ABSTRACT
How the function of microbial symbionts is affected by their population/consortium structure within a host remains poorly understood. The symbiosis established between Euprymna scolopes and Vibrio fischeri is a well-characterized host-microbe association in which the function and structure of V. fischeri populations within the host are known: V. fischeri populations produce bioluminescence from distinct crypt spaces within a dedicated host structure called the light organ. Previous studies have revealed that luminescence is required for V. fischeri populations to persist within the light organ and that deletion of the lux gene locus, which is responsible for luminescence in V. fischeri, leads to a persistence defect. In this study, we investigated the impact of bioluminescence on V. fischeri population structure within the light organ. We report that the persistence defect is specific to crypt I, which is the most developmentally mature crypt space within the nascent light organ. This result provides insight into the structure/function relationship that will be useful for future mechanistic studies of squid-Vibrio symbiosis. In addition, our report highlights the potential impact of the host developmental program on the spatiotemporal dynamics of host-microbe interactions.
IMPORTANCE Metazoan development and physiology depend on microbes. The relationship between the symbiotic function of microbes and their spatial structure within the host environment remains poorly understood. Here we demonstrate, using a binary symbiosis, that the host requirement for the symbiotic function of the microbial symbiont is restricted to a specific host environment. Our results also suggest a link between microbial function and host development that may be a fundamental aspect of the more complex host-microbe interactions.
INTRODUCTION
Microbes are a dominant form of life on Earth, and their associations with humans and other animals directly impact host development, physiology, and evolution (1–3). In many cases, the acquisition of microbes from the environment represents a key moment in the life history of the host. Studies of mammalian gut microbiota have suggested that the spatial distribution of these microbes within the host, i.e., their biogeography, is an important factor in the assemblage and maintenance of such associations (see reference 4 for a recent review). For instance, when microbial communities isolated from diverse habitats are transplanted into gnotobiotic mice, the resulting composition closely resembles that normally found in the mouse cecum (5). Body part-specific microbiota have also been observed in nonmammalian systems, such as that of the jellyfish Aurelia aurita, which shows significant differences in microbial composition between the gastric cavity and exumbrellar mucus (6). These studies suggest that hosts select certain biogeographical landscapes for their associated microbiota. However, examining how biogeography impacts the symbiotic function of microbes remains challenging for most associations, due in part to the variability in niche spaces among individual hosts.
Such relationships are typically easier to examine in certain coevolved animal-microbe symbioses, in which microbial symbionts occupy well-defined niche spaces and provide host-specific biological functions (7, 8). One such system is the light organ symbiosis that is established between the Hawaiian bobtail squid, Euprymna scolopes, and the bioluminescent bacterium Vibrio fischeri (9). Populations of V. fischeri are housed in a dedicated light organ that is located on the ventral side within the squid mantle cavity (Fig. 1A). In exchange for host-derived nutrients, V. fischeri provides the squid with bioluminescence, which serves as camouflage within the water column (10, 11). The binary nature of the squid-Vibrio symbiosis has provided insight into the molecular mechanisms that promote the successful acquisition of horizontally transmitted microbes during each host generation.
FIG 1.
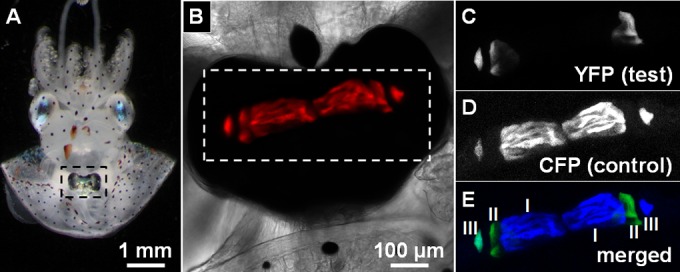
Crypt populations within squid light organ. (A) Juvenile squid with light organ (indicated by dashed box) exposed by mantle dissection. (B) Light organ colonized with V. fischeri expressing mCherry (red). (C to E) YFP (C), CFP (D), and merged (E) images of light organ region (indicated by dashed box in panel B). Crypt III (on the left side) is cocolonized with YFP- and CFP-labeled cells.
Juvenile squid hatch uncolonized and acquire V. fischeri symbionts from the seawater environment (12). The nascent light organ is a bilobed structure with each side containing three epithelium-lined crypt spaces (crypts I, II, and III) that house distinct V. fischeri populations (13). During embryogenesis, the crypt spaces of the light organ emerge sequentially, such that they are at different developmental stages during the initial period of V. fischeri colonization (13). Crypt I is the largest crypt space in the nascent light organ and accounts for approximately 75% of the total volume available for occupancy by V. fischeri populations (14). In contrast, crypt III, in which V. fischeri cells have access to less than 5% of the total volume of crypt space, is the smallest space (14). The epithelial cells of crypt I respond to V. fischeri colonization within 48 h postinoculation (p.i.) by changing cell shape from columnar to cuboidal (15). This symbiont-induced change in epithelial cell shape is reduced in crypt II and absent in crypt III. Colonized E. scolopes squid also expel (i.e., “vent”) up to 95% of the resident V. fischeri populations into the environment every dawn, which promotes the transmission of symbiotic strains of V. fischeri (16). The percentages of V. fischeri cells vented from the juvenile light organ differ among the three crypts during the first 72 h of colonization (15). With each expulsion, crypt I almost completely empties, crypt II partially empties, and crypt III expels hardly any bacterial cells. Together, the results of these studies highlight differences, e.g., in sizes, symbiont-induced host responses, and venting activities, among the distinct niches of V. fischeri within the nascent light organ.
Competition assays, which directly compare the abilities of two V. fischeri strains to colonize the squid light organ, have identified bacterial factors, such as quorum sensing and bioluminescence, that contribute to the symbiosis (17–19). To initiate such assays, juvenile squid are first exposed to a V. fischeri mixed inoculum with differentially labeled strains and, at a later time, are homogenized to determine the relative strain abundances by counting the number of CFU corresponding to each strain type. Competition assays using mutants with altered luminescence profiles have led to the hypothesis that light production is the primary function of V. fischeri during symbiosis (20). Luminescence is a by-product of the mixed-function oxidation of a long-chain aldehyde and reduced flavin mononucleotide by the enzyme luciferase (21). The luxA and luxB genes that encode the two subunits of luciferase are cotranscribed with five other genes (luxI, luxC, luxD, luxE, and luxG), and, together, they are commonly referred to as the lux operon. A mutant that has been particularly useful for studying the role of luminescence in squid-Vibrio symbiosis as a nonluminous strain is the Δlux mutant, which lacks the luxCDABEG gene cluster but retains the autoinducer synthase encoded by luxI, so it can still regulate other genes via quorum sensing (19, 22). Colonization assays competing the Δlux mutant and its luminous parental strain have revealed that the abundance of the Δlux mutant within cocolonized animals begins to decrease by 48 h p.i. and reaches the lower limit of detection around 2 weeks p.i. (19, 23), suggesting that the squid host actively selects for light-producing symbiont populations, or against nonluminous ones, after the initial infection has occurred. However, how this CFU-based persistence defect of the Δlux strain is related to the population structures within the crypt spaces is unclear.
Here we report our results from studying the crypt-specific V. fischeri population structure within the juvenile light organ. We found that the crypt I populations generally dictate the overall abundance of V. fischeri strains within each animal and that the luminescence requirement for light organ persistence of V. fischeri cells is limited to crypt I. These results suggest that the fitness of different bacterial strains depends on the developmental state of the host's symbiont-containing tissue.
MATERIALS AND METHODS
Media and growth conditions.
V. fischeri strains were grown aerobically at 28°C in LBS medium without glycerol (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 2% [wt/vol] NaCl, 50 mM Tris-HCl [pH 7.5]) (24). Strains harboring plasmids were grown in the presence of 2.5 μg/ml chloramphenicol.
Strains.
The V. fischeri strains used in this study were wild-type strain ES114 (25) and ES114-derived Δlux mutant EVS102 (19).
Plasmids.
Plasmid pSCV38 (26) was used to label V. fischeri cells with mCherry and yellow fluorescent protein (YFP). To construct pSCV37, which labels V. fischeri cells with cyan fluorescent protein (CFP) and mCherry, the cfp gene was amplified from pNS2-sigmaVL (27) using primers 5′-GGTCTAGATTTAAGAAGGAGATATACATATGACTAGCAAAAGAAGCAAAGGT-3′/5′-GCATGCATCAAACAACAGATAAAACGAAAGGC-3′ and cloned into the vector fragment of pSCV26 (28) generated by XbaI/NsiI.
Squid colonization assays.
Juvenile E. scolopes squid contained in filter-sterilized seawater (FSSW) were exposed to an inoculum containing a 1:1 mixture of two strains labeled with pSCV37 (YFP) or pSCV38 (CFP). Every 24 h, animals were washed by transfer into fresh FSSW. At 72 h p.i., animals were anesthetized in 5% ethanol–FSSW and dissected to expose the light organ. A Zeiss 780 confocal microscope (Carl Zeiss AG, Jena, Germany) equipped with a C-Apochromat 10×/0.45 W objective was used to acquire Z-stacks of mCherry, CFP, and YFP fluorescence. Crypts exhibiting mCherry fluorescence were further scored for CFP (control only), YFP (test only), or both CFP and YFP. After imaging, animals were homogenized and plated onto LBS medium and incubated at 28°C. CFU counts were scored for CFP and YFP fluorescence using an Olympus SX16 fluorescence dissecting microscope (Olympus Corp., Tokyo, Japan).
Statistical analyses.
GraphPad Prism software version 6.04 (GraphPad Software, Inc., La Jolla, CA, USA) was used for all statistical tests. The two-tailed Student's t test was used to determine whether log-transformed, CFU-based competitive indices were significantly different (alpha = 0.05) from the hypothetical mean (0.0). To test for correlation between CFU- and crypt-based competitive indices, linear regression was used for calculating values corresponding to the Pearson coefficient and significance of slope.
RESULTS
Correlation between CFU counts and crypt colonization in wild-type cells.
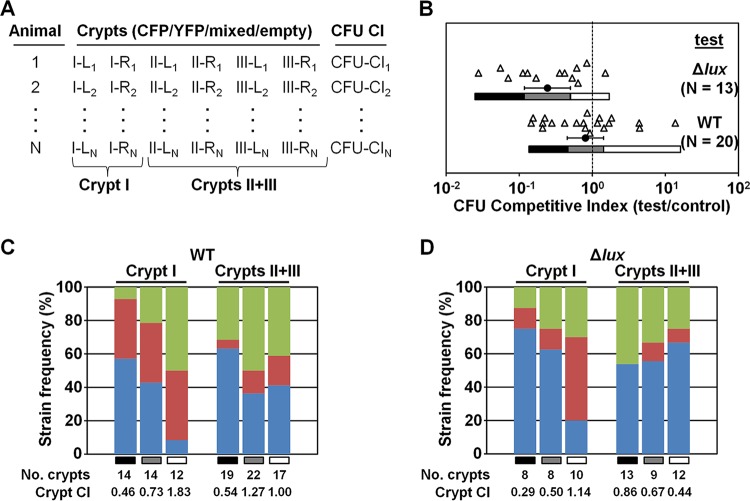
Past experiments designed to directly compare the abilities of two V. fischeri strains to colonize juvenile squid used different fluorescent proteins to distinguish strain types within the light organ (26, 29, 30). In the colonization experiments described here, isogenic plasmids pSCV37 and pSCV38 were used to differentially label strains with CFP and YFP, respectively. Both plasmids also constitutively express mCherry, which enables the unbiased scoring of crypt colonization within the host environment. To determine how the V. fischeri population structure within the light organ impacts the CFU counts obtained by plating light organ homogenates, we combined live-host imaging with CFU-plating techniques. Juvenile E. scolopes squid were exposed to a total inoculum of between 4,000 and 30,000 CFU/ml containing a 1:1 mixture of wild-type (WT) strain ES114 (23) harboring either pSCV37 or pSCV38. At 72 h p.i., each animal was anesthetized and dissected to reveal the light organ, which was immediately examined by confocal microscopy. The mCherry fluorescence was used to determine which crypts were colonized (Fig. 1B), and the CFP and YFP fluorescence signals indicated which strain was present within each crypt (Fig. 1C to E). Following imaging, animals were homogenized and plated onto LBS medium for CFU counting. Figure 2A highlights the scoring approach used to track crypt contents and CFU counts associated with individual animals. Because crypts II and III were difficult to accurately label in the instances when only one was colonized, we labeled bacterium-containing crypts “crypt I” or “crypts II and III.”
FIG 2.
Strain frequency of crypts in cocolonized light organs. (A) Scoring system for individual squid from a cocolonization assay. Crypts of colonized animal are scored as CFP/YFP/mixed/empty. I, II, and III refer to crypt numbers, and L and R indicate left and right sides of light organ, respectively. CFU-based competitive index (CFU CI) values were determined by calculating the test CFU/control CFU ratio using homogenized animals. (B) CFU-based competitive index values determined for individual squid cocolonized with the CFP-labeled wild-type strain (control) and the indicated YFP-labeled test strain. Each triangle represents an individual animal. Data sets correspond to trial 1 of the respective test strain (Table 1). Circles and error bars represent means and 95% confidence intervals. For each trial, gray bars represent the 95% confidence interval, and black and white bars represent the corresponding lower and upper confidence bounds, respectively. (C and D) Strain frequencies of crypts in animals determined as described for panel B. The test strains were the WT strain (C) and the Δlux mutant (D). Blue, green, and red represent the control strain (CFP), the test strain (YFP), and both strain types, respectively. Grayscale bars indicate animal subgroups defined by 95% confidence intervals as described for panel B. Crypt competitive indices (crypt CI) were calculated as numbers of crypts containing the test strain/numbers of crypts containing the control strain.
From three independent trials, CFU plating revealed that the majority of animals exposed to the mixed WT inoculum were cocolonized (Table 1). For each trial, the mean CFU competitive index value (test CFU/control CFU) was not significantly different from the hypothetical mean of 1.0 (Table 1), which is similar to previous reports of competitions between isogenic test and control strains (18, 31). By examining the corresponding sets of light organ images, we counted the number of crypts colonized with the control strain, the test strain, or both strains. Across the three trials, 19% (± 4.0%) of colonized crypts were cocolonized, which is a frequency comparable to that previously reported from studying the V. fischeri population structure within the light organ using differentially labeled strains (29). Within each cocolonized crypt space, we observed that the cell types were mixed (see, e.g., Fig. 1E) and not segregated. Using the crypt counts described above, we also determined the crypt competitive index (number of crypts containing any test cells/number of crypts containing any control cells). Across the three trials involving the use of the WT as both the test strain and the control strain, the mean crypt competitive index was 0.83, indicating that the likelihoods of either labeled strain being present within a colonized crypt were equivalent.
TABLE 1.
Light organ colonization assays
| Test straina | Trial no. | Total no. of animals | No. (%) of cocolonized animalsb | CFU competitive index log (CI) ± 95% confidence interval | Total no. of colonized crypts | No. (%) of cocolonized animals with cocolonized cryptsc |
|---|---|---|---|---|---|---|
| WT | 1 | 20 | 20 (100) | −0.09 ± 0.25 | 98 | 15 (75) |
| 2 | 21 | 14 (63) | 0.18 ± 0.32 | 68 | 8 (57) | |
| 3 | 15 | 14 (93) | −0.01 ± 0.29 | 74 | 9 (64) | |
| Δlux mutant | 1 | 14 | 13 (93) | −0.62 ± 0.31 | 60 | 6 (46) |
| 2 | 26 | 23 (88) | −0.38 ± 0.36 | 103 | 14 (61) | |
| 3 | 17 | 15 (88) | −1.05 ± 0.42 | 69 | 8 (53) |
For the inoculum of each trial, the indicated YFP-labeled test strain was mixed evenly with the WT control strain labeled with CFP.
Data represent animals with light organ homogenates that yielded both CFP- and YFP-labeled CFU were scored as cocolonized. Percentages were calculated on the basis of the number of animals in each trial.
Data represent animals with at least one crypt containing both CFP- and YFP-labeled cells. Percentages were calculated on the basis of the number of animals determined to be cocolonized by CFU plating.
The CFU competitive indices that we observed with differentially labeled WT cells varied by up to 2 orders of magnitude (Fig. 2B). Such variation is a common outcome for squid cocolonization assays (see, e.g., references 18 and 31). We hypothesized that the CFU competitive index value for an individual squid is skewed by the V. fischeri populations within the crypt I pair. To test this hypothesis, we investigated the V. fischeri population structures specific to crypt I and to crypts II and III. To facilitate this analysis, we defined three subgroups of cocolonized animals for each trial using the 95% confidence interval of the CFU competitive indices (see, e.g., Fig. 2B, gray bars) and the corresponding lower and upper confidence bounds (see, e.g., Fig. 2B, black and white bars, respectively). For trial 1 (Table 1), the mean crypt competitive index value for crypt I within the 95% confidence interval was 0.73 (Fig. 2C). The subgroups within the lower and upper confidence bounds yielded crypt I competitive indices below and above those of the middle subgroup (0.46 and 1.83, respectively). In contrast, the crypt competitive indices of crypts II and III in the same subgroups did not show the same increasing trend.
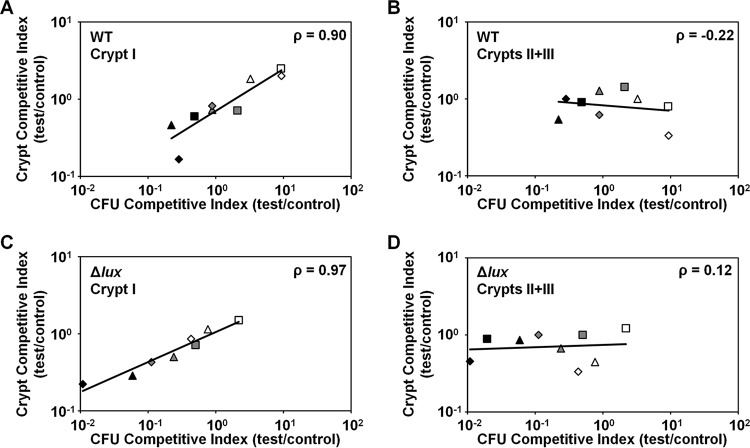
Using this approach, we examined the relationship between the CFU competitive indices and crypt competitive indices across the three trials. For crypt I, we found that the crypt competitive index is significantly correlated with the CFU competitive index (Fig. 3A; using log-transformed data, Pearson's ρ = 0.90 with P = 0.0011). In contrast, the competitive indices of crypts II and III were uncorrelated with the CFU competitive index (Fig. 3B; using log-transformed data, Pearson's ρ = −0.22 with P = 0.58). Together, these results suggest that the CFU competitive index value for each animal is generally determined by the population in crypt I and is independent of the population in crypt II or crypt III, i.e., the relatively large capacity of crypt I spaces appears to skew the CFU competitive index value for each animal toward the strain types within crypt I pairs.
FIG 3.
Correlation of CFU plating with crypt colonization. Competitive indices were determined based on CFU and colonization of crypt I or of crypts II and III in regions defined by the 95% confidence interval. Regions are indicated by the symbol color, with regions below (black), within (gray), or above (white) the mean CI ± 95% confidence intervals. For each test strain, the results of trials 1, 2, and 3 (Table 1) are indicated by triangle, diamond, and square shapes. Pearson's correlation (ρ) was calculated with log-transformed CFU- and crypt-based competitive indices. (A) Crypt I, test strain = WT. (B) Crypts II and III, test strain = WT. (C) Crypt I, test strain = Δlux mutant. (D) Crypts II and III, test strain = Δlux mutant.
Crypt specificity of the Δlux mutant persistence defect.
To examine the spatial structure of the Δlux mutant populations within light organs cocolonized with Δlux mutant and WT cells, we exposed juvenile squid to an inoculum containing 7,000 to 40,000 CFU/ml mixed 1:1 with Δlux strain EVS102 (19) (harboring pSCV38) and the WT strain (harboring pSCV37). At 72 h p.i., the imaging and plating protocol described above was performed in similar manners using these animals.
As shown by the results of three independent trials, the majority of animals were cocolonized with both the Δlux mutant and WT strains based on the results from CFU plating (Table 1). In contrast to the competitions involving only the WT, we found that the mean CFU competitive indices were significantly different from the theoretical mean of 1.0 (0.24, 0.41, and 0.088), which are similar to previous reports of competitions involving the Δlux mutant and WT strains (19, 23). The average crypt competitive index value across the three trials was 0.68, which was lower than that shown above for the WT (0.83) but not significantly different (P = 0.27 [Student's t test performed on log-transformed data]), suggesting that the lower abundance of Δlux mutant CFU was not simply due to a reduction in the number of Δlux cells accessing crypt spaces.
To determine how the reduction in Δlux mutant CFU counts was manifested within cocolonized light organs, we examined the populations of crypt I and crypts II and III within the three subgroups defined by the 95% confidence interval of the CFU competitive indices. Relative to the Δlux mutant results, the occurrence of the WT in crypt I was more pronounced, particularly in animals with values determined within the lower confidence bound (Fig. 2D). Notably, within the lower confidence bound of trial 3, the Δlux mutant was not observed in crypt I in any of the animals, which precluded the calculation of a crypt competitive index value for this subgroup. Even without this subgroup, the competitive index value for crypt I showed a strong correlation with the CFU competitive index value across the three trials (Fig. 3C; using log-transformed data, Pearson's ρ = 0.97 with P < 0.0001). Remarkably, the crypt competitive indices for crypts II and III were poorly correlated with CFU competitive indices (Fig. 3D; using log-transformed data, Pearson's ρ = 0.12 with P = 0.77). Together, these results suggest that the Δlux mutant persistence defect is attributable to the lack of persistence in crypt I within the light organ.
DISCUSSION
The success of host-microbe interactions is often determined by calculating the number of CFU released by homogenizing bacterium-containing tissues or, in some cases, whole animals. However, protocols that use such homogenization steps inherently disrupt the spatial structures of microbial populations and consortia within the host. To understand how such spatial structures can impact the results of experiments that use homogenization steps, we investigated a competition assay that is routinely used to compare two V. fischeri strains during colonization of the squid light organ. Facilitating this study was the a priori knowledge that light organ embryogenesis yields six distinct infection sites (crypt spaces), within which only 1 or 2 bacterial cells serve as founders for the resulting infections (13, 29). With differentially labeled wild-type strains, we found that any CFU bias from cocolonized animals was correlated with the strain type present in crypt I, which is the largest of the three crypts in the nascent light organ (14). Using this approach, we subsequently determined that the rate of occurrence of the Δlux mutant, which has been a valuable tool for studying microbial symbiotic function (19), was lower in crypt I at 72 h p.i. In contrast, the occurrence of the Δlux mutant within crypt II or crypt III was comparable to that of wild-type cells. Together, these results suggest that the persistence defect of the Δlux mutant is limited to crypt I within 72 h p.i.
By showing that the luminescence requirement for the persistence of V. fischeri cells is limited to crypt I early in symbiosis, these findings tie the luminescence function to the most developmentally mature tissue that harbors the symbionts. This report also helps focus future investigations of bacterial persistence to a specific host niche. For example, V. fischeri persistence within crypt I must account for the increased level of bacterial expulsion, which is a physical mechanism that can drastically alter the composition of the corresponding population within the first 72 h p.i. In contrast, the reduced venting levels and delayed maturation of crypts II and III may enable nonluminous cells to persist for 2 weeks within the light organ, as shown by recent experiments that involved plating homogenates of reared animals colonized with the Δlux mutant (23). That is, as crypts II and III continue to mature, they too must discriminate against the Δlux mutant, resulting in a total loss of this mutant by 15 days p.i. (23). Future studies that track the morphological changes of each crypt type, e.g., shape changes in epithelial cells, in response to the presence of the Δlux mutant will be of particular interest. Correlations between such symbiont-induced host responses and symbiotic function (luminescence) will provide insight into the host-specific cellular processes that promote the maintenance of squid-Vibrio symbiosis.
Our report also underscores the complex interplay between the host developmental program and the acquisition and persistence of microbial symbionts. For instance, polyps of the freshwater cnidarian Hydra initially assemble a microbial consortium of high diversity that becomes attenuated over time as the animals mature (32). Analysis of compositions at different life stages of the moonlight jelly fish Aurelia aurita revealed stage-specific microbiota (6). The microbiota of the asexual polyp stage differed significantly from the microbiota seen in the subsequent development stages that led to the formation of individuals in the free-living, sexual medusa stage. Patterns of assembly of microbial consortia have been similarly linked to host development for the human gut microbiome, with massive alterations in composition correlating well with developmental transitions in eating behaviors (33). Finally, in bivalves at hydrothermal vents, the developmental stage of gill tissue is a factor that contributes to colonization by chemosynthetic symbionts (34).
The results presented here provide novel insight into the relationship between population structure and function of mixed V. fischeri populations within the squid light organ. Numerous studies of squid-Vibrio symbiosis have used the CFU-based competition assay to highlight the individual contributions of signaling pathways (18, 35), cell-envelope monomers (36), motility factors (37), and even strain phylogeny (38) to light organ colonization. The approach presented here can also be used to examine the spatial organization of these strains, thereby facilitating better understanding of the link between the bacterium-associated factors and the developmental stages of the light organ crypts. Recent microscopy-based techniques, such as BacSpace and multilabeled fluorescence in situ hybridization (MiL-FISH), have begun to provide insight into the spatial organization in the gut microbiome and other animal-microbe symbioses (39, 40). Such imaging-based approaches will complement sequencing-based studies of microbial consortia and populations and increase our understanding of the microbial world in which animals reside.
ACKNOWLEDGMENTS
We thank members of the Miyashiro laboratory and M. Wollenberg (Kalamazoo College) for constructive criticism of this project. We also thank three anonymous reviewers for their feedback, which significantly strengthened this report.
This work was supported by the NIH grant R00 GM 097032 (T.M.) and the Eberly College of Science at Penn State University (T.M.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert SF, Bosch TC, Ledón-Rettig C. 2015. Eco-Evo-Devo: developmental symbiosis and developmental plasticity as evolutionary agents. Nat Rev Genet 16:611–622. doi: 10.1038/nrg3982. [DOI] [PubMed] [Google Scholar]
- 3.Eloe-Fadrosh EA, Rasko DA. 2013. The human microbiome: from symbiosis to pathogenesis. Annu Rev Med 64:145–163. doi: 10.1146/annurev-med-010312-133513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donaldson GP, Lee SM, Mazmanian SK. 2016. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seedorf H, Griffin NW, Ridaura VK, Reyes A, Cheng J, Rey FE, Smith MI, Simon GM, Scheffrahn RH, Woebken D, Spormann AM, Van Treuren W, Ursell LK, Pirrung M, Robbins-Pianka A, Cantarel BL, Lombard V, Henrissat B, Knight R, Gordon JI. 2014. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell 159:253–266. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiland-Bräuer N, Neulinger SC, Pinnow N, Künzel S, Baines JF, Schmitz RA. 2015. Composition of bacterial communities associated with Aurelia aurita changes with compartment, life stage, and population. Appl Environ Microbiol 81:6038–6052. doi: 10.1128/AEM.01601-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandel MJ. 2010. Models and approaches to dissect host-symbiont specificity. Trends Microbiol 18:504–511. doi: 10.1016/j.tim.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Ruby EG. 2008. Symbiotic conversations are revealed under genetic interrogation. Nat Rev Microbiol 6:752–762. doi: 10.1038/nrmicro1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McFall-Ngai MJ. 2014. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol 68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graf J, Ruby EG. 1998. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci U S A 95:1818–1822. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones BW, Nishiguchi MK. 2004. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca:Cephalopoda). Mar Biol 144:1151–1155. doi: 10.1007/s00227-003-1285-3. [DOI] [Google Scholar]
- 12.McFall-Ngai MJ, Ruby EG. 1991. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science 254:1491–1494. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 13.Montgomery MK, McFall-Ngai M. 1993. Embryonic development of the light organ of the sepiolid squid Euprymna scolopes Berry. Biol Bull 184:296–308. doi: 10.2307/1542448. [DOI] [PubMed] [Google Scholar]
- 14.Montgomery MK, McFall-Ngai M. 1994. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120:1719–1729. [DOI] [PubMed] [Google Scholar]
- 15.Sycuro LK, Ruby EG, McFall-Ngai M. 2006. Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their morphological complexity and dynamic function in symbiosis. J Morphol 267:555–568. doi: 10.1002/jmor.10422. [DOI] [PubMed] [Google Scholar]
- 16.Lee K-H, Ruby EG. 1994. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol 60:1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol 182:4578–4586. doi: 10.1128/JB.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. 2010. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol 77:1556–1567. doi: 10.1111/j.1365-2958.2010.07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bose JL, Rosenberg CS, Stabb EV. 2008. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol 190:169–183. doi: 10.1007/s00203-008-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, Harvie EA. 2012. The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol 24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meighen EA. 1993. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J 7:1016–1022. [DOI] [PubMed] [Google Scholar]
- 22.Antunes LC, Schaefer AL, Ferreira RB, Qin N, Stevens AM, Ruby EG, Greenberg EP. 2007. Transcriptome analysis of the Vibrio fischeri LuxR-LuxI regulon. J Bacteriol 189:8387–8391. doi: 10.1128/JB.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koch EJ, Miyashiro T, McFall-Ngai MJ, Ruby EG. 2014. Features governing symbiont persistence in the squid-vibrio association. Mol Ecol 23:1624–1634. doi: 10.1111/mec.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graf J, Dunlap PV, Ruby EG. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol 176:6986–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg EP. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci U S A 102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y, LaSota ED, Cecere AG, LaPenna KB, Larios-Valencia J, Wollenberg MS, Miyashiro T. 2016. Intraspecific competition impacts Vibrio fischeri strain diversity during initial colonization of the squid light organ. Appl Environ Microbiol 82:3082–3091. doi: 10.1128/AEM.04143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunlop MJ, Cox RS III, Levine JH, Murray RM, Elowitz MB. 2008. Regulatory activity revealed by dynamic correlations in gene expression noise. Nat Genet 40:1493–1498. doi: 10.1038/ng.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Verma SC, Bogale H, Miyashiro T. 2015. NagC represses N-acetyl-glucosamine utilization genes in Vibrio fischeri within the light organ of Euprymna scolopes. Front Microbiol 6:741. doi: 10.3389/fmicb.2015.00741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wollenberg MS, Ruby EG. 2009. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl Environ Microbiol 75:193–202. doi: 10.1128/AEM.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Septer AN, Stabb EV. 2012. Coordination of the arc regulatory system and pheromone-mediated positive feedback in controlling the Vibrio fischeri lux operon. PLoS One 7:e49590. doi: 10.1371/journal.pone.0049590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks JF II, Gyllborg MC, Cronin DC, Quillin SJ, Mallama CA, Foxall R, Whistler C, Goodman AL, Mandel MJ. 2014. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci U S A 111:17284–17289. doi: 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franzenburg S, Fraune S, Altrock PM, Kunzel S, Baines JF, Traulsen A, Bosch TC. 2013. Bacterial colonization of Hydra hatchlings follows a robust temporal pattern. ISME J 7:781–790. doi: 10.1038/ismej.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wentrup C, Wendeberg A, Schimak M, Borowski C, Dubilier N. 2014. Forever competent: deep-sea bivalves are colonized by their chemosynthetic symbionts throughout their lifetime. Environ Microbiol 16:3699–3713. doi: 10.1111/1462-2920.12597. [DOI] [PubMed] [Google Scholar]
- 35.Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. 2006. The symbiosis regulator rscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol 62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adin DM, Phillips NJ, Gibson BW, Apicella MA, Ruby EG, McFall-Ngai MJ, Hall DB, Stabb EV. 2008. Characterization of htrB and msbB mutants of the light organ symbiont Vibrio fischeri. Appl Environ Microbiol 74:633–644. doi: 10.1128/AEM.02138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Millikan DS, Ruby EG. 2002. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl Environ Microbiol 68:2519–2528. doi: 10.1128/AEM.68.5.2519-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wollenberg MS, Ruby EG. 2012. Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME J 6:352–362. doi: 10.1038/ismej.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, Sonnenburg JL. 2015. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schimak MP, Kleiner M, Wetzel S, Liebeke M, Dubilier N, Fuchs BM. 16 October 2015. MiL-FISH: multi-labelled oligonucleotides for fluorescence in situ hybridization improve visualization of bacterial cells. Appl Environ Microbiol doi: 10.1128/AEM.02776-15. [DOI] [PMC free article] [PubMed] [Google Scholar]