation of probiotics in health and disease. Gut 62:787–796. doi: 10.1136/gutjnl-2012-302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kristensen NB, Bryrup T, Allin KH, Nielsen T, Hansen TH, Pedersen O. 2016. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med 8:52. doi: 10.1186/s13073-016-0300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goulet O. 2015. Potential role of the intestinal microbiota in programming health and disease. Nutr Rev 73(Suppl 1):S32–S40. [DOI] [PubMed] [Google Scholar]
- 6.Ferrario C, Taverniti V, Milani C, Fiore W, Laureati M, De Noni I, Stuknyte M, Chouaia B, Riso P, Guglielmetti S. 2014. Modulation of fecal Clostridiales bacteria and butyrate by probiotic intervention with Lactobacillus paracasei DG varies among healthy adults. J Nutr 144:1787–1796. doi: 10.3945/jn.114.197723. [DOI] [PubMed] [Google Scholar]
- 7.Veiga P, Pons N, Agrawal A, Oozeer R, Guyonnet D, Brazeilles R, Faurie JM, van Hylckama Vlieg JE, Houghton LA, Whorwell PJ, Ehrlich SD, Kennedy SP. 2014. Changes of the human gut microbiome induced by a fermented milk product. Sci Rep 4:6328. doi: 10.1038/srep06328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derrien M, van Hylckama Vlieg JE. 2015. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol 23:354–366. doi: 10.1016/j.tim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Turroni F, Duranti S, Bottacini F, Guglielmetti S, Van Sinderen D, Ventura M. 2014. Bifidobacterium bifidum as an example of a specialized human gut commensal. Front Microbiol 5:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guglielmetti S, Tamagnini I, Mora D, Minuzzo M, Scarafoni A, Arioli S, Hellman J, Karp M, Parini C. 2008. Implication of an outer surface lipoprotein in adhesion of Bifidobacterium bifidum to Caco-2 cells. Appl Environ Microbiol 74:4695–4702. doi: 10.1128/AEM.00124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serafini F, Strati F, Ruas-Madiedo P, Turroni F, Foroni E, Duranti S, Milano F, Perotti A, Viappiani A, Guglielmetti S, Buschini A, Margolles A, van Sinderen D, Ventura M. 2013. Evaluation of adhesion properties and antibacterial activities of the infant gut commensal Bifidobacterium bifidum PRL2010. Anaerobe 21:9–17. doi: 10.1016/j.anaerobe.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Turroni F, Serafini F, Foroni E, Duranti S, O'Connell Motherway M, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sanchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Piazza L, Palanza P, Bolchi A, Guglielmetti S, van Sinderen D, Ventura M. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci U S A 110:11151–11156. doi: 10.1073/pnas.1303897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.López P, Gonzalez-Rodriguez I, Sanchez B, Ruas-Madiedo P, Suarez A, Margolles A, Gueimonde M. 2012. Interaction of Bifidobacterium bifidum LMG13195 with HT29 cells influences regulatory-T-cell-associated chemokine receptor expression. Appl Environ Microbiol 78:2850–2857. doi: 10.1128/AEM.07581-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guglielmetti S, Zanoni I, Balzaretti S, Miriani M, Taverniti V, De Noni I, Presti I, Stuknyte M, Scarafoni A, Arioli S, Iametti S, Bonomi F, Mora D, Karp M, Granucci F. 2014. Murein lytic enzyme TgaA of Bifidobacterium bifidum MIMBb75 modulates dendritic cell maturation through its cysteine- and histidine-dependent amidohydrolase/peptidase (CHAP) amidase domain. Appl Environ Microbiol 80:5170–5177. doi: 10.1128/AEM.00761-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, Yamaguchi M, Kumagai H, Katayama T, Yamamoto K. 2008. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol 74:3996–4004. doi: 10.1128/AEM.00149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, Sanchez B, Ferrario C, Viappiani A, Mangifesta M, Mancino W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2015. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ Microbiol 17:2515–2531. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- 17.Cardona S, Eck A, Cassellas M, Gallart M, Alastrue C, Dore J, Azpiroz F, Roca J, Guarner F, Manichanh C. 2012. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol 12:158. doi: 10.1186/1471-2180-12-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milani C, Hevia A, Foroni E, Duranti S, Turroni F, Lugli GA, Sanchez B, Martín R, Gueimonde M, van Sinderen D, Margolles A, Ventura M. 2013. Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS One 8:e68739. doi: 10.1371/journal.pone.0068739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huda-Faujan N, Abdulamir AS, Fatimah AB, Anas OM, Shuhaimi M, Yazid AM, Loong YY. 2010. The impact of the level of the intestinal short chain fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem J 4:53–58. doi: 10.2174/1874091X01004010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Royston P. 1993. A pocket-calculator algorithm for the Shapiro-Francia test for non-normality: an application to medicine. Stat Med 12:181–184. doi: 10.1002/sim.4780120209. [DOI] [PubMed] [Google Scholar]
- 22.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, MetaHIT Consortium, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, et al. . 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turroni F, Milani C, van Sinderen D, Ventura M. 2011. Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human-microbe co-evolution. Gut Microbes 2:183–189. doi: 10.4161/gmic.2.3.16105. [DOI] [PubMed] [Google Scholar]
- 24.Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. 2009. Two distinct alpha-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19:1010–1017. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- 25.González-Rodríguez I, Sánchez B, Ruiz L, Turroni F, Ventura M, Ruas-Madiedo P, Gueimonde M, Margolles A. 2012. Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl Environ Microbiol 78:3992–3998. doi: 10.1128/AEM.08024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ministero della Salute. 2013. Guidelines on probiotics and prebiotics. Revision May 2013. Ministero della Salute, Rome, Italy: http://www.salute.gov.it/imgs/C_17_pubblicazioni_1016_ulterioriallegati_ulterioreallegato_0_alleg.pdf. [Google Scholar]
- 27.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davenport ER, Mizrahi-Man O, Michelini K, Barreiro LB, Ober C, Gilad Y. 2014. Seasonal variation in human gut microbiome composition. PLoS One 9:e90731. doi: 10.1371/journal.pone.0090731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knights D, Ward TL, McKinlay CE, Miller H, Gonzalez A, McDonald D, Knight R. 2014. Rethinking “enterotypes.” Cell Host Microbe 16:433–437. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorvitovskaia A, Holmes SP, Huse SM. 2016. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome 4:15. doi: 10.1186/s40168-016-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Linnenbrink M, Künzel S, Fernandes R, Nadeau MJ, Rosenstiel P, Baines JF. 2014. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc Natl Acad Sci U S A 111:E2703–E2710. doi: 10.1073/pnas.1402342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. 2016. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 65:57–62. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SW, Suda W, Kim S, Oshima K, Fukuda S, Ohno H, Morita H, Hattori M. 2013. Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res 20:241–253. doi: 10.1093/dnares/dst006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dotterud CK, Avershina E, Sekelja M, Simpson MR, Rudi K, Storro O, Johnsen R, Oien T. 2015. Does maternal perinatal probiotic supplementation alter the intestinal microbiota of mother and child? J Pediatr Gastroenterol Nutr 61:200–207. doi: 10.1097/MPG.0000000000000781. [DOI] [PubMed] [Google Scholar]
- 36.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gosalbes MJ, Durban A, Pignatelli M, Abellan JJ, Jimenez-Hernandez N, Perez-Cobas AE, Latorre A, Moya A. 2011. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One 6:e17447. doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 39.Wang F, Yu T, Huang G, Cai D, Liang X, Su H, Zhu Z, Li D, Yang Y, Shen P, Mao R, Yu L, Zhao M, Li Q. 2015. Gut microbiota community and its assembly associated with age and diet in Chinese centenarians. J Microbiol Biotechnol 25:1195–1204. doi: 10.4014/jmb.1410.10014. [DOI] [PubMed] [Google Scholar]
- 40.Salonen A, Lahti L, Salojarvi J, Holtrop G, Korpela K, Duncan SH, Date P, Farquharson F, Johnstone AM, Lobley GE, Louis P, Flint HJ, de Vos WM. 2014. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J 8:2218–2230. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martínez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG, Louk JA, Rose DJ, Kyureghian G, Peterson DA, Haub MD, Walter J. 2013. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J 7:269–280. doi: 10.1038/ismej.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West CE, Ryden P, Lundin D, Engstrand L, Tulic MK, Prescott SL. 2015. Gut microbiome and innate immune response patterns in IgE-associated eczema. Clin Exp Allergy 45:1419–1429. doi: 10.1111/cea.12566. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, Guo J, Qian G, Fang D, Shi D, Guo L, Li L. 2015. Gut dysbiosis in acute-on-chronic liver failure and its predictive value for mortality. J Gastroenterol Hepatol 30:1429–1437. doi: 10.1111/jgh.12932. [DOI] [PubMed] [Google Scholar]
- 44.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, Jarnerot G, Tysk C, Jansson JK, Engstrand L. 2010. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139:1844.e1–1854.e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 45.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. 2012. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV, Wanke CA, Ward HD. 2015. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis 211:19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scaldaferri F, Lopetuso L, Pecere S, Petito V, Schiavoni E, Gerardi V, Laterza L, Pistone MT, D'Ambrosio D, Ricca O, D'Agostino A, Zambrano D, Paroni Sterbini F, Gaetani E, Franceschi F, Cammarota G, Sanguinetti M, Masucci L, Gasbarrini A. 2015. Gut microbiota molecular spectrum in healthy controls, diverticular disease, IBS and IBD patients: time for microbial marker of gastrointestinal disorders? J Crohns Colitis 9:S443. doi: 10.1093/ecco-jcc/jju027.842. [DOI] [Google Scholar]
- 48.Costello ME, Ciccia F, Willner D, Warrington N, Robinson PC, Gardiner B, Marshall M, Kenna TJ, Triolo G, Brown MA. 2015. Intestinal dysbiosis in ankylosing spondylitis. Arthritis Rheumatol 67:686–691. doi: 10.1002/art.38967. [DOI] [PubMed] [Google Scholar]
- 49.Clemente-Postigo M, Queipo-Ortuno MI, Boto-Ordonez M, Coin-Araguez L, Roca-Rodriguez MM, Delgado-Lista J, Cardona F, Andres-Lacueva C, Tinahones FJ. 2013. Effect of acute and chronic red wine consumption on lipopolysaccharide concentrations. Am J Clin Nutr 97:1053–1061. doi: 10.3945/ajcn.112.051128. [DOI] [PubMed] [Google Scholar]
- 50.Karl JP, Fu X, Wang X, Zhao Y, Shen J, Zhang C, Wolfe BE, Saltzman E, Zhao L, Booth SL. 2015. Fecal menaquinone profiles of overweight adults are associated with gut microbiota composition during a gut microbiota-targeted dietary intervention. Am J Clin Nutr 102:84–93. doi: 10.3945/ajcn.115.109496. [DOI] [PubMed] [Google Scholar]
- 51.Biedermann L, Brulisauer K, Zeitz J, Frei P, Scharl M, Vavricka SR, Fried M, Loessner MJ, Rogler G, Schuppler M. 2014. Smoking cessation alters intestinal microbiota: insights from quantitative investigations on human fecal samples using FISH. Inflamm Bowel Dis 20:1496–1501. doi: 10.1097/MIB.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 52.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, Serrazanetti DI, Di Cagno R, Ferrocino I, Lazzi C, Turroni S, Cocolin L, Brigidi P, Neviani E, Gobbetti M, O'Toole PW, Ercolini D. 2015. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 54.Scheperjans F, Aho V, Pereira PA, Koskinen K, Paulin L, Pekkonen E, Haapaniemi E, Kaakkola S, Eerola-Rautio J, Pohja M, Kinnunen E, Murros K, Auvinen P. 2015. Gut microbiota are related to Parkinson's disease and clinical phenotype. Mov Disord 30:350–358. doi: 10.1002/mds.26069. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, Krajmalnik-Brown R. 2009. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A 106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. 2013. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lucke K, Miehlke S, Jacobs E, Schuppler M. 2006. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J Med Microbiol 55:617–624. doi: 10.1099/jmm.0.46198-0. [DOI] [PubMed] [Google Scholar]
- 59.Chen W, Liu F, Ling Z, Tong X, Xiang C. 2012. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rigsbee L, Agans R, Shankar V, Kenche H, Khamis HJ, Michail S, Paliy O. 2012. Quantitative profiling of gut microbiota of children with diarrhea-predominant irritable bowel syndrome. Am J Gastroenterol 107:1740–1751. doi: 10.1038/ajg.2012.287. [DOI] [PubMed] [Google Scholar]
- 61.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, Huttenhower C, Littman DR. 2013. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Björck I, Bäckhed F. 2015. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab 22:971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 63.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip M, Hyoty H, Veijola R, Simell T, Simell O, Neu J, Wasserfall CH, Schatz D, Atkinson MA, Triplett EW. 2011. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One 6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kang DW, Park JG, Ilhan ZE, Wallstrom G, Labaer J, Adams JB, Krajmalnik-Brown R. 2013. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One 8:e68322. doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Son JS, Zheng LJ, Rowehl LM, Tian X, Zhang Y, Zhu W, Litcher-Kelly L, Gadow KD, Gathungu G, Robertson CE, Ir D, Frank DN, Li E. 2015. Comparison of fecal microbiota in children with autism spectrum disorders and neurotypical siblings in the Simons Simplex collection. PLoS One 10:e0137725. doi: 10.1371/journal.pone.0137725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ley RE. 2016. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol 13:69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 67.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. 2013. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bourdu S, Dapoigny M, Chapuy E, Artigue F, Vasson MP, Dechelotte P, Bommelaer G, Eschalier A, Ardid D. 2005. Rectal instillation of butyrate provides a novel clinically relevant model of noninflammatory colonic hypersensitivity in rats. Gastroenterology 128:1996–2008. doi: 10.1053/j.gastro.2005.03.082. [DOI] [PubMed] [Google Scholar]
- 69.Jeffery IB, O'Toole PW, Ohman L, Claesson MJ, Deane J, Quigley EM, Simren M. 2012. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- 70.Teixeira TF, Grzeskowiak L, Franceschini SC, Bressan J, Ferreira CL, Peluzio MC. 2013. Higher level of faecal SCFA in women correlates with metabolic syndrome risk factors. Br J Nutr 109:914–919. doi: 10.1017/S0007114512002723. [DOI] [PubMed] [Google Scholar]
- 71.Payne AN, Chassard C, Zimmermann M, Muller P, Stinca S, Lacroix C. 2011. The metabolic activity of gut microbiota in obese children is increased compared with normal-weight children and exhibits more exhaustive substrate utilization. Nutr Diabetes 1:e12. doi: 10.1038/nutd.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conlon MA, Bird AR, Clarke JM, Le Leu RK, Christophersen CT, Lockett TJ, Topping DL. 2015. Lowering of large bowel butyrate levels in healthy populations is unlikely to be beneficial. J Nutr 145:1030–1031. doi: 10.3945/jn.114.209460. [DOI] [PubMed] [Google Scholar]
- 73.Guglielmetti S, Riso P. 2015. Reply to Conlon et al. J Nutr 145:1031–1032. doi: 10.3945/jn.114.209890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.